Abstract
We report a sampling strategy based on Mendelian Breeding Units (MBUs), representing an interbreeding group of individuals sharing a common gene pool. The identification of MBUs is crucial for case-control experimental design in association studies. The aim of this work was to evaluate the possible existence of bias in terms of genetic variability and haplogroup frequencies in the MBU sample, due to severe sample selection. In order to reach this goal, the MBU sampling strategy was compared to a standard selection of individuals according to their surname and place of birth. We analysed mitochondrial DNA variation (first hypervariable segment and coding region) in unrelated healthy subjects from two different areas of Sardinia: the area around the town of Cabras and the western Campidano area. No statistically significant differences were observed when the two sampling methods were compared, indicating that the stringent sample selection needed to establish a MBU does not alter original genetic variability and haplogroup distribution. Therefore, the MBU sampling strategy can be considered a useful tool in association studies of complex traits.
Keywords: breeding units strategy, mtDNA haplogroup distribution, association studies
Introduction
Population definition, sample selection and choice of markers are crucial points in human population genetics studies, and the sampling strategy depends principally on the questions being asked. In addition to biological aspects, such studies should also take into account important socio-cultural parameters, such as language and religion, along with social and self-identity affiliation. If a human population is clearly ethnically-identified and recent admixture is negligible, sampling strategies based only on surname (whenever distinctive) and place of birth are preferred, since they allow exclusion of recent immigrants, not yet blended into the gene pool, from the analysis. Moreover, surname and place of birth criteria can be extended from the DNA donors to their ancestors, provided that genealogical information is available.
A more stringent sampling strategy is required in studies based on genome-wide association scans, which look for different allele distributions between individuals with (cases) or without (controls) a phenotype of interest. The case-control experimental design is expected to be appropriate in surveys on homogeneous populations, whereas both false-positive and false-negative results may occur in heterogeneous or substructured populations, if cases and controls are not carefully sampled according to their origin. This scenario is likely to occur in an island like Sardinia, where the majority of the present population is distributed among 363 isolated villages (Siniscalco et al., 1999) which, while sharing common ancestry, might have diversified during many centuries of isolation. Therefore, it is important to identify true Mendelian Breeding Units (MBUs), i.e. interbreeding groups of individuals sharing a common ancestral gene pool. In Sardinia, the most practical way to define a MBU is to derive a direct estimate of the percentage of endogamous mating occurring in the last 200 years. This information was obtained anonymously from municipal and ecclesiastical marriage registers (Siniscalco et al., 1999). However, rigorous sample selection for reconstructing MBUs led to a conspicuous reduction in sample size, which might have significantly skewed haplotypic or allelic frequencies. In a previous paper (Siniscalco et al, 1999), we reported a pilot study on 55 unrelated controls belonging to the MBU of Carloforte, who were genotyped at six markers. We showed there that the allele frequencies, and therefore the genomic profile, remained constant even when only a subset of 20 individuals was analysed.
The main goal of this work was to evaluate the reliability of the MBU approach in describing genetic variation in human populations, particularly regarding its application to association studies of complex traits.
We compared genetic variability in two sets of samples which included different individuals recruited from the same areas, using two diverse sampling strategies. With the Standard (STD) Method, individuals unrelated for at least two generations were selected on the basis of the surname and place of birth of their grandparents, depicting present-day genetic variation, with the sole exclusion of the most recent immigrants. Using the MBU Method, the selected DNA donors were proven to be descendants of individuals present in the 17th century archives, with no common ancestors for up to at least five generations. This was ascertained by means of a complete genealogical history checking, based on the official records made available to us by the City Halls. Samples collected using the latter method, being representative of population settlements before the migratory events of the last few centuries, allow an extension of the temporal resolution of genetic variability. Therefore, comparison of the two sampling methods might also reveal possible occurrences of diachronic genetic variation in the analysed areas, due to micro-evolutionary dynamics such as drift or gene flow from neighbouring populations.
The analysed samples belong to two different socio-cultural areas, Cabras and western Campidano, whose cultural traits differentiated around the second half of the 19th century: the former, and its neighbouring area, became a flourishing fishing centre, while the latter consists of rural villages whose economy is based on farming and sheep raising.
We studied mitochondrial DNA (mtDNA), since it has been extensively used as a molecular marker during the past 20 years, is maternally inherited, does not recombine and is in a haploid state; thus it is more sensitive than nuclear DNA to the effects of genetic drift and gene flow, and any discrepancy between the two sampling methods is expected to be enhanced.
Materials and Methods
Sample selection
Using the MBU strategy, we analysed 85 unrelated healthy subjects from two areas located in southwestern Sardinia: 35 individuals from Cabras and 50 individuals from western Campidano (Figure 1). Using the STD strategy, we analysed 71 unrelated individuals coming from the same areas. Comparison was performed between 48 samples from Cabras and its neighbouring area (up to 50 km) and 23 samples from the western Campidano area.
Figure 1-.
Map showing the distribution of the two areas analysed in southwest Sardinia. Cas: Cabras. W Camp: western Campidano.
mtDNA analysis
Whole genomic DNA was extracted using standard procedures. For each individual, mitochondrial haplogroup affiliation was determined by both sequencing of the first hypervariable segment (HVS-I) of the control region from position 15997 to 16399 bp (Anderson et al., 1981) and RFLP (Restriction Fragment Length Polymorphism) analysis of the coding region for the presence/absence of haplo-group diagnostic markers (see Table 1 for details).
Table 1-.
Oligonucleotide pairs used and polymorphic sites investigated to classify mitochondrial coding regions into haplogroups H, V and U/K.
| Haplogroup | Primer sequences | Polymorphic site | Enzyme |
|---|---|---|---|
| H | L: aagcaatatgaaatgatctgc H: gcgtaggtttggtctag |
–7025 | AluI |
| V | L: gagcttaaacccccttattt H: gtattgattggtagtattggttatggttca |
–4577 | NlaIII |
| U/K | L: ctcaaccccgacatcattacc H: attacttttatttggagttgcaccaagatt |
+12308 | HinfI |
Data analysis
BioEdit software 7.0.5.2 (Hall, 1999) was used to align the sequences obtained. To characterise genetic variation among sampling sites, estimates of the number of polymorphic sites (S), the number of haplotypes (h), the nucleotide diversity (Pi), and the haplotype diversity (Hd) were obtained using the DnaSP 4.10 software (Rozas and Rozas, 1999). Pearson chi-square (χ2) values (Pearson, 1900) were calculated in order to assess whether there was any difference between the haplotype frequency distributions obtained for the same areas by means of different sampling strategies (MBU and STD). Principal Coordinate Analysis (PCoA) was carried out on the matrix of DNA pairwise differences, using the Genalex 6.3 software (Peakall and Smouse, 2006). The method based on the covariance matrix with data standardisation was applied. In order to assess the occurrence of significant genetic structuring among samples, analysis of molecular variance (AMOVA) was performed on the matrix of pairwise DNA distances among haplotypes, using the Arlequin 3.1 computer package (Excoffier et al., 2005). Furthermore, genetic differentiation between pairs of samples was estimated by pairwise ΦST values, computed from the matrix of haplotype DNA pairwise differences. The significance of variance components and F-statistic was assessed by a random permutation test (10,000 replicates).
A Median-Joining network was drawn for each sampling strategy using Network 4.2.0.1 software (http://www.fluxus-engineering.com).
Results
Nucleotide sequence analysis of HVS-I (GenBank accession numbers: HM584611-HM584695 for MBU samples, and HM594952-HM595022 for STD samples) combined with RFLP analysis allowed the clustering of samples from both MBU and STD strategies into nine main haplogroups. They increased to eleven when sub-haplo-groups K and U5b3 were also considered (Table 2). Haplogroup H, which includes the Cambridge Reference Sequence (CRS) (Anderson et al., 1981), proved to be the most common. Haplogroup U5b3, reported as Sardinian-specific (Fraumene et al., 2006; Pala et al., 2009), was found in Cabras MBU, western Campidano MBU and Cabras STD, missing in western Campidano STD only. The values of genetic diversity, calculated for the dataset of HVS-I, were similar for all regions and sampling strategies considered, showing a high level of variability (Table 3). Furthermore, we found a total of 82 different haplotypes. Those whose occurrence was detected by both sampling methods (MBU and STD) showed comparable relative frequency distributions, with no significant Pearson chi-square values (Table 4).
Table 2-.
MtDNA haplogroup distribution obtained using the two sampling methods (values are expressed as relative distribution frequencies). Cas: Cabras; W Camp: western Campidano; B: MBU method; S: STD method.
| Haplogroup | Cas-B | Cas-S | W Camp-B | W Camp-S |
|---|---|---|---|---|
| V | 2.9 | 6.3 | 10.0 | 4.4 |
| H tot | 37.1 | 29 | 50.0 | 56.5 |
| T tot | 17.1 | 14.6 | 6.0 | 21.7 |
| J tot | 5.7 | 16.7 | 14.0 | 8.7 |
| U1 (x U5b3, K) | 17.1 | 16.7 | 8.0 | 8.7 |
| U5b3 | 5.7 | 8.3 | 10.0 | - |
| K | 2.9 | - | - | - |
| I | 11.4 | - | - | - |
| W | - | 4.2 | - | - |
| X | - | 4.2 | - | - |
| M tot | - | - | 2.0 | - |
This haplogroup does not include the sub-haplogroups U5b3 and K.
Table 3-.
Estimates of genetic diversity among samples analysed. S: segregating sites; h: number of haplotypes; Hd: haplotype diversity; Pi: nucleotide diversity; Uh: frequency of unique haplotypes per total number of haplotypes; Sh: frequency of individuals with shared haplotypes; Rh: frequency of haplotypes observed in more than one individual.
| S | h | Hd | Pi | Uh | Sh | Rh | |
|---|---|---|---|---|---|---|---|
| Cas-B | 38 | 22 | 0.946 | 0.019 | 0.629 | 0.486 | 0.114 |
| Cas-S | 45 | 37 | 0.976 | 0.021 | 0.771 | 0.333 | 0.104 |
| W Camp-B | 55 | 32 | 0.937 | 0.018 | 0.640 | 0.460 | 0.100 |
| W Camp-S | 35 | 16 | 0.913 | 0.018 | 0.696 | 0.391 | 0.087 |
Cas: Cabras; W Camp: western Campidano; B: MBU method; S: STD method.
Table 4-.
MtDNA haplotype distribution obtained using the two sampling methods (values are expressed as relative distribution frequencies). N: number of individuals; Cas: Cabras; W Camp: western Campidano; B: MBU method; S: STD method; χ2: values of Pearson chi-square (significance level with p values ≤0.05).
| Haplotype | Haplogroup | Cas-B N = 35 | Cas-S N = 48 | χ2 | W Camp-B N = 50 | W Camp-S N = 23 | χ2 | GenBank accession n° |
|---|---|---|---|---|---|---|---|---|
| Hap1 | J | - | 4.2 | 1.46 | 2.0 | - | 0.46 | HM594952 |
| Hap2 | X | - | 2.1 | 0.73 | - | - | - | HM594953 |
| Hap3 | V | 2.9 | 2.1 | 0.05 | 2.0 | - | 0.46 | HM594954 |
| Hap4 | U | - | 2.1 | 0.73 | - | - | - | HM594955 |
| Hap5 | J | - | 2.1 | 0.73 | - | - | - | HM594956 |
| Hap6 | U5b3 | 2.9 | 6.2 | 0.48 | 8.0 | - | 1.84 | HM594957 |
| Hap7 | W | - | 2.1 | 0.73 | - | - | - | HM594958 |
| Hap8 | U | - | 2.1 | 0.73 | - | - | - | HM594960 |
| Hap9 | H | 17.1 | 14.6 | 0.08 | 24.0 | 30.4 | 0.25 | HM594961 |
| Hap10 | H | - | 2.1 | 0.73 | - | - | - | HM594962 |
| Hap11 | X | - | 2.1 | 0.73 | - | - | - | HM594963 |
| Hap12 | T | 14.3 | 2.1 | 0.30 | - | - | - | HM594964 |
| Hap13 | U5b3 | - | 2.1 | 0.73 | - | - | - | HM594965 |
| Hap14 | T | - | 2.1 | 0.73 | - | - | - | HM594966 |
| Hap15 | H | - | 2.1 | 0.73 | - | - | - | HM594967 |
| Hap16 | U | - | 4.2 | 1.46 | - | - | - | HM594969 |
| Hap17 | H | 2.9 | 4.2 | 0.10 | - | 4.3 | 2.17 | HM594972 |
| Hap18 | H | - | 2.1 | 0.73 | - | - | - | HM594973 |
| Hap19 | U | - | 2.1 | 0.73 | - | - | - | HM594974 |
| Hap20 | T | - | 2.1 | 0.73 | - | - | - | HM594976 |
| Hap21 | U | - | 2.1 | 0.73 | - | 4.3 | 2.17 | HM594978 |
| Hap22 | J | 2.9 | 2.1 | 0.05 | 6.0 | - | 1.38 | HM594979 |
| Hap23 | T | - | 2.1 | 0.73 | - | - | - | HM594980 |
| Hap24 | J | - | 2.1 | 0.73 | 2.0 | 4.3 | 0.32 | HM594981 |
| Hap25 | V | - | 2.1 | 0.73 | - | - | - | HM594982 |
| Hap26 | V | - | 2.1 | 0.73 | - | - | - | HM594984 |
| Hap27 | T | 2.9 | 2.1 | 0.05 | - | 4.3 | 2.17 | HM594986 |
| Hap28 | W | - | 2.1 | 0.73 | - | - | - | HM594987 |
| Hap29 | T | - | 2.1 | 0.73 | - | - | - | HM594988 |
| Hap30 | U | - | 2.1 | 0.73 | - | - | - | HM594989 |
| Hap31 | J | - | 2.1 | 0.73 | 2.0 | - | 0.46 | HM594990 |
| Hap32 | U | - | 2.1 | 0.73 | - | - | - | HM594991 |
| Hap33 | T | - | 2.1 | 0.73 | 2.0 | 4.3 | 0.32 | HM594993 |
| Hap34 | J | - | 2.1 | 0.73 | - | - | - | HM594994 |
| Hap35 | J | - | 2.1 | 0.73 | - | - | - | HM594997 |
| Hap36 | H | - | 2.1 | 0.73 | 4.0 | 4.3 | 0.005 | HM594998 |
| Hap37 | H | - | 2.1 | 0.73 | - | 4.3 | 2.17 | HM594999 |
| Hap38 | H | - | - | - | - | 4.3 | 2.17 | HM595001 |
| Hap39 | U | - | - | - | - | 4.3 | 2.17 | HM595002 |
| Hap40 | T | - | - | - | - | 8.7 | 4.351 | HM595003 |
| Hap41 | V | - | - | - | - | 4.3 | 2.17 | HM595005 |
| Hap42 | J | - | - | - | - | 4.3 | 2.17 | HM595010 |
| Hap43 | H | - | - | - | - | 4.3 | 217 | HM595014 |
| Hap44 | H | - | - | - | - | 4.3 | 2.17 | HM595015 |
| Hap45 | T | - | - | - | - | 4.3 | 2.17 | HM595018 |
| Hap46 | U | 5.7 | - | 2.74 | 4.0 | - | 0.92 | HM584612 |
| Hap47 | M | - | - | - | 2.0 | - | 0.46 | HM584613 |
| Hap48 | H | - | - | - | 2.0 | - | 0.46 | HM584615 |
| Hap49 | H | - | - | - | 2.0 | - | 0.46 | HM584621 |
| Hap50 | J | - | - | - | 2.0 | - | 0.46 | HM584622 |
| Hap51 | V | - | - | - | 2.0 | - | 0.46 | HM584623 |
| Hap52 | H | - | - | - | 2.0 | - | 0.46 | HM584624 |
| Hap53 | V | - | - | - | 2.0 | - | 0.46 | HM584626 |
| Hap54 | U | - | - | - | 2.0 | - | 0.46 | HM584629 |
| Hap55 | T | - | - | - | 2.0 | - | 0.46 | HM584631 |
| Hap56 | H | - | - | - | 2.0 | - | 0.46 | HM584632 |
| Hap57 | H | - | - | - | 2.0 | - | 0.46 | HM584633 |
| Hap58 | U | - | - | - | 2.0 | - | 0.46 | HM584634 |
| Hap59 | U5b3 | - | - | - | 2.0 | - | 0.46 | HM584635 |
| Hap60 | J | - | - | - | 2.0 | - | 0.46 | HM584639 |
| Hap61 | V | - | - | - | 2.0 | - | 0.46 | HM584644 |
| Hap62 | H | - | - | - | 2.0 | - | 0,.46 | HM584646 |
| Hap63 | H | - | - | - | 2.0 | - | 0.46 | HM584650 |
| Hap64 | V | - | - | - | 2.0 | - | 0.46 | HM584653 |
| Hap65 | H | - | - | - | 2.0 | - | 0.46 | HM584654 |
| Hap66 | T | - | - | - | 2.0 | - | 0.46 | HM584656 |
| Hap67 | H | - | - | - | 2.0 | - | 0.46 | HM584657 |
| Hap68 | H | - | - | - | 2.0 | - | 0.46 | HM584660 |
| Hap69 | H | 2.9 | - | 1.37 | - | - | - | HM584667 |
| Hap70 | I | 11.4 | - | 5.491 | - | - | - | HM584668 |
| Hap71 | U | 2.9 | - | 1.37 | - | - | - | HM584669 |
| Hap72 | U | 2.9 | - | 1.37 | - | - | - | HM584674 |
| Hap73 | U | 2.9 | - | 1.37 | - | - | - | HM584676 |
| Hap74 | H | 2.9 | - | 1.37 | - | - | - | HM584677 |
| Hap75 | U | 2.9 | - | 1.37 | - | - | - | HM584679 |
| Hap76 | H | 2.9 | - | 1.37 | - | - | - | HM584683 |
| Hap77 | J | 2.9 | - | 1.37 | - | - | - | HM584685 |
| Hap78 | H | 2.9 | - | 1.37 | - | - | - | HM584686 |
| Hap79 | K | 2.9 | - | 1.37 | - | - | - | HM584687 |
| Hap80 | H | 2.9 | - | 1.37 | - | - | - | HM584690 |
| Hap81 | U5b3 | 2.9 | - | 1.37 | - | - | - | HM584692 |
| Hap82 | H | 2.9 | - | 1.37 | - | - | - | HM584694 |
Significant values of χ2.
Nucleotide sequences from the control region were combined with RFLP data on the coding region to obtain a single dataset for the following analysis.
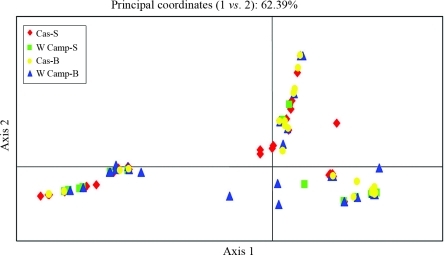
The first two coordinates of PCoA, which account for 62.39% of the total variability, identify two main groups of haplotypes. However, haplotypes were not grouped either according to the geographic area of origin (Cabras or western Campidano) or to the sampling strategy adopted (MBU versus STD) (Figure 2).
Figure 2-.
Principal Coordinate Analysis (PCoA) plot: the first PC accounts for 37.82% of variance, while the second PC accounts for 25.07% (Cas: Cabras; W Camp: western Campidano; B: MBU method; S: STD method).
Accordingly, the analysis of molecular variance (AMOVA) did not indicate significant genetic differentiation among samples (ΦST = 0.0096, p > 0.05). Indeed, nearly all variance was found within samples (99.04%), whereas differences among samples accounted for only 0.96% of the total variation. These results were further confirmed by the pairwise comparison of samples, which did not show any significant genetic differentiation (Table 5).
Table 5-.
Population pairwise ΦST values among samples obtained from MBU and STD strategies. Population codes are reported as in Table 2. Conventional ΦST values are shown below the diagonal and corresponding P values with significance level ≤0.05 are shown above the diagonal.
| Cas-B | Cas-S | W Camp-B | W Camp-S | |
|---|---|---|---|---|
| Cas-B | - | 0.4150 | 0.0940 | 0.2286 |
| Cas-S | –0.0003 | - | 0.1552 | 0.1082 |
| W Camp-B | 0.0150 | 0.0087 | - | 0.2020 |
| W Camp-S | 0.0096 | 0.0181 | 0.0098 | - |
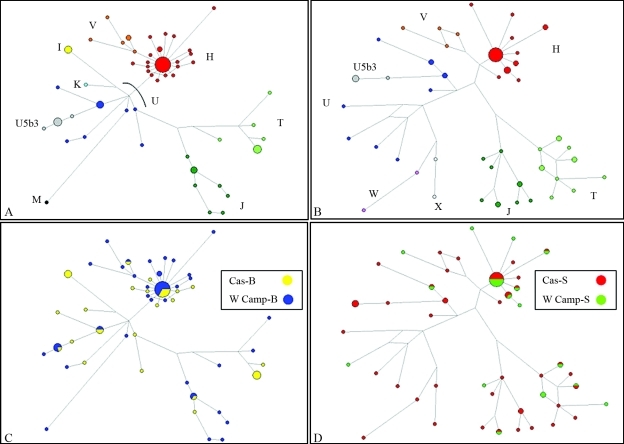
Furthermore, network analysis showed similar relationships among haplogroups without geographical structuring when the two sampling methods were compared (Figure 3).
Figure 3-.
Networks obtained from combined dataset (control region and coding region) for MBU (A and C) and STD (B and D) strategies. A and B: phylogenetic relationships among mitochondrial haplogroups; C and D: geographic distribution of mitochondrial haplogroups. Cas: Cabras; W Camp: western Campidano; B: MBU method; S: STD method.
Discussion
Estimates of genetic diversity (Table 2) obtained for the two sampling strategies were compatible with no occurrence of high levels of repeated haplotypes in the STD strategy, as could be expected. This finding supports the possible occurrence of a homogeneous population shared by both the western Campidano and Cabras areas, with a constant high level of genetic variability in the samples obtained by the two sampling methods and low levels of stochastic forces.
The similarity of genetic diversity values between areas and sampling strategies may be explained considering the lack of diachronic divergence between the present and past genetic settlement of the western Campidano and Cabras areas. Furthermore, this finding is attributable to the absence of genetic drift in the analysed areas. Indeed, this stochastic force, if present, could lead to genetic heterogeneity due to random loss of haplogroups and alteration of their frequencies. The absence of higher levels of identical haplotypes among the STD samples suggests that no significant founder effects affected the population recently. Consistently, the result of PCoA applied to the combined dataset (control region + coding region) (Figure 2) contributed to group MBU and STD samples without genetic structuring. Such similarity was also confirmed by the corresponding, not significant, P values of ΦST.
Network analysis was also consistent with the results above. The two sampling strategies displayed similar global relationships among mitochondrial haplogroups without geographical structuring, showing that mtDNA haplo-group frequencies and distribution obtained by the MBU method were not skewed by the severe sample selection of the method used.
Overall, these results suggest a lack of genetic variation in southwest Sardinia, probably due to a continuous gene flow between the areas, either in the past or more recently, which may have counterbalanced the development of microheterogeneity due to genetic drift.
Previous studies carried out on the paternal unilinear marker Y-chromosome pointed out a similar trend for the entire Sardinian population, suggesting an initial settlement of a relatively large number of individuals with a common origin (Contu et al., 2008) and conspicuous genetic variability.
The presence of genetic structuring is the major obstacle in association studies based on genome-wide scans searching for linkage disequilibrium (LD) between patients and controls (Risch and Botstein, 1996; Terwilliger and Weiss, 1998), even in isolated populations like Finns and Sardinians (Eaves et al. 2000; Taillon-Miller et al., 2000). Pooling individuals belonging to different breeding units may merge alleles that might have different frequencies in different villages, as we have previously reported for some common polymorphisms in Sardinian villages (Robledo et al., 2002).
As previously shown, in a well-defined breeding unit, a small sample was sufficient to describe the genomic profile of the population, which was not affected by severe reduction of sample size (Siniscalco et al., 1999). More importantly, the repeated application of our strategy in different MBUs offers the advantage of reducing the risk of false-positive results due to population stratification, since obtaining similar artifactual results in different MBUs is not anticipated.
In conclusion, the comparison of the variability detected by means of the MBU and STD sampling methods points to a diachronic continuity of the genetic structure of southwestern Sardinia. The benefit of the MBU sampling strategy lies in the possibility of: i) selecting the original population on the basis of written documents and not by inferring surname monophyletism, and ii) not excluding from the analysis unrelated individuals with polyphyletic surnames, when present, in the founder families.
Our results confirm that the MBU sampling strategy, despite the drastic reduction in sample size, does not introduce deviations in gene frequencies, even if haploid markers such as mtDNA are used. Therefore it can be considered a useful tool in association studies of complex traits, making it possible to infer the genetic settlement of the population, recovering the deepest branches of a genealogy and avoiding the recent contribution of foreign peopling.
Acknowledgments
We wish to thank all the participants who made this study possible. We are also grateful to Marcello Siniscalco and Marco Casu for helpful discussions and criticism and to Mary Ann Groeneweg for revising the manuscript. This work was supported by funds from the Fondazione Golinelli made available by Marcello Siniscalco, Compagnia di San Paolo (to A.T.) and grants from the Italian Ministry of Research, MIUR (funds ex 60% to R.R. and P.F.).
Footnotes
Associate Editor: Angela M. Vianna-Morgante
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Contu D, Morelli L, Santoni F, Foster JW, Francalacci P, Cucca F. Y-chromosome based evidence for pre-Neolithic origin of the genetically homogeneous but diverse Sardinian population: Inference for association scans. PLoS One. 2008;3:e1430. doi: 10.1371/journal.pone.0001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA. The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common disease genes. Nat Genet. 2000;25:320–323. doi: 10.1038/77091. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin v. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fraumene C, Belle EMS, Castri L, Sanna S, Mancosu G, Cosso M, Marras F, Barbujani G, Pirastu M, Angius A. High resolution analysis and phylogenetic network construction using complete mtDNA sequences in Sardinian genetic isolates. Mol Biol Evol. 2006;23:2101–2111. doi: 10.1093/molbev/msl084. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Pala M, Achilli A, Olivieri A, Hooshiar Kashani B, Perego UA, Sanna D, Metspalu E, Tambets K, Tamm E, Accetturo M, et al. Mitochondrial haplogroup U5b3: A distant echo of the epipaleolithic in Italy and the legacy of the early Sardinians. Am J Hum Genet. 2009;84:814–821. doi: 10.1016/j.ajhg.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Phil Mag Ser. 1900;5:157–175. [Google Scholar]
- Risch N, Botstein D. A manic depressive history. Nat Genet. 1996;12:351–353. doi: 10.1038/ng0496-351. [DOI] [PubMed] [Google Scholar]
- Robledo R, Orrù S, Sidoti A, Muresu R, Esposito D, Grimaldi MC, Carcassi C, Rinaldi A, Bernini L, Contu L, et al. A 9.1-kb gap in the genome reference map is shown to be a stable deletion/insertion polymorphism of ancestral origin. Genomics. 2002;80:585–592. doi: 10.1006/geno.2002.7014. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP v. 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Siniscalco M, Robledo R, Bender P, Carcassi C, Contu L, Beck J. Population genomics in Sardinia: A novel approach to hunt for genomic combinations underlying complex traits and diseases. Cytogenet Cell Genet. 1999;86:148–152. doi: 10.1159/000015369. (and Erratum 87:296). [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardiña I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY. Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet. 2000;25:324–328. doi: 10.1038/77100. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: Fantasy or reality? Curr Opin Biotechnol. 1998;9:578–594. doi: 10.1016/s0958-1669(98)80135-3. [DOI] [PubMed] [Google Scholar]