Abstract
Resistance (R) protein mediated recognition of pathogen avirulence effectors triggers signaling that induces a very robust form of species-specific immunity in plants. The soybean Rpg1-b protein mediates this form of resistance against the bacterial blight pathogen, Pseudomonas syringae expressing AvrBPgyrace4. Likewise, the Arabidopsis RPM1 protein also mediates species-specific resistance against AvrB expressing bacteria. RPM1 and Rpg1-b are non-orthologous and differ in their requirements for downstream signaling components. We recently showed that the activation of Rpg1-b derived resistance signaling requires two host proteins that directly interact with AvrB. These proteins share high sequence similarity with the Arabidopsis RPM1 interacting protein 4 (RIN4), which is essential for RPM1-derived resistance. The two soybean RIN4-like proteins (GmRIN4a and b) differ in their abilities to interact with Rpg1-b as well as to complement the Arabidopsis rin4 mutation. Because the two GmRIN4 proteins interact with each other, we proposed that they might function as a heteromeric complex in mediating Rpg1-b-derived resistance. Absence of GmRIN4a or b enhanced basal resistance against bacterial and oomycete pathogens in soybean. Lack of GmRIN4a also enhanced the virulence of avrB bacteria in plants lacking Rpg1-b. Our studies suggest that multiple RIN4-like proteins proteins mediate R-mediated signaling, in soybean.
Key words: AvrB, soybean defense, effector recognition, gaurdee, resistance protein, bacterial blight, gene silencing
Recognition of pathogens in a species-specific manner results in the generation of a very robust mode of resistance in plants. This form of protection termed resistance (R) protein-mediated or effector-triggered immunity is induced when a plant encoded R protein “perceives” the presence of a pathogen-derived avirulence (Avr) effector. “Perception” occurs either via direct or indirect interactions between the R and Avr proteins.1–7 One or more plant proteins, that themselves usually physically associate with the Avr and R proteins, mediate indirect R-Avr interactions. Such proteins have been termed “guardee” based upon the hypothesis that Avr-derived alterations of these proteins are guarded by R proteins.5–7 First proposed to explain the perception of AvrPtoPtoJL1065 from Pseudomonas syringae in tomato,8,9 the “guard” model has been extended to several other R-Avr interactions.5,7,10 This mode of interaction is typified in the recognition of the Pseudomonas syringae AvrB effector by the Arabidopsis R protein, RPM1 (resistance to P. syringae pv. maculicola 1). RPM1 mediates resistance against bacteria expressing either AvrRpm1PmaM6 or AvrBPgyrace4.11 However, direct interactions between RPM1 and its cognate Avr proteins have not been detected. Rather, RPM1 associates with the host protein, RIN4 (RPM1-interacting 4), which in turn interacts with AvrRpm1 and AvrB. Consistent with its role as a “guardee” protein, RIN4 is required for RPM1-induced resistance and is phosphorylated by AvrRpm1/AvrB, albeit only in the presence of a plant-derived factor.12 The phosphorylation status of RIN4 is likely monitored by RPM1 for the induction of resistance signaling. The “guard” model implies that unlike R proteins, “guardee” proteins are highly conserved. Indeed, RIN4-like proteins appear to be conserved in diverse plants including cowpea, lettuce, maize, potato, rice, tobacco and tomato.13–16 Additionally, the tomato and lettuce RIN4 proteins are known to mediate defense against microbial pathogens.14,15 However, due to the fact that “guardee” proteins have only been identified in the context of specific R-Avr pairs, their requirement in mediating responses to a common avirulence effector in diverse hosts has remained untested. We tested this corollary of the “guard” model in soybean since soybean too can induce resistance to AvrB expressing bacteria in an R gene-specific manner. We demonstrated that soybean does encode RIN4-like proteins and that these are important for mediating resistance to avrB P. syringae.17 This is an important finding since the soybean R protein Rpg1-b is non-orthologous to RPM1 and differs in its requirements for downstream signaling components.18–21 Furthermore, unlike RPM1, Rpg1-b does not provide resistance against bacteria expressing the AvrRpm1 effector.22
Genome sequence search identified four genes encoding RIN4-like proteins in soybean, designated GmRIN4a-d. Both in planta bimolecular florescence complementation (BiFC) and in vitro “pull-down” assays detected binding between AvrB and all four GmRIN4 proteins, indicating that these interactions did not require additional plant-derived factors. Interactions were further confirmed by co-immunoprecipitation (Co-IP, Fig. 1). AvrB tagged with the FLAG (3X) epitope and the various GmRIN4 isoforms tagged with the MYC epitope were transiently expressed in Nicotiana benthamiana. Total protein extracts from leaves expressing AvrB-FLAG, GmRIN4a/b/c/d-MYC or co-expressing AvrB-FLAG with GmRIN4a/b/c/d-MYC proteins were used for immunoprecipitation with anti-FLAG antibodies. Immunoprecipitated proteins were visualized using anti-MYC antibodies in western blots (Fig. 1). Three of these (GmRIN4b, c and d) also interacted with Rpg1-b directly. However, GmRIN4a was unable to interact with Rpg1-b in planta or in vitro. Although GmRIN4a and b share very high amino acid identity (∼94%), only GmRIN4b interacted with Rpg1-b. However, silencing either GmRIN4a or b abrogated resistance to avrB bacteria in Rpg1-b plants, suggesting that both proteins were essential for the activation of Rpg1-b derived signaling.17 This raised the possibility that GmRIN4a and b might oligomerize to function in Rpg1-b-derived signaling. Indeed, GmRIN4b interacted with GmRIN4a as well as with GmRIN4c and d.17 GmRIN4b also interacted with itself. GmRIN4a, c and d neither interacted with each other, nor themselves. Together, these results suggest that the GmRIN4 isoforms might oligomerize. Whether the oligomer exists in the presence or absence of AvrB, and whether binding of one or the other isoform alters the dynamics of the complex to change affinities for Rpg1-b and/or AvrB, remains to be examined. The fact that the GmRIN4c and d isoforms also interact with Rpg1-b and that they associate with GmRIN4b raises the untested possibility that these too might function in Rpg1-b mediated resistance.
Figure 1.
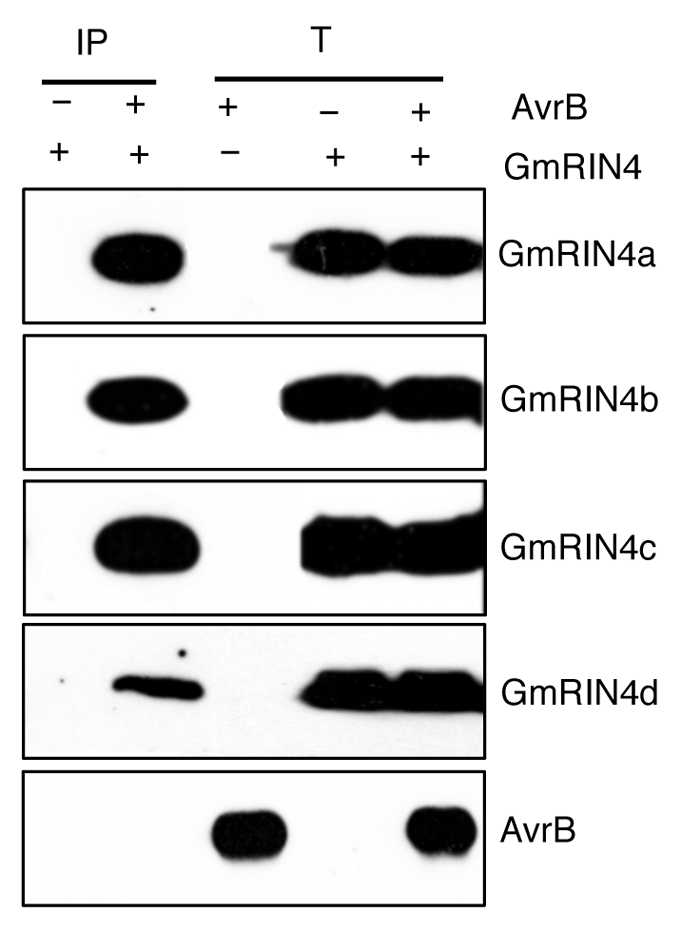
GmRIN4 proteins co-immunoprecipitate with AvrB. Agrobacterium cells expressing MYC-tagged GmRIN4a, b, c or d were expressed individually or together with FLAG (3X)-tagged AvrB in Nicotiana benthamiana. Proteins were immunoprecipitated (IP) from total extracts (T) using anti-FLAG antibodies, electrophoresed on SDS-PAGE and visualized using tagspecific antibodies (α-MYC for the various GmRIN4 proteins, α-FLAG for AvrB). Part showing AvrB is from the AvrB-GmRIN4a co-immunoprecipitation (Co-IP) experiment and is representative of Co-IPs with GmRIN4b, c and d.
In Arabidopsis, RIN4 also associates with the RPS2 (resistance to P. syringae 2) protein, which mediates resistance against P. syringae expressing avrRpt2. RPS2-mediated signaling is activated when AvrRpt2PtoJL1065, a cysteine protease, cleaves RIN4.23–25 Since absence of RIN4 results in the ectopic induction of RPS2 activity and thereby lethality, the rin4 mutation can be generated only in plants lacking RPS2 (rps2). Absence of RIN4 also activates residual RPM1 activity.13 Therefore, rin4 rps2 plants exhibit increased PR-1 (pathogenesis related 1) gene expression and enhanced basal resistance to virulent bacteria. The residual RPM1 activity is not however sufficient to provide resistance against avrB or avrRpm1 expressing bacteria. Thus, rin4 rps2 plants are compromised in RPM1-derived resistance against the avrB/avrRpm1 bacterial strains. Interestingly, overexpression of GmRIN4b, but not GmRIN4a, was able to restore RPM1 function in the Arabidopsis rin4 rps2 mutant. Pathogen inoculation of transgenic Arabidopsis rin4 rps2 mutant plants constitutively expressing GmRIN4a showed that these plants were as susceptible to avrB or avrRpm1 P. syringae as the rin4 rps2 mutant. In contrast, the 35S-GmRIN4b transgenic plants accumulated similar avrB or avrRpm1 bacteria as wild-type (ecotype Col-0) plants. Likewise, transgenic overexpression of GmRIN4b, but not GmRIN4a was able to complement the ecotopic induction of defenses in the rin4 rps2 mutant. The failure of GmRIN4a to complement the rin4 was not related to interaction with the R protein; both GmRIN4a and b associated with RPM1 as well as AvrRpm1 in BiFC17 as well as Co-IP assays (Fig. 2). Deciphering the reason underlying inability of GmRIN4a to complement the rin4 mutation should provide important insights into the RIN4 dependent activation of RPM1 activity.
Figure 2.
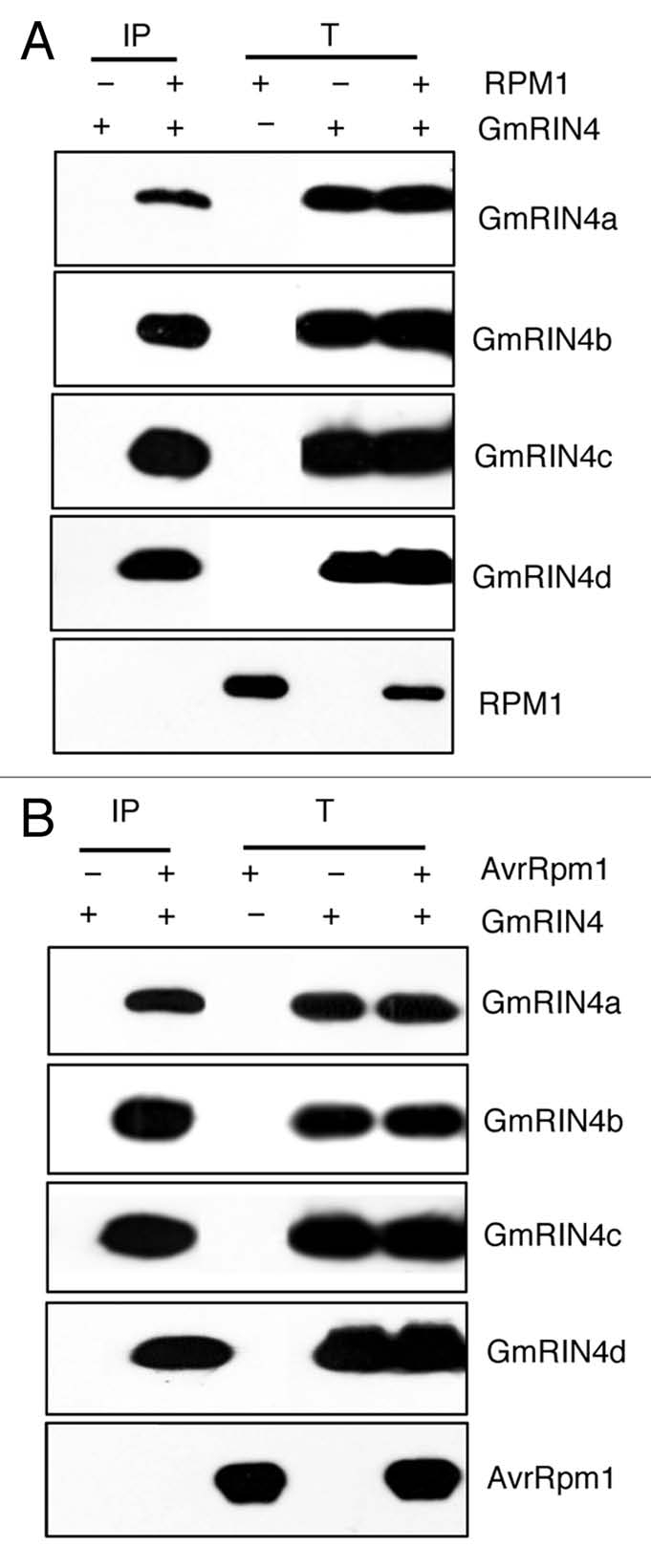
GmRIN4 proteins co-immunoprecipitate with RPM1 (A) and AvrRpm1 (B). Agrobacterium cells expressing MYC tagged GmRIN4a, b, c or d were expressed individually (GmRIN4) or together with 3XFLAG tagged AvrRpm1 (A) or RPM1 (B) in Nicotiana benthamiana. Proteins were immunoprecipitated (IP) from total extracts (T) using anti-FLAG antibodies. Proteins were visualized on western blots using tag-specific antibodies. Parts showing RPM1 and AvrRpm1 are from co-immunoprecipitation (Co-IP) experiments with GmRIN4a and are representative of Co-IPs with GmRIN4b, c and d.
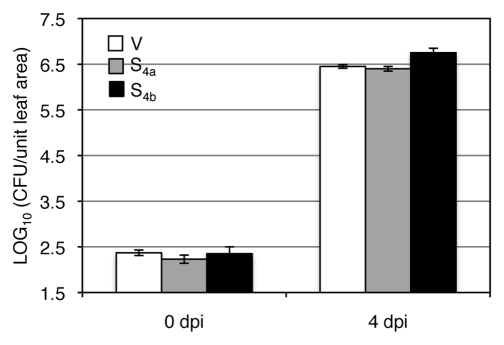
Interestingly, silencing either GmRIN4a or b enhanced resistance to virulent strains of P. syringae and the oomycete pathogen Phytophthora sojae in soybean. This suggested that both GmRIN4a and b contributed to basal defense in soybean. Increased basal defense in the GmRIN4a/b-silenced plants could not be attributed to residual Rpg1-b activity since the enhanced resistance phenotype was observed in the rpg1-b background (cv. Essex). Furthermore, the GmRIN4a- or b-silenced Rpg1-b plants (cv. Harosoy) accumulated similar levels of virulent bacteria as the control plants (Fig. 3). However, the possibility that loss of GmRIN4a or b activates other unidentified R proteins in the Essex cultivar cannot be ruled out. Assessing resistance in different genetic backgrounds lacking GmRIN4a and/or b will help clarify this. Inoculation of the GmRIN4a- or b-silenced rpg1-b plants with avrB bacteria showed that neither GmRIN4a nor b was required for the virulence function of AvrB; presence of AvrB enhanced bacterial growth on both GmRIN4a- and b-silenced rpg1-b plants. Interestingly, avrB bacteria were even more virulent on the GmRIN4a-silenced rpg1-b plants as compared to the control or GmRIN4b-silenced rpg1-b plants. These data suggest that GmRIN4a might negatively regulate the virulence function of AvrB. Analyzing the effects of GmRIN4a overexpression in the rpg1-b background will help clarify this.
Figure 3.
Silencing GmRIN4a or b does not enhance resistance to virulent Pseudomonas syrinage in Rpg1-b (cv. Harosoy) plants. Bacterial counts in plants silenced for GmRIN4a (S4a) or GmRIN4b (S4b) as compared to vector-inoculated (V) plants. LOG values of colony forming units (CFU) per unit leaf area from infected leaves at 0 or 4 days post-inoculation (dpi) are presented. Error bars indicate standard deviation (n = 5).
Conclusions and Future Prospects
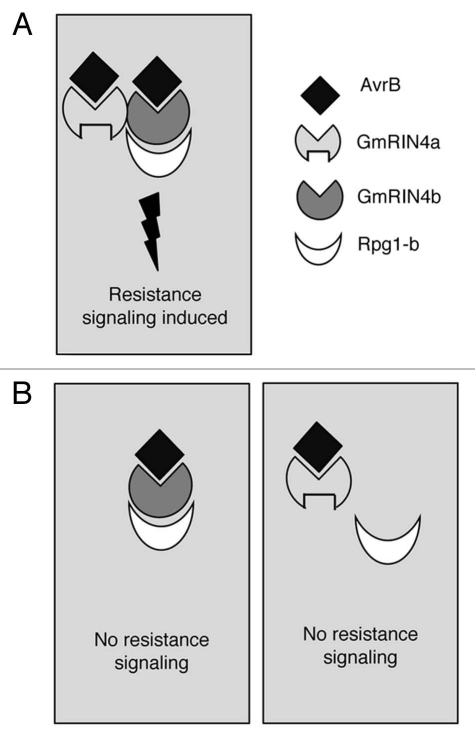
(i) Soybean encodes four RIN4-like proteins and at least two (GmRIN4a & b) of these are essential for Rpg1-b-mediated resistance signaling, even though only GmRIN4b directly interacts with Rpg1-b (Fig. 4). This emphasizes the need to examine the defense related roles of RIN4-like proteins in other plants reported to encode more than a single RIN4-like protein, including Arabidopsis, tomato and lettuce.14,15,26
Figure 4.
Role of soybean RIN4-like proteins (GmRIN4a & b) in Rpg1-b-derived resistance signaling. (A) GmRIN4a & b interact with AvrB and with each other. GmRIN4b, but not GmRIN4a, interacts with Rpg1-b. Binding of AvrB to GmRIN4a and/or b may be detected by Rpg1-b to induce resistance signaling. Whether activation of Rpg1-b requires binding of AvrB to GmRIN4a and/or b has not been demonstrated. (B) Absence of either GmRIN4a (right part) or b (left part) abrogates the induction of a resistance response.
(ii) GmRIN4b interacts with itself and the three other isoforms, raising the possibility that these proteins might function as heteromeric complexes whose dynamics may change in response to pathogen-derived factors including the Avr protein.
(iii) Only the Rpg1-b-interacting isoform, GmRIN4b, is able to complement RPM1 function in the Arabidopsis rin4 mutant, providing an exciting avenue for examining the precise mechanism of RPM1 activation via RIN4.
(iv) Both GmRIN4a and b participate in soybean basal defense, consistent with the predicted roles of “guardee” proteins.
(v) Similar to Arabidopsis RIN4, and consistent with the findings that RIN4 is not the only virulence target of Avr proteins13,27 neither GmRIN4a, nor b, is essential for the virulence function of AvrB. However, GmRIN4a likely act as a negative regulator of AvrB-dependent virulence in soybean. This is similar to AvrRpt2-dependent virulence in Arabidopsis, which is enhanced in rin4 mutant plants when delivered via the weak pathogen P. syringae pv. maculicola.13 Understanding the effects of the absence of GmRIN4a on AvrB could provide important insights into the mechanism via which Avr proteins exert virulence activity.
Acknowledgements
This work was supported by the United States Department of Agriculture-National Research Initiative (2006-01854) and the United Soybean Board (9244).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13462
References
- 1.Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, et al. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 2.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leister RT, Katagiri F. A resistance gene product of the nucleotide binding site—leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 2000;22:345–354. doi: 10.1046/j.1365-313x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 4.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Plant Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 6.Innes R. Guarding the goods; New insights into the central alarm system of plants. Plant Physiol. 2004;135:695–701. doi: 10.1104/pp.104.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 8.Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 9.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 10.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 11.Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 12.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 13.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeuken MJW, Zhang NW, McHale LK, Pelgrom K, den Boer E, Lindhout P, et al. Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell. 2009;21:3368–3378. doi: 10.1105/tpc.109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–2472. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Timko MP. Gene-for-gene resistance in Striga-cowpea associations. Science. 2009;325:1094. doi: 10.1126/science.1174754. [DOI] [PubMed] [Google Scholar]
- 17.Selote D, Kachroo A. RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol. 2010;153:1199–1211. doi: 10.1104/pp.110.158147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE. Regulatory role of SGT1 in early R gene-mediated plant defenses. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 19.Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu DQ, Ghabrial S, Kachroo A. GmRAR1 and GmSGT1 are required for basal, R gene-mediated and systemic acquired resistance in soybean. Mol Plant Microbe Interact. 2009;22:86–95. doi: 10.1094/MPMI-22-1-0086. [DOI] [PubMed] [Google Scholar]
- 22.Ashfield T, Keen NT, Buzzell RI, Innes RW. Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic or closely linked, at the RPG1 locus. Genetics. 1995;141:1597–1604. doi: 10.1093/genetics/141.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 24.Mackey D, Belkhadir Y, Alfonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 25.Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA. 2005;102:6496–6501. doi: 10.1073/pnas.0500792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Lim MTS, Kunkel BN. The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J. 2004;40:790–798. doi: 10.1111/j.1365-313X.2004.02251.x. [DOI] [PubMed] [Google Scholar]