Abstract
A method to detect binary interactions among SNAREs, membrane proteins mediating vesicle fusion, in Arabidopsis cells was established. In this method, a pair of recombinant SNAREs is first expressed within Arabidopsis protoplasts at levels similar to their endogenous proteins in 96-well plates. Changes of the interaction are then detected by luminescence. Here, we report that the interaction of SYP122 and VAMP721, a SNARE pair mediating exocytosis, is enhanced when Arabidopsis protoplasts are incubated in the dark. Microscopic observation of plants expressing GFP-SYP122 by the syp122 promoter suggests SYP122 is expressed in the root tip when the seedlings are grown in the dark but not in the light. In the identical dark-grown condition, the subcellular localization of SYP111/KNOLLE, specifically expressed in dividing cells, is altered. Together with our previous report, we hypothesize that expression, localization and interaction of SNAREs are selectively altered by light conditions to regulate cargo transports in Arabidopsis.
Key words: luciferase complementation assay, arabidopsis, SNARE, vesicle trafficking, light
Vesicle trafficking plays a crucial role in cellular development and adaptation in both animals and plants.1,2 SNAREs (N-ethylmaleimide sensitive factor attachment protein receptors) are membrane proteins that mediate fusion between vesicles and organelles to transport cargo molecules within the cells.3 At least one SNARE molecule is localized in a vesicle4 (designated v-SNARE). When the vesicle arrives at the target organelle, the SNARE in the vesicle physically interacts with SNAREs localized in the target (designated t-SNARE). Because selected members of the SNARE family proteins are localized in selected types of organelles, SNARE-SNARE interactions are thought to occur very selectively in the cells, though not necessarily selective in vitro,5–7 among the SNARE family members encoded in the genome.3 Arabidopsis genome encodes at least 64 SNARE family members,8 and their selective subcellular localizations were previously determined.9,10 We thought mapping changes of vesicle trafficking patterns/pathways in Arabidopsis would be useful to understand the mechanism of cargo transports during cellular development and adaptation in plants. To this end, we developed a method, based on a split-luciferase complementation assay,11,12 to assess SNARE-SNARE interactions in Arabidopsis cells in a high-throughput format.13 In this method, a pair of recombinant SNARE proteins, which are fused to the N- and C-terminal fragment of Renilla luciferase respectively, are expressed by constitutive promoters within Arabidopsis protoplasts in 96-well plates.
In a previous publication,13 we reported that accumulation levels of the recombinant SNAREs in protoplasts are similar to that of their endogenous proteins. The results from the method largely agreed with that from co-immunoprecipitation assays. We also demonstrated the capability of detecting changes of a SNARE-SNARE interaction by luminescence in real-time. Using the method, we found a novel phenomenon that the interaction between SYP121 and VAMP722, a SNARE pair mediating exocytosis,14 is enhanced when Arabidopsis protoplasts were incubated in the light. Furthermore, we showed that seedlings of transgenic Arabidopsis expressing GFP-SYP121 (green fluorescent protein tagged SYP121) by the syp121 promoter accumulate GFP-SYP121 in the vacuoles of the root tip when the seedlings are grown in the dark (24 h) for 5 days. On the other hand, when the seedlings are grown in the light (16 h light and 8 h dark) for 5 days, GFP-SYP121 is localized in the plasma membrane. Based on these findings, we suggested that light signaling may upregulate plasma membrane localization of SYP121 and enhance the interaction with VAMP722.
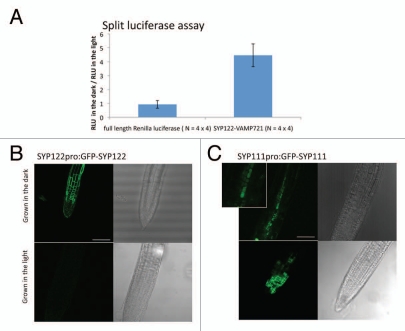
As an addendum, we examined the interaction between SYP122 and VAMP721, closest ortholog of SYP121 and VAMP722 respectively.8 Because several studies have suggested functional overlap among these orthologous SNARE proteins,15–17 we expected the interaction between SYP122 and VAMP721 to be also enhanced in the light. However, surprisingly for us, the interaction between SYP122 and VAMP721 is enhanced when Arabidopsis protoplasts are incubated in the dark but not in the light (Fig. 1A). Seedlings of transgenic Arabidopsis that express GFP-SYP122 by the syp122 promoter10 accumulate GFP-SYP122 in the plasma membranes in the root tip when the seedlings are grown in the dark (24 h) for five days (Fig. 1B). On the other hand, when the seedlings are grown in the light (16 h light and 8 h dark) for 5 days, GFP-SYP122 is not detected in the root tip (Fig. 1B). These suggest that, in contrast to SYP121 and VAMP722, the continuous dark may upregulate the expression of SYP122 and also enhance the interaction with VAMP721. This finding led us to examine the expressions of other SNARE family members in the continuous dark. We analyzed previously published microarray data in wild type seedlings (the microarray probes 61 SNARE family members).18 Our analysis by Genevestigator V3 with a default setting19 identified 11 SNAREs, including SYP122, that are upregulated (>2.0 fold) in dark-grown seedlings, compared to light-grown seedlings (Table 1). This suggested that not only SYP122 but at least 10 other SNAREs may be upregulated by the continuous dark at a transcription level in seedlings. The analysis also revealed that SYP111/KNOLLE, specifically expressed in dividing cells and localized in the cell division planes,20 is downregulated (<0.5 fold) in the dark-grown seedlings (Table 1). To examine the regulation of SYP111 at a cell level, we observed the root tip of transgenic Arabidopsis that expresses GFP-SYP111 by the syp111 promoter.10 When the seedlings are grown in the light (16 h light and 8 h dark) for 5 days, GFP-SYP111 is localized in the division planes in the root tip as others reported10,21 (Fig. 1C). However, when the seedlings are grown in the dark (24 h) for 5 days, GFP-SYP111 is localized in the vacuoles as well (Fig. 1C). This suggests that the continuous dark may not only downregulate the gene expression but also relocate SYP111 from the division planes to the vacuoles.
Figure 1.
Changes in expression, localization and interaction of SNARE proteins in Arabidopsis cells in the dark. (A) Enhancement of the SYP122-VAMP721 interaction in the split luciferase complementation assay. Recombinant SYP122 and VAMP721 proteins, genetically fused to the N- and C-terminal fragment of Renilla luciferase respectively, were expressed by constitutive promoters in Arabidopsis protoplasts. The protoplasts were then incubated in the light (100 µmol/m2/s) or the dark for 16 h before measuring relative luminescence units (RLU). As a control, full length Renilla luciferase was expressed. An average of the RLUs of four independent transformations in a 96-well plate incubated in the dark was divided by an average of the RLUs of four independent transformations in another 96-well plate incubated in the light. The results from four independent experiments are presented with the standard errors. The changes of the RLUs in SYP122-VAMP721 (4.46-fold) are significantly (p = 0.012) higher than that in full-length Renilla luciferase (0.93-fold). (B) GFP-SYP122 is expressed in the root tip in the dark. Seedlings of transgenic Arabidopsis expressing GFP-SYP122 by the syp122 promoter (SYP122PRO:GFP-SYP122) were grown in the dark (24 h) or light (100 µmol/m2/s, 16 h light and 8 h dark) for five days. Transmission images of root tips are shown on the right side of fluorescence images for reference. Bar = 500 µm. (C) GFP-SYP111/KNOLLE is localized in the vacuoles, instead of division planes, in the dark. Seedlings of transgenic Arabidopsis expressing GFP-SYP111 by the syp111 promoter (SYP111PRO:GFP-SYP111) were grown in the dark (24 h) or light (100 µmol/m2/s, 16 h light and 8 h dark) for five days. An enlarged fluorescence image for the dark-grown seedling is also shown to clarify the vacuole accumulation of GFP-SYP111. Bar = 500 µm.
Table 1.
Summary of changes in expression, interaction, and localization of SNAREs by the dark
| Annotation | SYP73 | SYP72 | VTI14 | VAMP711 | SYP23 | SYP51 | VAMP712 |
| Gene locus | AT3G61450 | AT3G45280 | AT5G39630 | AT4G32150 | AT4G17730 | AT1G16240 | AT2G25340 |
| Default localization# | ER/PM | ER/PM | Endosome | Vacuole | LE/Vacuole | Endosome | TGN/Vacuole |
| Expression* (fold change) | 7.69 | 5.56 | 5.00 | 3.45 | 3.33 | 3.12 | 2.63 |
| Interaction** (fold change) | - | - | - | - | - | - | - |
| Localization in the root tip*** | - | - | - | - | - | - | - |
| Annotation | SYP124 | SYP122 | SYP24 | MEMB11 | SYP111 | SYP121 | SYP132 |
| Gene locus | AT1G61290 | AT3G52400 | AT1G32270 | AT2G36900 | AT1G08560 | AT3G11820 | AT5G08080 |
| Default localization# | PM | PM | LE/Vacuole | ER/TGN | DP | PM | PM |
| Expression* (fold change) | 2.56 | 2.50 | 2.27 | 2.12 | 0.38 | 0.86 | 0.97 |
| Interaction** (fold change) | - | 4.46 | - | - | - | 0.28 | 0.73 |
| Localization in the root tip*** | - | PM | - | - | Vacuole | Vacuole | PM |
| Control |
Localizations defined by Bassham, et al. (2008) and Uemura, et al. (2004). ER, endoplasmic reticulum; PM, plasma membrane; LE, late endosome; TGN, trans Golgi network; DP, division plane.
Results of the analysis on microarray data (Dohmann, et al. 2008) by Genevestigator V3 with a default setting (Hruz, et al. 2008). Expression signals in Arabidopsis seedlings germinated in the dark for 7 days are divided by that in seedlings germinated in the light for 7 days. SNAREs that show higher than 2 fold or lower than 0.5 fold are listed. SYP121 and SYP132 are also listed for the comparison.
Results of the split luciferase complementation assays. RLU (relative luminescence unit) in Arabidopsis protoplasts incubated in the dark for 16 h is divided by that in Arabidopsis protoplasts incubated in the light for 16 h. The interaction between SYP122 and VAMP721 or between SYP121 and VAMP722 or between SYP132 and VAMP722 is measured. The data of SYP121 and SYP132 are from the previous publication (Kato, et al. 2010).
Results of the GFP observation in the dark. Transgenic Arabidopsis that expresses GFP-SYP122, GFP-SYP121 or GFP-SYP111 (by the promoter of syp122, syp121 or syp111 respectively) is observed with a confocal microscope. The seedlings are grown in the dark (24 h) or in the light (16 h light 8 h dark) for 5 days before the observation. Subcellular localizations in the root tip are shown. GFP-SYP121 and GFP-SYP111 are localized in the plasma membrane and division planes, respectively, in the root tip when the seedlings are grown in the light. GFP-SYP122 is not detectable in the root tip when the seedlings are grown in the light. The data of SYP121 and SYP132 are from the previous publication (Kato, et al. 2010). -, not analyzed.
Together with our previous finding13 (summarized in Table 1), we now hypothesize that light conditions regulate selected pathways of vesicle trafficking to define intracellular distribution of cargo molecules in Arabidopsis. The examples are seen in changes of intracellular distribution of selected membrane proteins in the dark.22 We believe the regulation is detected as changes in expression, localization and interaction in selected SNAREs. Studies to confirm our findings by other methods are the focus of our next investigation.
Acknowledgements
N.K. thanks Dr. M.H. Sato at Kyoto Prefectural University in Japan for providing the transgenic Arabidopsis and Irene M. Bognar for critical reading.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13480
References
- 1.Fisher RJ, Pevsner J, Burgoyne RD. Control of fusion pore dynamics during exocytosis by Munc18. Science. 2001;291:875–878. doi: 10.1126/science.291.5505.875. [DOI] [PubMed] [Google Scholar]
- 2.Surpin M, Raikhel N. Traffic jams affect plant development and signal transduction. Nat Rev Cell Biol. 2004;5:100–109. doi: 10.1038/nrm1311. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 4.van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R. One SNARE complex is sufficient for membrane fusion. Nat Struct Mol Biol. 2010;17:358–364. doi: 10.1038/nsmb.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duman JG, Forte JG. What is the role of SNARE proteins in membrane fusion? Am J Physiol. 2003;285:237–249. doi: 10.1152/ajpcell.00091.2003. [DOI] [PubMed] [Google Scholar]
- 6.Fasshauer D, Antonin W, Margittai M, Pabst S, Jahn R. Mixed and non-cognate SNARE complexes. Characterization of assembly and biophysical properties. J Biol Chem. 1999;274:15440–15446. doi: 10.1074/jbc.274.22.15440. [DOI] [PubMed] [Google Scholar]
- 7.Yang B, Gonzalez L, Jr, Prekeris R, Steegmaier M, Advani RJ, Scheller RH. SNARE interactions are not selective. Implications for membrane fusion specificity. J Biol Chem. 1999;274:5649–5653. doi: 10.1074/jbc.274.9.5649. [DOI] [PubMed] [Google Scholar]
- 8.Sanderfoot A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 2007;144:6–17. doi: 10.1104/pp.106.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassham Diane C, Brandizzi F, Otegui Marisa S, Sanderfoot Anton A. The Secretory System of Arabidopsis. The Arabidopsis Book: The American Society of Plant Biologists. 2008:1–29. doi: 10.1199/tab.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enami K, Ichikawa M, Uemura T, Kutsuna N, Hasezawa S, Nakagawa T, et al. Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:280–289. doi: 10.1093/pcp/pcn197. [DOI] [PubMed] [Google Scholar]
- 11.Fujikawa Y, Kato N. Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 2007;52:185–195. doi: 10.1111/j.1365-313X.2007.03214.x. [DOI] [PubMed] [Google Scholar]
- 12.Kato N, Jones J. Split luciferase complementation assay. In: Henning L, Kohler C, editors. Methods in Plant Development. NY: Humana Press Inc; 2010. pp. 359–376. [Google Scholar]
- 13.Kato N, Fujikawa Y, Fuselier T, Adamou-Dodo R, Nishitani A, Sato MH. Luminescence detection of SNARE-SNARE interaction in Arabidopsis protoplasts. Plant Mol Biol. 2010;72:433–444. doi: 10.1007/s11103-009-9581-z. [DOI] [PubMed] [Google Scholar]
- 14.Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 15.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 16.Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell. 2004;15:5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pajonk S, Kwon C, Clemens N, Panstruga R, Schulze-Lefert P. Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J Biol Chem. 2008;283:26974–26984. doi: 10.1074/jbc.M805236200. [DOI] [PubMed] [Google Scholar]
- 18.Dohmann EM, Levesque MP, De Veylder L, Reichardt I, Jurgens G, Schmid M, et al. The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development. 2008;135:2013–2022. doi: 10.1242/dev.020743. [DOI] [PubMed] [Google Scholar]
- 19.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, et al. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv Bioinform. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukowitz W, Mayer U, Jurgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- 21.Dhonukshe P, Baluska F, Schlicht M, Hlavacka A, Samaj J, Friml J, et al. Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell. 2006;10:137–150. doi: 10.1016/j.devcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Laxmi A, Pan J, Morsy M, Chen R. Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS ONE. 2008;3:1510. doi: 10.1371/journal.pone.0001510. [DOI] [PMC free article] [PubMed] [Google Scholar]