Abstract
Cleidocranial dysplasia (CCD) is an autosomal dominant human skeletal disorder comprising hypoplastic clavicles, wide cranial sutures, supernumerary teeth, short stature, and other skeletal abnormalities. It is known that mutations in the human RUNX2 gene mapped at 6p21 are responsible for CCD. We analyzed the mutation patterns of the RUNX2 gene by direct sequencing in six Taiwanese index cases with typical CCD. One of the patients was a familial case and the others were sporadic cases. Sequencing identified four mutations. Three were caused by single nucleotide substitutions, which created a nonsense (p.R391X), two were missense mutations (p.R190W, p.R225Q), and the forth was a novel mutation (c.1119delC), a one-base deletion. Real time quantitative PCR adapted to determine copy numbers of the promoter, all exons and the 3’UTR region of the RUNX2 gene detected the deletion of a single allele in a sporadic case. The results extend the spectrum of RUNX2 mutations in CCD patients and indicate that complete deletions of the RUNX2 gene should be considered in those CCD patients lacking a point mutation detected by direct sequencing.
Keywords: cleidocranial dysplasia, CCD, RUNX2, RUNX2 deletion mutation
Cleidocranial dysplasia, (CCD, MIM#119600), is an autosomal dominant skeletal dysplasia comprising defective skull ossification with frontal bossing, open fontanels, clavicular hypoplasia, delayed ossification of the pelvis, late eruption of permanent teeth, malformed dental roots, supernumerary teeth, and normal intelligence (Mundlos, 1999).
Mutations in the core-binding factor A1 gene (CBFA1) located on chromosome 6p21 were identified as the cause of CCD (Gelb et al., 1995; Mundlos et al., 1997). This gene is composed of eight exons and spans a region of more than 220 kb in 6p21. The CBFA1 protein contains a highly conserved runt domain and therefore, the gene is also referred to as RUNX2 (GenBank accession number AF001450) (Yoshida et al., 2002). The runt domain interacts with the core binding factor consensus sequence TGT/CGGT located in the promoter region of target genes. RUNX2 regulates the expression of multiple genes expressed in osteoblasts and it is a cis-acting element in the osteocalcin promoter (Ducy et al., 1997). RUNX2 knockout mice show a complete lack of bone formation, whereas heterozygous RUNX2 mutated mice display some of the hallmarks of human CCD, including open fontanelles and hypoplastic clavicles, but not the dental anomalies (Otto et al., 1997).
The RUNX2 protein contains two regions divided by the runt domain. On the N-terminal side of the runt domain are glutamine-alanine repeats (Q/A domain) and on the C-terminal side is a region rich in proline-serine-threonine (PST domain). The PST domain is thought to be the transactivation domain. The last five amino acids of RUNX2 (VWRPY) compose a conserved motif in all runt proteins (Quack et al., 1999; Zhang et al., 2000; Otto et al., 2002).
Several mutations of RUNX2 were identified in patients with CCD (Otto et al., 2002). In this study, we screened six patients with typical CCD for mutations in the RUNX2 gene. Mutations were detected in five cases, these including an entire deletion of the RUNX2 coding region.
The six unrelated patients included in this study were first evaluated because of short stature. They received the clinical diagnosis of CCD (for review of diagnostic criteria, see Mundlos, 1999). Patient 4 was a familial case, whereas the others were sporadic. Except for Patients 5 and 6, their height was below the 5th centile for the Taiwanese population. All patients showed brachycephaly, mild frontal bossing, late tooth eruption with multiple crowded and supernumerary teeth, as well as clavicular hypoplasia. The upper part of the thorax was small and a funnel chest was noted. Scoliosis of the thoracic spine with convexity to the right and mild lordosis were also found. Chest radiographs showed hypoplasia of the clavicles with absence of the outer half. Three-dimensional head CT revealed enlarged anterior fontanelles beyond two years of age. Physical data for the six patients are summarized in Table 1.
Table 1-.
Summary of clinical manifestations and RUNX2 gene mutation sites in five Taiwanese cases with cleidocranial dysplasia.
| Case | Type | Age, years | Sex | Stature, cm (centile) | Clavicular hypoplasia | Dental anomalies | Open fontanelle | Mutation site on RUNX2 | Note |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Sporadic | 4 | M | 91 (< 3rd) | Y | Y | Y | c.1171 C → T, R391X | |
| 2 | Sporadic | 11 | M | 127 (< 5th) | Y | Y | Y | c.1119delC, frameshift | |
| 3 | Sporadic | 2 | F | 78 (< 3rd) | Y | Y | Y | exon 0∼7, 3’UTR deletion | |
| 4 | Familial | 10 | M | 122 (< 5th) | Y | Y | Y | c.568 C → T, R190W | Child |
| 40 | F | 144 (< 5th) | Y | Y | Y | c.568 C → T, R190W | Mother | ||
| 5 | Sporadic | 12 | M | 141 (10∼25 th) | Y | Y | Y | N.D. 4 | |
| 6 | Sporadic | 8 | F | 127 (75 th) | Y | Y | Y | c.674 G → A, R225Q |
Sex: M, male; F, female. Y: present; N: not present. N.D.: not detected.
Informed consent was obtained from the patients or the guardians of these patients. Genomic DNA was extracted from peripheral blood leukocytes using a MagNA Pure LC DNA Isolation Kit (Roche, Mannheim, Germany). The complete RUNX2 coding region, including intron/exon boundaries, promoter region and 3’UTR were amplified according to Quack et al. (1999) and Napierala et al. (2005). PCR products were purified from the agarose gel using a QIAEX II kit (Qiagen, Hilden, Germany) and subjected to direct sequencing using a BigDye 3.1 Terminator cycle sequencing kit (Applied Biosystems, Forst City, CA, U.S.A.) with ABI Prism 377 DNA Sequencer or ABI 3100 Genetic Analyzer (Applied Biosystems, Forst City, CA, U.S.A.). To determine carrier rates for the novel mutations in the Taiwanese population, the RUNX2 gene profile of 100 healthy individuals was analyzed using the same procedures. Reference sequence and base-pair numbers refer to GenBank accession numbers AF001443-AF001450 and NM_001024630.
RUNX2 mutations were detected in four patients. Patient 1 was heterozygous for a C-to-T transition at cDNA nucleotide position 1171 in exon 7 of RUNX2, this creating a stop codon at amino acid position 391 (p.R391X). The nonsense mutation, which caused early termination and produced a truncated protein lacking the PST-domain and the VWRPY motif (Thirunavukkarasu et al., 1998), has been previously reported (Tsai et al., 2000; Zhang et al., 2000). The mutation seen in Patient 4 was in heterozygosis and represented a C-to-T transition at nucleotide 568 in exon 1 of RUNX2, this causing the substituation of arginine by tryptophan at amino acid position 190 (p.R190W) in the runt domain (Otto et al., 2002). An arginine stretch located in the runt domain plays an important role in DNA binding. Therefore, disruption of this region may lead to the loss of DNA binding ability (Yoshida et al., 2002). As the side chain of arginine is the major part that interacts with DNA, replacing it by another amino acids of different hydrophobicity and charge, such as tryptophan, disrupts the structure of the runt domain and decreases DNA binding affinity.
Patient 6 was heterozygous for a G-to-A transition at cDNA nucleotide position 674 in exon 3 of RUNX2, this causing arginine to be substituted by glutamine at amino acid position 225 (p.R225Q) at the C-terminal end of the runt domain (Otto et al., 2002). This amino acid substitution abolishes the positive charge of the residue at this position and might affect the structure stability and interaction with other proteins.
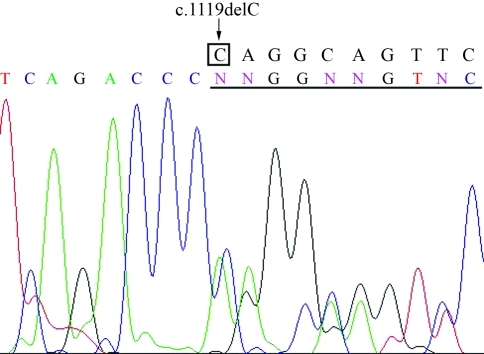
In Patient 2, the cytosine at cDNA nucleotide position 1119 was deleted (c.1119delC), which led to a frameshift at amino acid 373, creating a stop codon at position 483 (Figure 1). This deletion has not been reported previously and was not present in the patient’s parents. It disrupts both the repression subdomain and the VWRPY motif and is predicted to abolish RUNX2 function. The C-terminal region of the PST-domain, which contains a region that interacts with Smad proteins, is considered necessary for RUNX2-mediated transcriptional regulation (Zhang et al., 2000). The VWRPY motif is an interaction partner for the binding of transcriptional corepressors of the TLE/Grg family (Aronson et al., 1997; Levanon et al., 1998). Thus, even in caseswhere the runt domain is complete, the mutated RUNX2 protein may fail to interact with other transcription factors (Thirunavukkarasu et al., 1998; Zhang et al., 2000).
Figure 1-.
Sequence analyses of genomic DNA of the RUNX2 from Patient 2. A heterozygous cytosine deletion at cDNA position 1119 was detected (c.1119delC). The normal sequence is shown above the electropherogram.
In Patient 2, we furthermore found a single nucleotide polymorphism at cDNA nucleotide 240 (p.A80A), a substitution of G for A. This polymorphism site was described previously in another population (Machuca-Tzili et al., 2002).
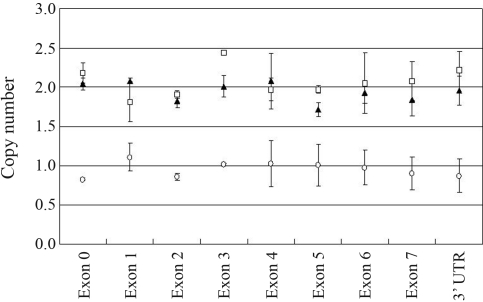
Patients 3 and 5 did not show any mutation in the promoter and the coding regions of RUNX2, similar to previous results showing that only 33∼50% of patients with an unambiguous clinical and radiographic diagnosis of CCD carry mutations in the RUNX2 gene (Quack et al., 1999). Large deletions or duplication of the RUNX2 gene are rarely seen in CCD patients (Lee et al., 2008). Thus, to investigate the possibility of a gross RUNX2 deletion, the genomic DNA of Patients 3 and 5 were subjected to real time quantitative PCR (RT-qPCR), following the analytical conditions described by Lee et al. (2008). Briefly, three independent experiments were performed to determine copy number variation in CCD patients and normal individuals. The RT-qPCR primers were designed to detect the promoter, exons, and 3’UTR regions of RUNX2. The glyceral-dehyde-3-phosphate dehydrogenase (GAPDH) gene was used as control. The formula for calculating copy number was: copy number = 2 * 2–(ΔCtp – ΔCtn) where Ct is the threshold cycle defined as the mean cycle at which the fluorescence curve reached an arbitrary threshold; ΔCt is calculated as Ct of RUNX2 - Ct of GAPDH, ΔCtp is the ΔCt for patients, and ΔCtn that for normal individuals. While two copies of the tested RUNX2 regions were present in Patient 5, Patient 3 had only one copy of theses regions (Figure 2). Thus, we concluded that Patient 3 carried a deletion of one of the RUNX2 alleles.
Figure 2-.
Analysis of RUNX2 copy number using RT-qPCR. Copy number was determined for the RUNX2 promoter region, all of the exons and the 3’-UTR. Patient 3 had only one copy of these regions. ▴: normal; ¡ : Patient 3; o : Patient 5. Each bar indicates mean ± SD of three independent experiments.
Although no mutation was found in Patient 5, following sequencing and RT-qPCR analysis, these result do not exclude other mutation types in the RUNX2 gene, such as inversions, translocations, mutations within introns, etc. On the other hand, a report showed that cytogenetic anomalies at chromosomal band 8q22 have been associated with a CCD-like phenotype case (Brueton et al., 1992). Thus, more detailed analyses are required to identify the genetic abnormality seen in Patient 5.
The patients with any of the four point mutations in the RUNX2 gene were similar in their clinical symptoms, independent of whether their RUNX2 protein retained the normal runt domain or not. The patients presented here lacking the PST domain and VWRPY motif exhibited typical CCD. This suggests that the C-terminal region plays an important role in RUNX2 function, with different genotypes seemingly generating a similar CCD phenotype. From the findings of Patient 3, who carried the deletion of one allele, we conclude that a one-allele deletion affects the gene expression level and produces clinical symptoms similar to those of point mutations. We furthermore inferred that CCD patients not showing a RUNX2 mutation in directed sequence data should be screened for a gene deletion or duplication, and RT-qPCR appears as a sensitive approach to this end.
Acknowledgments
The work was supported by grants from the National Science Council (NSC-93-2314-B-039-011) and the China Medical University Hospital (CMU98-S-02).
Footnotes
Associate Editor: John M. Opitz
References
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueton LA, Reeve A, Ellis R, Husband P, Thompson EM, Kingston HM. Apparent cleidocranial dysplasia associated with abnormalities of 8q22 in three individuals. Am J Med Genet. 1992;43:612–618. doi: 10.1002/ajmg.1320430322. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Cooper E, Shevell M, Desnick RJ. Genetic mapping of the cleidocranial dysplasia (CCD) locus on chromosome band 6p21 to include a microdeletion. Am J Med Genet. 1995;58:200–205. doi: 10.1002/ajmg.1320580222. [DOI] [PubMed] [Google Scholar]
- Lee MT, Tsai AC, Chou CH, Sun FM, Huang LC, Yen P, Lin CC, Liu CY, Wu JY, Chen YT, et al. Intragenic micro-deletion of RUNX2 is a novel mechanism for cleidocranial dysplasia. Genomic Med. 2008;2:45–49. doi: 10.1007/s11568-008-9024-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca-Tzili L, Monroy-Jaramillo N, Gonzalez-del Angel A, Kofman-Alfaro S. New mutations in the CBFA1 gene in two Mexican patients with cleidocranial dysplasia. Clin Genet. 2002;61:349–353. doi: 10.1034/j.1399-0004.2002.610505.x. [DOI] [PubMed] [Google Scholar]
- Mundlos S. Cleidocranial dysplasia: Clinical and molecular genetics. J Med Genet. 1999;36:177–182. [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Napierala D, Garcia-Rojas X, Sam K, Wakui K, Chen C, Mendoza-Londono R, Zhou G, Zheng Q, Lee B. Mutations and promoter SNPs in RUNX2, a transcriptional regulator of bone formation. Mol Genet Metab. 2005;86:257–268. doi: 10.1016/j.ymgme.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Otto F, Kanegane H, Mundlos S. Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum Mutat. 2002;19:209–216. doi: 10.1002/humu.10043. [DOI] [PubMed] [Google Scholar]
- Quack I, Vonderstrass B, Stock M, Aylsworth AS, Becker A, Brueton L, Lee PJ, Majewski F, Mulliken JB, Suri M, et al. Mutation analysis of core binding factor A1 in patients with cleidocranial dysplasia. Am J Hum Genet. 1999;65:1268–1278. doi: 10.1086/302622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FJ, Wu JY, Lin WD, Tsai CH. A stop codon mutation in the CBFA 1 gene causes cleidocranial dysplasia. Acta Paediatr. 2000;89:1262–1265. doi: 10.1080/080352500750027673. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kanegane H, Osato M, Yanagida M, Miyawaki T, Ito Y, Shigesada K. Functional analysis of RUNX2 mutations in Japanese patients with cleidocranial dysplasia demonstrates novel genotype-phenotype correlations. Am J Hum Genet. 2002;71:724–738. doi: 10.1086/342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]