Abstract
Phylogenies of Adh1 and Adh2 genes suggest that a widespread Mediterranean peony, Paeonia officinalis, is a homoploid hybrid species between two allotetraploid species, Paeonia peregrina and a member of the Paeonia arietina species group. Three phylogenetically distinct types of Adh sequences have been identified from both accessions of P. officinalis, of which two types are most closely related to the two homoeologous Adh loci of the P. arietina group and the remaining type came from one of the two Adh homoeologs of P. peregrina. The other Adh homoeolog of P. peregrina was apparently lost from the hybrid genome, possibly through backcrossing with the P. arietina group. This is a documentation of homoploid hybrid speciation between allotetraploid species in nature. This study suggests that hybrid speciation between allotetraploids can occur without an intermediate stage of genome diploidization or a further doubling of genome size.
Hybridization is a widely documented mode of speciation in flowering plants (1). Two models of hybrid speciation, homoploid hybrid speciation and allopolyploidization, have been described (2). Although examples of allopolyploidy are found throughout angiosperms, conformed cases of homoploid hybrid species are rare (3). The rarity of homoploid hybrid species may be due to a combination of factors such as hybrid sterility, hybrid breakdown, difficulty of evolving reproductive isolation in sympatry, and difficulties in unambiguous identification of homoploid hybrid species (2, 4).
Documented examples of homoploid hybrid species in nature have so far been limited to diploids (3). However, there are no theoretical reasons why this mode of speciation could not occur among polyploid species. In fact, the classic experimental demonstration of this mode involved tetraploid species of Gilia (5, 6). Here we report a natural example of a homoploid hybrid species, Paeonia officinalis, that has arisen following hybridization between allotetraploid peony species.
The genus Paeonia comprises approximately 35 species of shrubs and perennial herbs distributed in disjunct areas of the northern temperate region (7). The Mediterranean region accommodates nearly 20 herbaceous Paeonia species, of which two-thirds are tetraploids (2n = 20). Although previous cytogenetic studies suggested that the majority of the tetraploid species were allotetraploids (8, 9), the origin of the putative allotetraploids had not been reconstructed until recent analyses of DNA sequences, in particular sequences of low-copy nuclear gene Adh (10–12).
The Adh genes constitute a small gene family with two to three loci in diploid angiosperms (13). This is one of the best-studied low-copy nuclear gene families in plants and has been used for phylogenetic inference of interspecific relationships in a number of flowering plant groups (14–17). Peonies have two Adh genes, Adh1 and Adh2, that were duplicated before diversification of the genus Paeonia. Previous phylogenetic analyses indicated that each of the Adh1 and Adh2 genes, except for the Adh2 of Paeonia veitchii, is orthologous among the diploid Paeonia species (16). Phylogenetic analyses of Adh gene sequences have led to the reconstruction of origins of several allotetraploid Paeonia species (12). In this study, Adh phylogenies provided evidence for the homoploid hybrid origin of P. officinalis from two allotetraploid parental species.
Materials and Methods
The same accessions used in the previous phylogenetic studies of Paeonia (11) were included in this study. Paeonia officinalis2 was a new accession collected from Serra S. Antonio, Italy. Classification of Paeonia species included in this study is as follows: section Paeonia includes diploid species P. anomala, P. cambessedesii, P. lactiflora, P. mlokosewischi, P. tenuifolia, and P. veitchii, and tetraploid species P. arietina, P. humilis, P. officinalis, P. parnassica, and P. peregrina; section Oneapia includes P. californica; section Moutan includes P. lutea, P. rockii, P. suffruticosa, and P. szechuanica.
DNA extraction followed a standard CTAB protocol (18). Adh1 and Adh2 genes were amplified with the gene-specific primers: primers AdhF2 and Adh1R for the Adh1 gene and primers Adh2F and Adh2R for the Adh2 gene (12). PCR was conducted under reaction conditions reported previously (16). PCR products were cloned into plasmids by using TOPO TA cloning kits (Invitrogen). At least 15 clones with the correct insert (determined by digestion with EcoRI) were screened for each PCR. Adh1 clones were screened by comparing restriction fragments of EcoRV and AseI, and Adh2 clones were digested with HaeIII and AseI. All clones that were unique were sequenced in both directions. Sequencing was completed on an ABI373 automated sequencer using a DYEnamic ET Terminator Cycle Sequencing Premix Kit (Amersham Pharmacia). Sequences were edited in SeqEd (Perkin–Elmer Applied Biosystems) and aligned manually. Clones that clearly resulted from PCR recombination (16, 19) were not included in the analyses.
Parsimony, as implemented in PAUP* Version 4.0 (20), was used to infer phylogenies based on nucleotide substitutions in aligned sequences. Parsimony analyses were performed by heuristic search with TBR branch swapping, MULPARS option, ACCTRAN optimization, and 100 random addition replicates for the Adh data sets. Bootstrap analyses (21) were carried out with 1,000 replications of heuristic search with simple taxon addition and “maxtrees” set to 500. The trees were rooted between section Moutan and the other two sections based on intersectional relationships determined previously (16). The Kishino–Hasegawa test (22) was used to compare topologies of the Adh1 and Adh2 trees as well as the likelihood of the most parsimonious solutions to alternative hypotheses of relationships within each tree. The model of sequence evolution for both Adh1 and Adh2 data sets was HKY + G, as determined by MODELTEST Version 3.0 (23).
Results
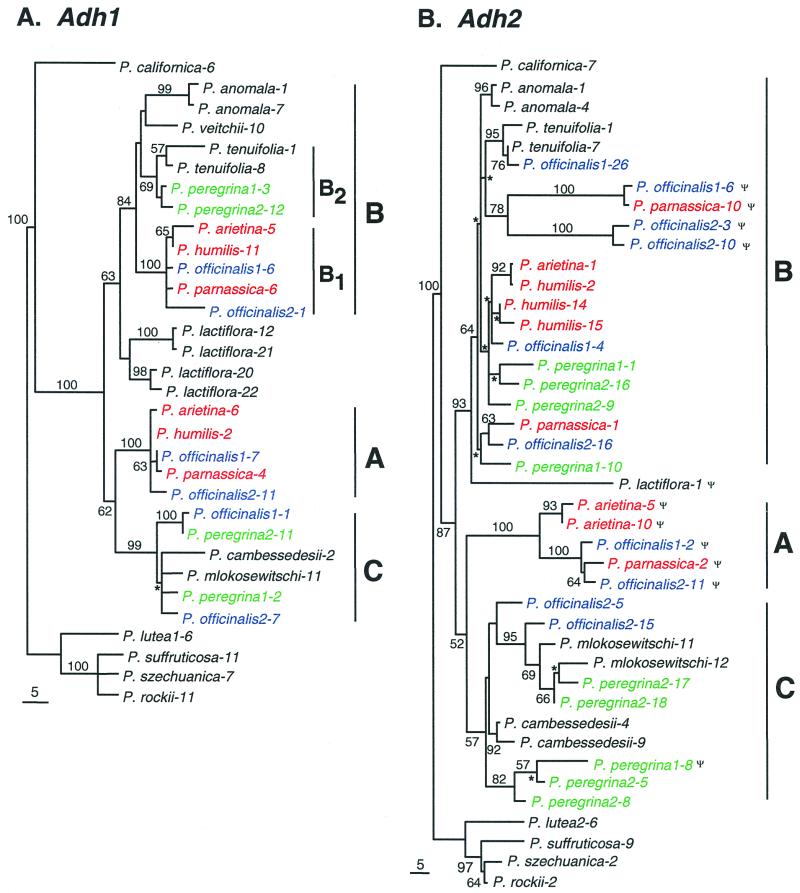
The aligned sequences of the Adh1 gene were 1214-bp long, of which 162 nucleotide sites were variable and 83 were phylogenetically informative. The Adh2 data set contained 1,186 nucleotide sites, of which 204 sites were variable and 175 were phylogenetically informative. Analysis of the Adh1 data set yielded 11 equally most parsimonious trees with a tree length of 196, a consistency index (CI) of 0.88, and a retention index (RI) of 0.93. One of the most parsimonious trees was randomly chosen and shown in Fig. 1A. Analysis of the Adh2 data set yielded 312 equally most parsimonious trees with a tree length of 427, CI of 0.79, and RI of 0.85. A randomly chosen Adh2 tree is shown in Fig. 1B.
Figure 1.
Adh gene phylogenies of Paeonia. Diploids are in black, and tetraploids are in color: green, P. peregrina; blue, P. officinalis; red, P. arietina species group. (A) One of 11 most parsimonious Adh1 trees chosen at random (tree length = 196, 114 without uninformative characters; consistency index = 0.88, 0.79 without uninformative characters; retention index = 0.93). (B) One of 312 most parsimonious Adh2 trees chosen at random (tree length = 427, 291 without uninformative characters; consistency index = 0.79, 0.69 without uninformative characters; retention index = 0.85). Numbers associated with branches are bootstrap percentages above 50%. Branch lengths are proportional to the number of nucleotide substitutions (scale represents five substitutions). Asterisks denote clades that collapse in the strict consensus. Pseudogenes are identified with Ψ. The A, B (B1, B2), and C sequence types are indicated. A number following a species name represents an accession number. A number following a hyphen represents a clone number.
Two distinct types of sequences of Adh1 and Adh2 genes were cloned from three closely related tetraploid species, P. arietina, P. humilis, and P. parnassica (Fig. 1). One type, which forms a well supported monophyletic group with diploid species P. anomala and P. tenuifolia, is designated as type B. The other type, consisting of only pseudogenes for Adh2, is designated as type A. The A-type Adh2 sequence was not recovered from P. humilis probably because of mutations at the PCR priming sites of the pseudogene (12). These three tetraploid species are referred to here as the P. arietina group. Forcing A and B-type sequences of the P. arietina group to be monophyletic was rejected by both the Adh1 (P = 0.04) and Adh2 (P = 0.002) data based on the Kishino–Hasegawa test (22).
Two distinct types of Adh1 and Adh2 sequences were also identified from both accessions of P. peregrina (Fig. 1). One type is nested within the B clade of the Adh phylogenies. The other type is clustered with diploid species P. cambessedesii and P. mlokosewischi in the C clade. Forcing type B and C sequences of P. peregrina into a monophyletic group was rejected by the Adh1 (P < 0.0001) and Adh2 (P < 0.0001) data.
In the B clade of the Adh1 phylogeny, sequences of the P. arietina group form a strongly supported clade which is further designated as B1. The sequences of P. peregrina and a diploid species P. tenuifolia form another clade designated as B2. Therefore, the P. arietina group has A and B1 types of Adh1 sequences, and P. peregrina has B2 and C types. On the Adh2 tree, however, B-type sequences of the P. arietina group or P. peregrina do not form their own groups (Fig. 1B).
Three types of Adh1 sequences were cloned from both accessions of P. officinalis, and fell into three strongly supported clades, A, B1, and C (Fig. 1A). On the Adh2 phylogeny, the accession P. officinalis2 also has the A, B, and C types (Fig. 1B). Only the A and B types of Adh2 sequences were recovered from P. officinalis1.
Relationships in the B clade of the Adh2 tree are obscured by a lack of resolution. This appears to be a result of dynamic gene duplication and deletion at the Adh2 locus, as indicated by the presence of a larger number of clones, as well as pseudogenes. Nevertheless, creating a B1 clade by forcing B-type sequences of the P. arietina group and P. officinalis (except for P. officinalis1-26) to be monophyletic was not rejected by the Adh2 data (P = 0.26). Placing P. officinalis1-26 within the B1 clade, however, was rejected by the Kishino–Hasegawa test (P = 0.013). Including P. officinalis1-26 with P. peregrina and P. tenuifolia, or the presence of a B2 clade in the Adh2 tree, was not rejected (P = 0.22). Therefore, the B1 and B2 types of Adh2 sequences seem to be hidden by a lack of resolution in the Adh2 phylogeny. The sequence P. officinalis1-26 is most likely to represent the B2 type, whereas the remaining P. officinalis sequences in the B clade are likely the B1 type. Therefore, P. officinalis2 has three types of Adh2 sequences, A, B1, and C, which are the same as the Adh1 types of both accessions. P. officinalis1, however, has A, B1, and B2 types of Adh2 sequences.
Discussion
Both accessions of P. officinalis have three distinct types of Adh1 sequences, of which A and B1 types are most closely related to the P. arietina group. The previous cytogenetic evidence and molecular phylogenetic data suggested that the P. arietina group has an allotetraploid origin, and the A and B1 types of Adh1 sequences represent the two Adh1 homoeologs derived from the diploid parents (12). The existence of an additional C type of Adh1 gene in P. officinalis could be explained by two alternative hypotheses. One suggests that P. officinalis had the same allotetraploid origin as the P. arietina group, and the C-type sequence is a result of a gene duplication. The other considers P. officinalis to be a homoploid hybrid species with one parent being the P. arietina group that contributed the A- and B1-type sequences and the other tetraploid parent that donated the C-type sequence.
The gene duplication hypothesis is very unlikely because it requires many additional assumptions. This hypothesis assumes that the gene was duplicated before diversification of the A and C clades, and one of the copies was subsequently deleted independently from all six other species except P. officinalis in the A and C clades (Fig. 1). Moreover, identification of the same three types of sequences, A, B1, and C, at both Adh1 and Adh2 loci of P. officinalis2 requires another hypothesis that the same pattern of gene duplication and deletion occurred between the two loci.
The hybridization hypothesis is favored not only because it requires fewer hypotheses, but also because both parents of the hybrid can be identified based on the Adh phylogenies. Whereas the P. arietina group is recognized as one of the parents of P. officinalis, the other parent is most likely P. peregrina. The C-type sequences of P. officinalis are closely related to those from P. peregrina. Particularly, the clone P. officinalis1-1 forms a strongly supported sister group with P. peregrina2-11 on the Adh1 tree. In addition, the monophyly of all P. peregrina and P. officinalis clones in the C clade is among the most parsimonious solutions of the Adh1 data. Formation of a monophyletic group of the C-type sequences of P. officinalis2 and P. peregrina is not rejected by the Adh2 data (P = 0.088).
P. peregrina was previously considered to be an allotetraploid based on cytogenetic evidence and sequence additivity in the internal transcribed spacers of nuclear ribosomal DNA (9, 10). Identification of the B and C types of sequences of Adh1 and Adh2 genes from both accessions of P. peregrina strongly supported its allotetraploid origin. These results, thus, suggest that P. officinalis is a homoploid hybrid species derived from two allotetraploid parents.
To better understand the origin of P. officinalis, it is necessary to further characterize the genome types of the allotetraploid parents. According to the Adh1 phylogeny, one of the diploid parents of the P. arietina group was from a basal lineage B1 of the B clade. The other parent was from the A clade, which is not closely related to any diploid species. The diploid parents of P. peregrina are closely related to P. tenuifolia of the B2 clade and P. cambessedesii and P. mlokosewitchi of the C clade. On the Adh2 phylogeny, the monophyly of B-type Adh2 sequences of P. peregrina and P. tenuifolia, which corresponds to the B2 clade of the Adh1 tree, was not rejected by the Kishino–Hasegawa test. Therefore, the Adh gene phylogenies suggested that P. arietina group has a genome type AAB1B1, and that P. peregrina has a genome type B2B2CC.
All diploid species of section Paeonia that are not of hybrid origin are included in this study (11). The remaining four diploid species of the section that are not included in this analysis had cloned sequences falling into distinct clades on Adh1 or Adh2 phylogeny (data not shown), supporting possible hybrid origins of these species (11). Furthermore, none of the excluded diploid species is more closely related to the diploid progenitors of the P. arietina group or P. peregrina than the ones included in this study. Therefore, the lack of a diploid species in the A clade is most likely due to the extinction of the diploid parent of the P. arietina group.
Given the inferred genome types of two allotetraploid parents, AAB1B1 and B2B2CC, the F1 homoploid hybrid should have a genome type AB1B2C. The sampled accessions of P. officinalis, however, have maintained only three of the four homoeologs for each Adh gene. For the Adh1 gene, both accessions have the A and B1 types from the P. arietina group and the C type from P. peregrina. For the Adh2 gene, the accession P. offcinalis2 has also maintained the A, B1, and C types, whereas P. officinalis1 possesses A, B1, and B2 types. In an attempt to isolate all Adh loci through PCR cloning, more than 30 clones of each Adh gene were screened from two runs of PCR for each accession of P. officinalis. In most cases, screening 15 clones is sufficient to identify both homoeologous Adh loci from an allotetraploid peony (ref. 12 and unpublished data). Furthermore, a C-type Adh2-specific primer, Adh2C (5′-CTTCTCTTTGATCTAATAAGT), was designed and used together with the reverse Adh2-specific primer Adh2R to amplify this genes from both accessions of P. officinalis. The C-type Adh2 gene was amplified from P. officinalis2 but not from P. officinalis1, indicating that this type of the Adh2 gene is indeed absent from P. officinalis1.
A stable homoploid hybrid species can be formed through recombinational speciation. According to this model, backcrossing or interbreeding among partially sterile F1 individuals may give rise to certain novel genotypes that have restored fertility and established at least partial reproductive isolation from the parental species (24, 25). The model has been rigorously tested at the genomic level through comparison of linkage maps of a homoploid hybrid species Helianthus anomalus and its diploid parents (26–29). Understanding of the genetic outcome of recombinational speciation at individual nuclear loci came from comparison of isozyme, RAPD, AFLP, and ISSR profiles of diploid hybrids and their parents (1, 30, 31). A homoploid hybrid species tends to have a combination of alleles and/or loci that are specific to either parent. However, it would be more difficult to identify a homoploid hybrid of allotetraploids by using these molecular markers because of the complex genome composition and possible gene silencing and deletion in both hybrid and parental species. The lack of suitable markers may have impeded the identification of this type of homoploid hybrid speciation in the past. Phylogenetic analysis of low-copy nuclear gene sequences, which has proven an effective approach to reconstruct allotetraploidization (12, 15, 32–34), may also contribute to the future documentation of homoploid hybrid species derived from allotetraploid parents.
It is probably impossible to predict genome composition of a homoploid hybrid species between allotetraploid parents even though the parental genome types are known. A tetraploid hybrid that integrates four sets of more or less diverged diploid genomes may have to undergo extensive genome reorganization. This process coupled with segregation, gene deletion (due to genetic redundancy), and possible backcrossing is most likely to yield a variety of combinations of the homoeologs from the allotetraploid parents in the hybrid genome. P. officinalis contains Adh genes from three of the four types of the genes from both parents. In all cases, the P. officinalis accessions have both Adh homoeologs (A and B1) from the P. arietina group and one of the homoeologs (B2 in P. officinalis1 or C in P. officinalis2) from P. peregrina. The other Adh2 homoeolog of P. peregrina was lost from the hybrid genome possibly through backcrossing with the P. arietina group.
Previous cytogenetic studies found that P. officinalis had abnormal chromosomal pairing at the meiotic metaphase I where many univalents and some multivalents were observed (35, 36). This suggests that the homoploid hybrid genome is not yet completely stabilized, and thus has not recovered full fertility (2). Apparently, P. officinalis is a well established species both morphologically and ecologically. It is among the most widely distributed and most abundant peony species in the Mediterranean region. Having two similar copies of the B genome, B1 and B2, in the F1 generation may have facilitated meiotic chromosomal pairing and helped the initial establishment of the hybrid population. Vegetative reproduction through rhizomes in peonies may also have facilitated the expansion of the hybrid populations despite potentially low fertility of hybrid individuals. Integration of genes from multiple genomes provided genetic and phenotypic variation that might facilitate the colonization of new habitats (4).
Future investigations of population and reproductive biology of P. officinalis should help determine hybrid fertility as well as other genetic and ecological factors involved in hybrid speciation. Increase in sampling of P. officinalis populations and the number of nuclear loci examined will contribute to a better characterization of this type of hybrid speciation at both populational and genomic levels.
It has been suggested that up to 70% of angiosperms are polyploids (37). Many angiosperms may have gone through several cycles of allopolyploidization followed by diploidization that results in a reduction in genome size through gene silencing or other means of genome rearrangement (38–40). This study suggests that hybrid speciation between allotetraploids can occur without an intermediate stage of genome diploidization given that the A, B, and C types of Adh1 sequences are not silenced (at least at the level of gene integrity; the expression status of Adh copies was not examined). Furthermore, homoploid hybrid speciation between allotetraploids allows an integration of multiple diploid genomes without a further doubling of genome size, and consequently a greater opportunity of reunion and interaction of diverged diploid genomes during the evolution of flowering plants. The frequency of this type of hybrid speciation during angiosperm evolution remains unknown, and awaits to be assessed especially based on low-copy nuclear gene phylogenies.
Acknowledgments
We thank Daniel Crawford, Loren Rieseberg, and David Tank for helpful discussion and critical reading of the manuscript. The manuscript was much improved by comments and suggestions from two anonymous reviewers. This research was supported by the National Science Foundation through a grant to T.S.
Footnotes
References
- 1.Arnold M L. Natural Hybridization and Evolution. New York: Oxford Univ. Press; 1997. [Google Scholar]
- 2.Grant V. Plant Speciation. 2nd Ed. New York: Columbia Univ. Press; 1981. [Google Scholar]
- 3.Rieseberg L H. Annu Rev Ecol Syst. 1997;28:359–389. [Google Scholar]
- 4.Buerkle C A, Morris R J, Asmussen M A, Rieseberg L H. Heredity. 2000;84:441–451. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 5.Grant V. Genetics. 1966;53:757–775. doi: 10.1093/genetics/53.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant V. Genetics. 1966;54:1189–1199. doi: 10.1093/genetics/54.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stern F C. A Study of the Genus Paeonia. London: Royal Horticultural Society; 1946. [Google Scholar]
- 8.Stebbins G L. Genetics. 1938;23:83–110. doi: 10.1093/genetics/23.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stebbins G L. Madrono. 1948;9:193–199. [Google Scholar]
- 10.Sang T, Crawford D J, Stuessy T F. Proc Natl Acad Sci USA. 1995;92:6813–6817. doi: 10.1073/pnas.92.15.6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang T, Crawford D J, Stuessy T F. Am J Bot. 1997;84:1120–1136. [PubMed] [Google Scholar]
- 12.Sang T, Zhang D. Syst Bot. 1999;24:148–163. [Google Scholar]
- 13.Gottlieb L D. Science. 1982;216:373–380. doi: 10.1126/science.216.4544.373. [DOI] [PubMed] [Google Scholar]
- 14.Ge S, Sang T, Lu B, Hong D. Proc Natl Acad Sci USA. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small R L, Ryburn J A, Cronn R C, Seelanan T, Wendel J F. Am J Bot. 1998;85:1301–1315. [PubMed] [Google Scholar]
- 16.Sang T, Donoghue M J, Zhang D. Mol Biol Evol. 1997;14:994–1007. doi: 10.1093/oxfordjournals.molbev.a025716. [DOI] [PubMed] [Google Scholar]
- 17.Gaut B S, Peek A S, Morton B R, Clegg M T. Mol Biol Evol. 1999;16:1086–1097. doi: 10.1093/oxfordjournals.molbev.a026198. [DOI] [PubMed] [Google Scholar]
- 18.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 19.Bradley R D, Hillis D M. Mol Biol Evol. 1997;14:247–277. doi: 10.1093/oxfordjournals.molbev.a025797. [DOI] [PubMed] [Google Scholar]
- 20.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer Associates; 1998. , Version 4.0. [Google Scholar]
- 21.Felsenstein J. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 22.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 23.Posada D, Crandall K A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 24.Grant V. Cold Spring Harbor Symp Quant Biol. 1958;23:337–363. doi: 10.1101/sqb.1958.023.01.034. [DOI] [PubMed] [Google Scholar]
- 25.Stebbins G L. Cytologia. 1957;36:336–340. (Suppl.). [Google Scholar]
- 26.Rieseberg L H, Van Fossen C, Desrochers A M. Nature (London) 1995;375:313–316. [Google Scholar]
- 27.Rieseberg L H, Sinervo B, Linder C R, Ungerer M C, Arias D M. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. [DOI] [PubMed] [Google Scholar]
- 28.Ungerer M C, Baird S J E, Pan J, Rieseberg L H. Proc Natl Acad Sci USA. 1998;95:11757–11762. doi: 10.1073/pnas.95.20.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rieseberg L H, Baird S J E, Gardner K A. Plant Mol Biol. 2000;42:205–224. [PubMed] [Google Scholar]
- 30.Rieseberg L H, Ellstrand N C. Crit Rev Plant Sci. 1993;12:213–241. [Google Scholar]
- 31.Wolfe A D, Xiang Q-Y, Kephart S R. Proc Natl Acad Sci USA. 1998;95:5112–5115. doi: 10.1073/pnas.95.9.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cronn R C, Small R L, Wendel J F. Proc Natl Acad Sci USA. 1999;96:14406–14411. doi: 10.1073/pnas.96.25.14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford V S, Gottlieb L D. Evolution. 1999;53:1060–1067. doi: 10.1111/j.1558-5646.1999.tb04521.x. [DOI] [PubMed] [Google Scholar]
- 34.Doyle J J, Doyle J L, Brown A H D, Pfeil B E. Syst Bot. 2000;25:437–448. [Google Scholar]
- 35.Dark S O S. J Genet. 1936;32:353–372. [Google Scholar]
- 36.Schwarzacher-Robinson T. Plant Syst Evol. 1986;154:259–274. [Google Scholar]
- 37.Masterson J. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 38.Leitch I J, Bennett M D. Trends Plant Sci. 1997;2:470–476. [Google Scholar]
- 39.Soltis D E, Soltis P S. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- 40.Wendel J F. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]