Abstract
Plants are able to perform photosynthesis and cannot escape from environmental stresses, so they therefore developed sophisticated, highly responsive and dynamic physiology. Others' and our results indicate that plants solve their optimal light acclimation and immune defenses, photosynthesis and transpiration by a computational algorithm of the cellular automation. Our recent results however suggest that plants are capable of processing information encrypted in light intensity and in its energy. With the help of nonphotochemical quenching and photoelectrophysiological signaling (PEPS) plants are able to perform biological quantum computation and memorize light training in order to optimize their Darwinian fitness. Animals have their network of neuron synapses, electrophysiological circuits and memory, but plants have their network of chloroplasts connected by stromules, PEPS circuits transduced by bundle sheath cells and cellular light memory. It is suggested that plants could be intelligent organisms with much higher organism organization levels than it was thought before.
Key words: excess excitation energy, cellular automation, cellular light memory, Darwinian fitness, nonphotochemical quenching, photoelectrophysiological signaling, SAA, SAR
Light, Photosynthesis and Life
Without light there are very limited life possibilities. Plants are unique organisms capable of photosynthesis that support non-photosynthetic life on our planet. Due to photosynthesis and oxygenic atmosphere, reach heterotrophic live forms could evolved on the Earth. Plants cannot escape from a stress situation. Therefore, they have developed a highly responsive, flexible and dynamic physiology, which allows them to function under short- and long-term fluctuations and rapid changes in their natural environment. In plants evolved natural capacity to absorb more light energy than that required for photosynthetic CO2 assimilation.1 Therefore plants have non-photochemical quenching (NPQ) and photochemical quenching (qp), mechanisms that control dissipation of absorbed light energy in excess2–5 and reactive oxygen species (ROS) metabolism.6–12 There are living trees that germinated long before Jesus Christ was born. What sort of life wisdom evolved in plants to make it possible to survive and propagate for so long a time in the same place they germinated?
When photosystem II (PSII) is exposed to excess light, acidification of the lumen increases and singlet oxygen stages (1O2) are generated in the higher extend among of the many other singlet stages of chlorophyll and carotenoids molecules. Due to basic quantum physics law, the Pauli exclusion principle, higher levels of 1O2 increase probability to generate other ROS, like superoxide radical (O2.-) and hydrogen peroxide (H2O2). It has to be noted here that absolute half-life time of 1O2 is several orders of magnitude shorter than absolute half-life time of O2.-, and absolute half-life time of O2.- is several orders of magnitude shorter than absolute half-life time of H2O2. Therefore 1O2 generation in excess light exposure is a part of the immediate NPQ mechanism, while further ROS metabolism like Mehler reaction, water-water cycle and photorespiration are part of qp mechanisms and beyond. Excess light exposure decreases the lumenal pH value that in turn triggers a change in PSII light harvesting antenna function from light absorption to light dissipation by the means of NPQ.4,5,13,14 Therefore during highly variable light, various components of the photosynthetic electron transport chain become transiently more reduced or oxidized and deregulate ROS metabolism. This in turn trigger changes in the chloroplast stromal redox state (reviewed in refs. 1, 9 and 15) and activate cellular mechanisms for induction of various light acclimatory and defense responses, such as systemic acquired acclimation and resistance (SAAR),9,10,12,16,17 shifting of transcription programs in the chloroplasts and in the nucleus,7,8,10,12,18–21 inducing state transitions and other light acclimatory mechanisms.11,13,22–24
Plants are able to integrate and simultaneously process multiple stimuli and prioritize their responses.12,16,19,20,25,26 Light acclimation processes in plants act to dissipate excess of absorbed light energy and optimize photosynthesis under variable light, temperature and humidity conditions. Plant leaves solve their problem of optimal gas exchange, transpiration and photosynthesis, in a way similar to that defined by the algorithm of the cellular automation.27,28 Cellular automation, proposed by John von Neumann and Oskar Morgenstern in 1944,27 is a mathematical model for a dynamic system that is discrete in time and in space. It depends on local interactions, but global phenomena emerge due to predefined local interactions. Therefore the finding of Peak and colleagues28 implies that there must be a molecular mechanism (hardware) that coordinates functioning of signaling networks that govern light acclimation, immune defenses, photosynthesis, transpiration and subsequent developmental processes in plants. Despite the importance of this information, surprisingly little is known about these signaling interactions and theirs coordination. Recently, however, we have identified parts of this hardware and described the mechanism of signaling interactions and theirs systemic coordination.17
Foliar NPQ Pattern, Photoelectrochemical Signaling and Light Memory
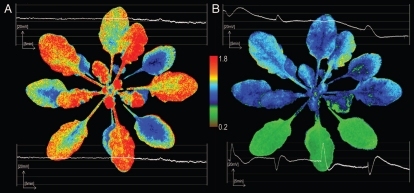
NPQ is immediate mechanisms that convert excess of absorbed light energy (absorbed in a form of electron resonance induced in chlorophyll molecules organised in light harvesting complexes) into a heat and subsequent hormonal signaling. In Arabidopsis thaliana rosettes partially exposed to excess light, changes in NPQ were observed and indicated for higher photooxidative stress, in both directly exposed leaves and leaves undergoing SAAR (Fig. 1A and B). Changes in NPQ varied, in some leaves with the lowest NPQ values in the central vein region and the highest values towards the edge of the leaf (Fig. 1A and B). Furthermore, we detected sectors of different NPQ values in leaves undergoing SAAR, but in leaves that were directly absorbing light energy in excess, NPQ was strongly reduced and its foliar gradient was flattened (Fig. 1B). Such a pattern of NPQ in leaves undergoing SAAR is, however, similar to that observed in control plants, but it is characterized by a higher gradient differences and lower average NPQ.
Figure 1.
Partial exposure to excess light and induction of SAAR is associated with systemic gradient-like changes of foliar NPQ and changes in PEPS. Arabidopsis thaliana Col-0 rosettes were grown at low-light conditions (LL, 100 µmol photons m−2s−1) and were partially exposed to excess light (EL, 2,000 µmol photons m−2s−1). (A) Left, controls that were LL grown with no excess light exposure. (B) Right, the same rosette partially exposed to EL for 60 min (three bottom leaves with the lowest NPQ value). NPQ estimated the nonphotochemical quenching from Fm to F′m is monitoring the apparent heat losses from PSII and is calculated from (Fm/F′m) − 1, where Fm is maximal chlorophyll fluorescence of dark-adapted PSII (all QA molecules are reduced), F′m = is maximal chlorophyll fluorescence of light-adapted PSII (all QA molecules involved in photosynthetic electron transport from water in provided light condition are reduced). Changes in PEPS (mV) were measured in bundle sheath cells of EL exposed leaves and in leaves undergoing SAAR. One bundle sheath cell of directly exposed leaf and another in a leaf undergoing SAAR were measured during whole experiment (60 min with 15 min periods of light on and off). Strong plasma membrane electrical potential changes were observed directly after switching on and off EL. Pattern of PEPS changes in directly EL exposed leaves and leaves undergoing SAAR in LL are symmetric. When depolarization is observed in EL exposed leaves, hyperpolarization is induced in systemic leaves. Other experimental data are presented in Szechynska-Hebda and colleagues.17
Acidification of the chloroplast lumen is prerequisite to induce NPQ changes.4,5,13 Therefore, pattern of changes in NPQ exactly reflects pattern of prior changes in ΔpH across thylakoid membrane in chloroplasts. This requires a very precise signaling mechanism that regulates and coordinates which chloroplasts in which group of cells or leaf sector should have lower or higher pH value in the chloroplast lumen. It is also known that different pH values in the chloroplast lumen differently change the redox status of the photosynthetic electron carriers and differently regulate/deregulate retrograde chloroplast to nucleus signaling that depends on the e.g., redox status of the plastoquinone and glutathione pools (reviewed in refs. 8, 9, 12, 18 and 19). Taking into consideration the foliar pattern of NPQ and its regulatory role for the redox status of the photosynthetic electron transport components it regulates/deregulates nuclear gene expression in a similar pattern as observed for NPQ.17 Peak and colleagues28 demonstrated that changes in chlorophyll fluorescence are dynamic, emergent and propagating across the leaf with speed of circa 0.6 cm per sec. Our results indicate that light wavelength-specific photoelectrophysiological signaling (PEPS) induced in response to excess light and induced by transthylakoid ΔpH and NPQ changes is propagated systemically by bundle sheath cells with similar or faster speed and is regulated, at least in part, by the same mechanism as for NPQ.17 Thes results17 (Fig. 1A and B) indicate that PEPS can regulate and coordinate specific, emergent and dynamic in time ΔpH and NPQ local and systemic changes (Fig. 1A and B) that trigger further concomitant changes in the pattern of gene expression.8–12,17 Cellular automation is an algorithm that can regulate pattern of these changes, but quantum-redox light-sensing mechanism in PSII with PEPS circuits and cellular light memory define algorithm of changes (like cellular automation programing). Several seconds of excess light illumination is sufficient to induce PEPS with maximal action potential and speed.17
Plants Intelligence
A definition of memory and intelligence for plants was proposed by Trewavas:29 adaptively variable growth and development during the lifetime of the individual. In animals, memory is connected with intelligence in such a way that the more intelligent the organism is, the greater the degree of individual adaptively variable behavior. Because this definition was used to describe intelligence in other organisms than humans, we used this definition in our experimental system.17 Do plants exhibit memory and behavior that result from memorized previous events? Our recent data indicate that plants possess memory of previous light incidents, called cellular light memory, which is used for optimization of future light acclimatory and immune defense responses (SAAR).17 In other words, plants can store and use information from the spectral composition of light for several days or more to anticipate changes that might appear in the near future in the environment, for example, for anticipation of pathogen attack. Therefore, plants have to possess a mechanism for processing the memorized information. This mechanism has to convert quantum information encrypted in the light intensity and energy (“color of light” or wavelength) into PEPS that specifically regulate physiological responses (cellular light memory and SAAR) in the whole organism. Our recent results17 allow us to suggest that plants actually work as a biological quantum computing device that is capable to process quantum information encrypted in light intensity and in its energy with help of NPQ and quenched singlet stages, transmute it into analogous (PEPS) information and finally is capable to physiologically memorize it with help of qp, O2.- and H2O2.
Taking into consideration the above and that representative reaction towards stimuli from internal chemical reactions or external environmental factors are in fact describing thinking process in leaving organisms, plants can actually think and remember. This is clearly illustrated in Figure 1A and B where emergent, dynamic and independently distributed changes in transthylakoid ΔpH, NPQ in different sectors of non-stressed leaves, leaves exposed to excess light and leaves undergoing SAAR are observed. Rephrasing this information, we can say that different group of chloroplasts and cells in the same leaf under identical constant and stable light, temperature and relative humidity condition have different opinion “what to do” in such conditions (Fig. 1A) and tests different scenarios of possible future development (like different military exercisers during peace time). But cells and leaves exposed to excess light represent one clear opinion: use as much NPQ as possible (Fig. 1B) (like in a real war zone). Moreover, leaves that are still in shadow (Fig. 1B) also have a different opinion that they had just one hour before (Fig. 1A) of the same Arabidopsis thaliana rosette that was partially exposedto excess light and are undergoing SAAR (like mobilization for war). Is this a sort of physiological game or exercisers or behavior of plant leaves?
Here we have to ask why do plants evolve mechanisms in which excess light and its spectral composition regulate SAAR? In dense canopy, light intensities are strongly reduced, therefore the majority of leaves are in shade and prone to pathogen attack.12,17 SAAR is in fact a mechanism in which plants use the disadvantages of being partly exposed to excess light to strengthen, for example, immune defenses in the dense canopy zone. Another possible answer to the above question is a light training of young naïve leaves. Let's imagine when young leaf or flower is emerging out of a plant, it would be nice for that leaf or flower to know about the conditions in which it is going to emerge. Older, more experienced leaves that actually are acclimated to outside conditions can train naïve emerging young leaves with the PEPS and cellular light memory mechanisms. This explains why plants possess a natural capacity to absorb more light energy than that required for photosynthetic CO2 assimilation. They need this absorbed energy in excess for optimization and training of light acclimatory and immune defenses.
In summary, plants solve their optimal light acclimation and immune defenses (SAAR), photosynthesis, gas exchange and transpiration with help of a mathematical algorithm (cellular automation)27,28 in which input, output and processing of the data are all accomplished using the same hardware. Our experiments17 identified some parts of this hardware, which includes quantum-redox sensing and changes in PSII (e.g., changes in transthylakoid ΔpH, in NPQ and redox status of the glutathione and plastoquinone pools), PEPS, ROS/hormonal circuits and finally the cellular light memory. Probably, this is the most elegant system that evolved in complex photosynthetic organisms, since it uses absorbed photons energy in excess by some leaves to improve survival chances of a whole plant. Animals have their network of neuron synapses, electrophysiological and PEPS circuits and memory. Plants however have their network of chloroplasts connected by stromules,30–33 electrophysiological and PEPS circuits transduced by bundle sheath cells and cellular light memory that regulates SAAR. Our results suggest that plants are intelligent organisms capable of performing a sort of thinking process (understood as at the same time and non-stress conditions capable of performing several different scenarios of possible future definitive responses), and capable of memorizing this training.17 Indeed leaves in the dark are able to not only “see” the light,8,34 but also are able to differently remember its spectral composition and use this memorized information to increase their Darwinian fitness.17
Acknowledgements
The authors are grateful to the Polish Science Foundation for support from the Welcome 2008/1 project operated within the Foundation for Polish Science Welcome Program co-financed by the European Regional Development Fund.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13243
References
- 1.Asada K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 2.Niyogi KK. Safety valves for photosynthesis. Curr Opin Plant Biol. 2000;3:455–460. doi: 10.1016/s1369-5266(00)00113-8. [DOI] [PubMed] [Google Scholar]
- 3.Holt NE, Fleming GR, Niyogi KK. Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry. 2004;43:8281–8289. doi: 10.1021/bi0494020. [DOI] [PubMed] [Google Scholar]
- 4.Ruban AV, Berera R, Ilioaia C, van Stokkum IHV, Kennis JTM, Pascal AA, et al. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature. 2007;450:575–578. doi: 10.1038/nature06262. [DOI] [PubMed] [Google Scholar]
- 5.Baker NR. Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu Rev Plant Biol. 2008;59:89–113. doi: 10.1146/annurev.arplant.59.032607.092759. [DOI] [PubMed] [Google Scholar]
- 6.Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, et al. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 1997;16:4806–4816. doi: 10.1093/emboj/16.16.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 8.Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- 9.Karpinska B, Wingsle G, Karpinski S. Antagonistic effects of hydrogen peroxide and glutathione on acclimation to excess excitation energy in Arabidopsis. IUBMB Life. 2000;50:21–26. doi: 10.1080/15216540050176548. [DOI] [PubMed] [Google Scholar]
- 10.Mateo A, Mühlenbock P, Rustérucci C, Chi-Chen C, Miszalski Z, Karpinska B, et al. The LESION SIMULATING DISEASE (LSD1) gene is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 2004;136:2818–2830. doi: 10.1104/pp.104.043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:672–677. doi: 10.1073/pnas.0609063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mühlenbock P, Szechynska-Hebda M, Płaszczyca M, Baudo M, Mullineaux PM, Parker JE, et al. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell. 2008;20:2339–2356. doi: 10.1105/tpc.108.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascal AA, Liu Z, Broess K, van Oort B, van Amerongen H, Wang C, et al. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature. 2005;436:134–137. doi: 10.1038/nature03795. [DOI] [PubMed] [Google Scholar]
- 14.Johnson MP, Davison PA, Ruban AV, Horton P. The xanthophyll cycle pool size controls the kinetics of non-photochemical quenching in Arabidopsis thaliana. FEBS Letts. 2008;582:262–266. doi: 10.1016/j.febslet.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Kruk J, Karpinski S. An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochem Biophys Acta. 2006;1757:1669–1675. doi: 10.1016/j.bbabio.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Mühlenbock P, Plaszczyca M, Mellerowicz E, Karpinski S. Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell. 2007;19:3819–3830. doi: 10.1105/tpc.106.048843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szechynska-Hebda M, Kruk J, Gorecka M, Karpinska B, Karpinski S. Evidence for light wavelength-specific systemic photoelectrophysiological signalling and cellular light memory of excess light episode in Arabidopsis. Plant Cell. 2010;22:1–18. doi: 10.1105/tpc.109.069302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfannschmidt T, Nilsson A, Allen J. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- 19.Pfannschmidt T, Braeutigam K, Wagner R, Dietzel L, Schroeter Y, Steiner S, et al. Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann Bot. 2009;103:599–607. doi: 10.1093/aob/mcn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullineaux P, Ball L, Escobar C, Karpinska B, Creissen G, Karpinski S. Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos Trans R Soc Lond B Biol Sci. 2000;355:1531–1540. doi: 10.1098/rstb.2000.0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fey V, Wagner R, Brautigam K, Wirtz M, Hell R, Dietzmann A, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2004;280:5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- 22.Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–906. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- 23.Bellafiore S, Bameche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- 24.Barneche F, Winter V, Crèvecur M, Rochaix J-D. ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 2006;25:5907–5918. doi: 10.1038/sj.emboj.7601472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullineaux P, Karpinski S. Signal transduction in response to excess light: getting out of the chloroplast. Curr Opin Plant Biol. 2002;5:43–48. doi: 10.1016/s1369-5266(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 26.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 27.Von Neumann J, Morgenstern O. Theory of Games and Economic Behavior. Princeton University Press; 1944. (ISBN 978-0-691-13061-3) [Google Scholar]
- 28.Peak D, West JD, Messinger SM, Mott KA. Evidence for complex, collective dynamics and emergent, distributed computation in plants. Proc Natl Acad Sci USA. 2004;101:918–922. doi: 10.1073/pnas.0307811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trewavas A. Aspects of plant intelligence. Ann Bot. 2003;92:1–20. doi: 10.1093/aob/mcg101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senn G. Die Gestalts- und Lageveränderung der Pflanzen-Chromatophoren. Engelmann, Leipzig: 1908. (Ger). [Google Scholar]
- 31.Hanson MR, Sattarzadeh A. Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environm. 2008;31:646–657. doi: 10.1111/j.1365-3040.2007.01768.x. [DOI] [PubMed] [Google Scholar]
- 32.Kwok EY, Hanson MR. Microfilaments and microtubules control the morphology and movement of nongreen plastids and stromules in Nicotiana tabacum. Plant J. 2003;35:16–26. doi: 10.1046/j.1365-313x.2003.01777.x. [DOI] [PubMed] [Google Scholar]
- 33.Kwok EY, Hanson MR. Stromules and the dynamic nature of plastid morphology. J Microsc. 2003;214:124–137. doi: 10.1111/j.0022-2720.2004.01317.x. [DOI] [PubMed] [Google Scholar]
- 34.Foyer CH, Noctor G. Leaves in the dark see the light. Science. 1999;284:599–601. doi: 10.1126/science.284.5414.599. [DOI] [PubMed] [Google Scholar]