Abstract
We recently reported that the cwa1 mutation disturbed the deposition and assembly of secondary cell wall materials in the cortical fiber of rice internodes. Genetic analysis revealed that cwa1 is allelic to bc1, which encodes glycosylphosphatidylinositol (GPI)-anchored COBRA-like protein with the highest homology to Arabidopsis COBRA-like 4 (COBL4) and maize Brittle Stalk 2 (Bk2). Our results suggested that CWA1/BC1 plays a role in assembling secondary cell wall materials at appropriate sites, enabling synthesis of highly ordered secondary cell wall structure with solid and flexible internodes in rice. The N-terminal amino acid sequence of CWA1/BC1, as well as its orthologs (COBL4, Bk2) and other BC1-like proteins in rice, shows weak similarity to a family II carbohydrate-binding module (CBM2) of several bacterial cellulases. To investigate the importance of the CBM-like sequence of CWA1/BC1 in the assembly of secondary cell wall materials, Trp residues in the CBM-like sequence, which is important for carbohydrate binding, were substituted for Val residues and introduced into the cwa1 mutant. CWA1/BC1 with the mutated sequence did not complement the abnormal secondary cell walls seen in the cwa1 mutant, indicating that the CBM-like sequence is essential for the proper function of CWA1/BC1, including assembly of secondary cell wall materials.
Key words: carbohydrate-binding module, COBRA-LIKE, CWA1/BC1, glycosylphosphatidylinositol-anchored protein, secondary cell wall formation
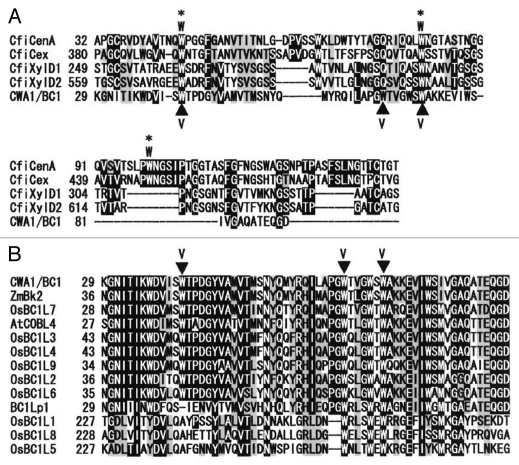
The main function of carbohydrate-binding modules (CBMs) of microbes and plants is to attach the enzyme to a variety of cell surface glycans and thereby increase the local concentration of substrate, leading to more efficient catalysis.1–4 Almost all CBMs studied to date contain surface-exposed aromatic rings, which have been shown to be the main sites of interaction with polysaccharides. These residues form face-to-face hydrophobic stacking interactions in which a Trp residue or ring of a Tyr residue interacts with the non-polar face of a sugar ring.5–9 CBMs have been classified into families based on amino acid sequence similarity. Currently, there are 59 defined families of CBMs and these CBMs display substantial variation in ligand specificity (http://www.cazy.org/Carbohydrate-Binding-Modules.html). Among these CBM families, the large family of CBM2 has been further classified into two subgroups, CBM2a and 2b, which have shown to bind cellulose and xylan, respectively.10–12 CBM2a characteristically possess three exposed Trp residues,13 whereas CBM2b have two Trp residues,14 which are conserved among the CBM2 members (Fig. 1A).
Figure 1.
Sequence alignment of the CBM-like sequence of CWA1/BC1, the BC1L proteins and bacterial CBM2 members. (A) Sequence alignment between bacterial CBM2a, 2b and CWA1/BC1. The three surface-exposed Trp residues of CBM2a members are indicated by asterisks and W. The CBM sequences of CBM2a are: CfiCenA, Cellulomonas fimi endo-1,4-glucanase; CfiCex, C. fimi exo-beta-1,4-glucanase. Those of CBM2b are: CfiXylD1, C. fimi endo-1,4-beta-xylanase D; CfiXylD2, C. fimi endo-1,4-beta-xylanase. CWA1/BC1 shows weak similarity to CBM2, and some Trp residues are conserved with bacterial CBM2 members. (B) Sequence alignment of CWA1/BC1, the BC1L proteins and CWA1/BC1 orthologs, Zea maiz Brittle Stalk 2 (ZmBk2) and Arabidopsis thaliana COBRA-LIKE 4 (AtCOBL4). The CBM-like sequence of CWA1/BC1, especially the Trp residues, is highly conserved among the analyzed sequences. Substituted Trp (W) residues to Val (V) in CWA1/BC1 are indicated by closed triangles. Numbers at the left are the positions of the amino acids in each protein, with gaps (dashes) included to maximize alignments. Identical and similar amino acids are shaded and gray, respectively.
Our recent study showed that the defect of the rice CWA1/BC1 (CELL WALL ARCHITECTURE 1/BRITTLE CULM 1) gene induced abnormal secondary cell wall formation with amorphous and bulky structures at the cytoplasm side and CWA1/BC1 encodes one of COBRA-like glycosylphosphatidylinositol (GPI)-anchored proteins, which are specifically found in plants, suggesting that CWA1/BC1 regulates assembly of secondary cell wall materials in rice sclerenchyma. Furthermore, several reports have shown that the N-terminus of rice CWA1/BC1 and other COBRA-like GPI-anchored proteins in Arabidopsis (12 members) and maize Brittle Stalk 2 (Bk2) share weak similarity to a CBM2 in several bacterial cellulases.15,16 However, the importance of CBM-like sequence in COBRA family members has not been clarified. To investigate the nature of CWA1/BC1, we compared the CBM-like sequence in rice CWA1/BC1 with bacterial CBM2, 10 members of the BC1-like (BC1L) protein in rice and CWA1/BC1 orthologs, Arabidopsis COBL4 and maize Bk2. Furthermore, we constructed three-point mutated CWA1/BC1, in which three conserved Trp residues in CBM-like sequence were substituted for Val residues (CWA1/BC1W→V), and introduced it into the cwa1 mutant to evaluate the necessity of the CBM-like sequence for proper function of CWA1/BC1. We discuss a putative explanation, based on our results, of the properties and possible functions of CWA1/BC1.
The Trp Residues in CBM-Like Sequence in CWA1/BC1 is Conserved between Bacterial CBM2, CWA1/BC1 Orthologs and BC1-Like Proteins
A database search for the amino acid sequence of CWA1/BC1 has shown weak similarity of the amino acids from 30 to 91 with those of bacterial CBM2 (www.sbg.bio.ic.ac.uk/phyre/index.cgi).17 To investigate the relationship between this CBM-like sequence within CWA1/BC1 and bacterial CBM2, sequence alignment was analyzed to determine their homology. CBM2a and 2b have been shown to bind cellulose and xylan, respectively.10–12 The three exposed Trp residues of CBM2a13 and the two on CBM2b14 were conserved among the CBM2 members (Fig. 1A). The CBM-like sequence of CWA1/BC1 shows weak similarity with those of CBM2 members and a few Trp residues were conserved among the CWA1/BC1 and CBM2 members (Fig. 1A). To confirm whether the CBM-like sequence of CWA1/BC1 is conserved among CWA1/BC1 orthologs (Arabidopsis COBL4 and maize Bk2) and BC1L proteins in rice, N-terminal amino acid sequences of these proteins were compared. The CBM-like sequence of each protein shared high homology; most of the Trp residues were especially conserved (Fig. 1B). Therefore, it is possible that the CBM-like sequence in CWA1/BC1 plays a role in carbohydrate binding, which may be essential for the CWA1/BC1 function in assembling secondary cell wall materials in rice sclerenchyma.
The Trp Residue in the CBM-like Sequence is Essential for Proper Function of CWA1/BC1
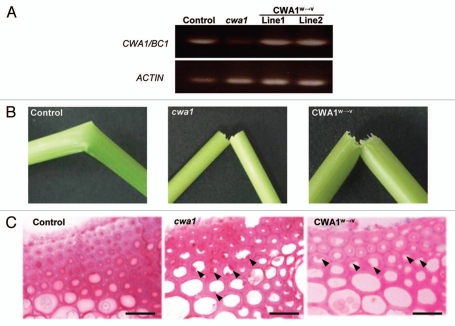
To evaluate the importance of conserved Trp residues in the CBM-like sequence of CWA1/BC1, we constructed mutated CWA1/BC1 with three substituted Trp residues to Val residues (designated as CWA1/BC1W→V) as shown in Figure 1. For constructing CWA1/BC1W→V, we used QuikChange II Site-Directed Mutagenesis Kit (Stratagene) and cloned the sequence into the binary vector, pBI121Hm. The mutations were confirmed by sequencing. Wild-type CWA1/BC1 or mutated CWA1/BC1W→V genes were expressed in the cwa1 mutant plants under the control of the 1973-bp CWA1/BC1 promoter. Transgenic cwa1 plants are produced by Agrobacterium tumefaciens-mediated transformation.18 To confirm the expression of these introduced genes in the cwa1 plants, we performed RT-PCR of the CWA1/BC1 gene. The expression of the CWA1/BC1 gene in the cwa1 mutant was hardly detected (Fig. 2A), whereas transgenic cwa1 plants introducing CWA1/BC1 (Control) or CWA1/BC1W→V (CWA1W→V) expressed each gene in their young internodes (Fig. 2A), confirming that the transgenic cwa1 plants express each introduced gene.
Figure 2.
Evaluation of transgenic and cwa1 mutant plants. (A) Expression of the CWA1/BC1 gene in the transgenic cwa1 plant expressing the wild-type CWA1/BC1 gene (control), cwa1 mutant and transgenic cwa1 plants expressing mutated CWA1/BC1W→V (CWA1W→V, lines 1 and 2). Expression of the CWA1/BC1 gene is rarely detected in the cwa1 mutant, whereas obvious expression is detected in the transgenic cwa1 plants introduced with each construct. (B) Mature internodes of transgenic cwa1 plants expressing wild-type CWA1/BC1 (Control), the cwa1 mutant (cwa1) and transgenic cwa1 plant expressing mutated CWA1/BC1W→V (CWA1W→V) after hand flexing. The brittle culm phenotype of transgenic cwa1 plant was rescued by expression of wild-type CWA1/BC1 (Control), but not by expression of CWA1/BC1W→V (CWA1W→V). (C) Transverse section at the cortical fiber of mature internodes from transgenic cwa1 plant expressing wild-type CWA1/BC1 (Control), the cwa1 mutant (cwa1) and transgenic cwa1 plant expressing mutated CWA1/BC1W→V (CWA1W→V). The transgenic cwa1 plant expressing CWA1/BC1W→V has abnormal and amorphous cell wall structures (arrowheads) as seen in the cwa1 mutant. Scale bars = 20 µm.
The cwa1 plants showed a brittle culm phenotype (Fig. 2B) and abnormal and amorphous secondary cell wall structures in the cortical fiber of rice internodes (Fig. 2C and arrowheads). In contrast, the brittle phenotype (Fig. 2B and control) and abnormal secondary cell wall structures in the cortical fiber (Fig. 2C and control) of transgenic cwa1 plants were rescued by introduction of wild-type CWA1/BC1. On the other hand, the cwa1 plants introduced with CWA1/BC1W→V still exhibited the brittle phenotype and abnormal secondary cell walls with uneven thickness and a rough surface on the cytoplasmic side of the cortical fiber as seen in the cwa1 mutant (Fig. 2B and C, CWA1W→V). These results strongly suggest that the Trp residues in the CBM-like sequence of CWA1/BC1 is essential for its function, especially for proper assembly of secondary cell wall materials at appropriate sites. Therefore, at least one of the three mutated Trp residues should have the capacity to binding polysaccharides in the cell walls, such as cellulose or hemicelluloses in microbes.8,14
How Does CWA1/BC1 Regulate Secondary Cell Wall Assembly via CBM?
GPI-anchored proteins including the COBRA family are predicted to be localized in the outer surface of the plasma membrane or cell wall.19,20 Arabidopsis COBRA protein has also been shown to be abundantly localized in the cell wall.21 Therefore, it is possible that the proper assembly of secondary cell wall materials by the CWA1/BC1 requires binding between the CBM-like sequence within CWA1/BC1 and cell wall polysaccharides such as cellulose and/or hemicelluloses. Furthermore, it was proposed that the nascent cellulose microfibrils are oriented by binding to a scaffold of cell wall polysaccharides or plasma membrane proteins.22 Our previous study revealed that CWA1/BC1 is expressed before the start of secondary cell wall formation. Consequently, CWA1/BC1 may play an important role at the initial stage of secondary cell wall formation and act as a scaffolding protein for regulating the orientation of cellulose microfibrils and/or reserving space for secondary cell wall thickening between the plasma membrane and the cell wall or between cell wall polysaccharides, by binding cell wall polysaccharides via the CBM-like sequence. In order to elucidate the role of CWA1/BC1 in the assembly of secondary cell wall materials, the target polysaccharide of the CBM-like sequence and cellular localization of the CWA1/BC1 protein need to be investigated.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13342
References
- 1.Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, et al. Pseudomonas cellulose-binding domains mediate their effects by increasing substrate proximity. Biochem J. 1998;331:775–781. doi: 10.1042/bj3310775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill J, Rixon JE, Bolam DN, McQueen-Mason S, Simpson PJ, Williamson MP, et al. The type II and X cellulose-binding domains of Pseudomonas xylanase A potentiate catalytic activity against complex substrates by a common mechanism. Biochem J. 1999;342:473–480. [PMC free article] [PubMed] [Google Scholar]
- 3.Black GW, Rixon JE, Clarke JH, Hazlewood GP, Theodorou MK, Morris P, et al. Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem J. 1996;319:515–520. doi: 10.1042/bj3190515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Casado G, Urbanowicz BR, Damasceno CMB, Rose JKC. Plant glycosyl hydrolases and biofuels: a natural marriage. Curr Opin Plant Biol. 2008;11:329–337. doi: 10.1016/j.pbi.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, Shoham Y, et al. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy T, Simpson P, Williamson MP, Hazlewood GP, Gilbert HJ, Orosz L. All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 7.Din N, Forsythe IJ, Burntnick LD, Gilkes NR, Miller R, Jr, Warren RA, et al. The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol Microbiol. 1994;11:747–755. doi: 10.1111/j.1365-2958.1994.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 8.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urbanowicz BR, Catalá C, Irwin D, Wilson DB, Ripoll DR, Rose JKC. A tomato endo-β-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49) J Biol Chem. 2007;282:12066–12074. doi: 10.1074/jbc.M607925200. [DOI] [PubMed] [Google Scholar]
- 10.Gilkes NR, Jervis E, Henrissat B, Tekant B, Miller R, Jr, Warren RA, et al. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992;267:6743–6749. [PubMed] [Google Scholar]
- 11.Dupont C, Roberge M, Shareck F, Morosoli R, Kluepfel D. Substrate-binding domains of glycanases from Streptomyces lividans: characterization of a new family of xylan-binding domains. Biochem J. 1998;330:41–45. doi: 10.1042/bj3300041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black GW, Hazlewood GP, Millward-Sadler SJ, Laurie JI, Gilbert HJ. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem J. 1995;307:191–195. doi: 10.1042/bj3070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu GY, Ong E, Gilkes NR, Kilburn DG, Muhandiram DR, Harris-Brandts M, et al. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 14.Simpson PJ, Xie H, Bolam DN, Gilbert HJ, Williamson MP. The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J Biol Chem. 2000;275:41137–41142. doi: 10.1074/jbc.M006948200. [DOI] [PubMed] [Google Scholar]
- 15.Roudier F, Schindelman G, DeSalle R, Benfey PN. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol. 2002;130:538–548. doi: 10.1104/pp.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, et al. Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol. 2007;145:1444–1459. doi: 10.1104/pp.107.102582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocol. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 18.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 19.Sherrier DJ, Prime TA, Dupree P. Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis. 1999;20:2027–2035. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2027::AID-ELPS2027>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 20.Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 2002;129:486–499. doi: 10.1104/pp.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GH, Schindelman G, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskin TI. On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma. 2001;215:150–171. doi: 10.1007/BF01280311. [DOI] [PubMed] [Google Scholar]