Abstract
VERNALIZATION INSENSITIVE 3 (VIN3) is required for vernalization-mediated repression of FLOWERING LOCUS C (FLC) in Arabidopsis. The induction of VIN3 by long-term exposure to cold is one of earliest events in vernalization response. However, molecular mechanisms underlying for the VIN3 induction are poorly understood. Recently, we reported that the constitutive repression of VIN3 in the absence of the cold exposure is due to multiple repressive chromatin modifying components, including a transposable element (TE)-derived sequence, LIKE-HETEROCHROMATIN PROTEIN 1 (LHP1) and POLYCOMB REPRESSION COMPLEX 2 (PRC2). In addition, the maximum level of VIN3 induction requires EARLY FLOWERING 7 (ELF7) and EARLY FLOWERING IN SHORDAYS (EFS), which are components of activating chromatin modifying complexes. Furthermore, dynamic changes in histone modifications at VIN3 chromatin are observed during the course of vernalization. Thus, mechanisms underlying the induction of VIN3 include changes at the level of chromatin.
Key words: vernalization, flowering, chromatin
Vernalization can be defined as “the acquisition or acceleration of the ability to flower by a chilling treatment.”1 To maximize floral reproductive capability, plants in temperate climates have evolved a vernalization requirement to prevent flowering before the winter season and to ensure flowering in the spring. Vernalization response provides plants with competence to flower, rather than to induce flowering itself, through changes that remain stable even after cold exposure. This process is an epigenetic switch, whereby molecular changes remain stable throughout subsequent mitotic divisions despite the absence of initiating stimulus, cold exposure.2–4 Vernalization response can be thought of as being comprised of two phases. The first is a cold perception system that measures the cumulative time of exposure to cold. The second phase is essentially the output of cold perception system: when a sufficient duration of cold has been perceived, a series of changes of gene expression ensue, ultimately leading to the epigenetic repression of FLC.
Measurement of Duration of Cold in Arabidopsis
Although a vernalization response may only be elicited at near or above freezing temperatures, the exact temperatures and length of cold exposure required to efficiently prompt a response varies among plant species.5 Since cold perception by plants involves the measurement of the length of exposure, it is the duration of cold that creates two distinct responses—cold acclimation and vernalization. Entering a state of cold acclimation requires a short period of time, typically within hours of low and non-freezing temperatures.6 The low threshold required to elicit a cold acclimation response is quite critical in dealing with fluctuating temperatures. Conversely, achieving a vernalized state in plants occurs only after a sufficient and prolonged exposure to cold (e.g., weeks to months) has occurred, indicative of the passing of winter. Notably, the molecular components tested to date that affect cold acclimation (a short-term cold response) have no effect on vernalization, suggesting that the cold perception system during vernalization response represents a distinct regulatory process specifically responsible for long-term cold sensing.
Regulation of VIN3 at the Level of Chromatin
As of yet, the most direct link between measurement of the duration of cold and the output of vernalization response is the induction of VIN3 expression, which is tightly linked to prolonged cold exposure and whose expression is completely abrogated upon return to warm temperatures.7 VIN3 encodes a PHD (plant homeodomain) protein, a motif routinely found in a variety of proteins involved in modifying chromatin.8 After induction, VIN3 expression initiates a series of repressive histone modifications such as methylations at Histone H3 Lys 9 (H3K9) and Histone H3 Lys 27 on FLC, a potent floral repressor.2,3 FLC repression by vernalization requires components of Ploycomb Repression Complex 2 (PRC2) and LIKE-HETEROCHROMATIN PROTEIN 1 (LHP1).9–12 Interestingly, VIN3 is also de-repressed in the absence of LHP1 and components of PRC2 prior to the cold exposure, suggesting that common components are necessary for the repression of both VIN3 and its target, FLC. Both LHP1 and PRC2 increase their associations with FLC chromatin to repress FLC during the course of vernalization.9–13 On the other hand, LHP1 and PRC2 are constantly associated with VIN3 chromatin during the course of vernalization; LHP1 and PRC2 are enriched at VIN3 chromatin before vernalization and remain associated even when VIN3 is induced during vernalization. Thus, VIN3 induction overcomes the repressive effects of LHP1 and PRC2.
Screens for rapidly flowering mutants in Arabidopsis revealed ELF7 and ELF8, two homologs of the yeast PAF1 complex, which are required for high FLC expression and Histone H3 Lys 4 trimethylation (H3K4me3) enrichment at FLC chromatin.14,15 Induced levels of VIN3 by vernalization are significantly lower in elf7 and efs mutants compared to the wild-type, implicating their function in VIN3 activation. Concomitant with ELF7 involvement in VIN3 induction, H3K4me3 is enriched during vernalizing cold treatment, when VIN3 is induced. Thus, activating complex components (e.g., PAF1 and Trithorax-like proteins) are likely required for the full extent of VIN3 induction by vernalization.
Although H3K27me3 and H3K9me2 are constantly enriched at VIN3 chromatin during the course of vernalization, an activation mark, H3K4me3 becomes enriched when VIN3 is induced under the cold exposure, creating a bivalent state characterized by both repressive and active histone marks concomitantly. Such bivalent chromatin states are typical of genes that are prepared to be activated or repressed, thus providing elasticity for gene expression in undifferentiated cells.16,17 It is considerable that this bivalent state of repressive marks and active marks of VIN3 chromatin could contribute to a rapid re-repression of VIN3 when plants return to warm growth temperature.
Common Machineries for Histone Modifications of Genes Involved in Arabidopsis Flowering
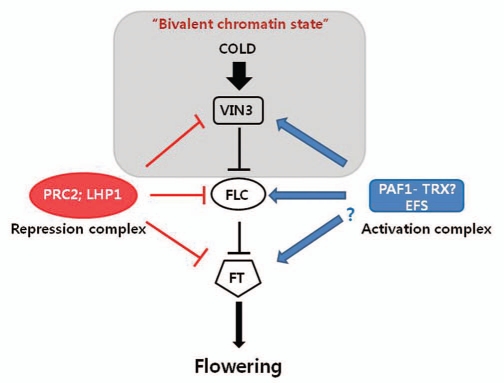
The direct association of PRC2 and LHP1 with VIN3 chromatin suggests these common regulators play roles in the repression of both VIN3 and its target, FLC. Furthermore, PRC2 and LHP1 are also required for the repression of FLOWERING LOCUST (FT), a target of FLC.18–20 In addition, common activators, including PAF1 and EFS, are also necessary for the full extent of both VIN3 and FLC. Thus, the VIN3-FLC-FT regulatory network to control flowering by vernalization has common regulatory components as well as specific regulatory mechanisms to shift the status of genes regulated by these common regulators (Fig. 1). One example is vernalization-mediated repression of FLC. Prolonged cold exposure induces VIN3 expression, which over-comes repressive activity of PRC2-LHP1 on VIN3 locus. After induction, VIN3 is apparently required for the repression of FLC by PRC2. VIN3 biochemically co-purifies with PRC2, suggesting that the formation of VIN3-PRC2 complex enhances the repressive activity of PRC2 on FLC. Therefore, the induced nature of VIN3 by vernalization shifts the activity of the VIN3-FLC-FT network towards the repression of FLC and the activation of FT, which results in promotion of floral transition (Fig. 1).
Figure 1.
Common regulatory networks to control the VIN3-FLC-FT pathway to control flowering time by vernalization. Common repression and activation complexes are likely involved in regulations of VIN3, FLC and FT. Bivalent chromatin status at VIN3 chromatin is achieved by vernalizing cold, resulting in transient induction of VIN3.
Components Specific for the Induction of VIN3 by Vernalization Are Not Known
VIN3 induction by vernalization still occurs in the absence of either PAF1 or EFS and de-repression of VIN3 in the absence of repressive complexes (i.e., PRC2 and LHP1) is not sufficient for the complete induction. Thus, there must be unknown cold-induced activators and/or cold-repressed repressors responsible for maximum VIN3 induction during vernalization. Together, these unknown factors along with PAF1 and EFS are likely responsible for the full extent of VIN3 induction during prolonged cold exposure, which overcomes repression by PRC2 and LHP1. Since VIN3 is required for vernalization response, it is conceivable that we would recover mutants that affect induction of VIN3 from vernalization mutant screens. To date, however, no mutant that affects vernalization response impairs VIN3 induction by vernalization. Although it is possible that the lack of such mutants is simply a matter of lack of saturating mutagenesis, it is also possible that involvement of common regulators in the VIN3-FLC-FT network impedes the recovery of such mutants based on flowering times. Thus, the efforts to study the VIN3 regulation may need to focus on the VIN3 expression itself during the course of vernalization rather than on conventional flowering mutant screens. Further study on this unknown initial factor is necessary to fully understand on mechanisms underlying vernalization-mediated VIN3 induction and it will shed insights on long-term cold sensing mechanism of plants.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13465
References
- 1.Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Physiol. 1960;11:191–238. [Google Scholar]
- 2.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 3.Dennis ES, Peacock WJ. Epigenetic regulation of flowering. Curr Opin Plant Biol. 2007;10:520–527. doi: 10.1016/j.pbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Wu C, Morris JR. Genes, genetics and epigenetics: a correspondence. Science. 2001;293:1103–1105. doi: 10.1126/science.293.5532.1103. [DOI] [PubMed] [Google Scholar]
- 5.Lang A. Physiology of flower initiation. Berlin: Springer-Verlag; 1965. [Google Scholar]
- 6.Thomashow MF. So what's new in the field of plant cold acclimation? Lots! Plant Physiol. 2001;125:89–93. doi: 10.1104/pp.125.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- 8.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 9.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, et al. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 12.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh S, Zhang H, Ludwig P, van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- 19.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang D, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One. 2008;3:3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]