Abstract
Basic and molecular cytogenetic analyses were performed in specimens of Characidium cf. zebra from five collection sites located throughout the Tietê, Paranapanema and Paraguay river basins. The diploid number in specimens from all samples was 2n = 50 with a karyotype composed of 32 metacentric and 18 submetacentric chromosomes in both males and females. Constitutive heterochromatin was present at the centromeric regions of all chromosomes and pair 23, had additional interstitial heterochromatic blocks on its long arms. The nucleolar organizer regions (NORs) were located on the long arms of pair 23, while the 5S rDNA sites were detected in different chromosomes among the studied samples. One specimen from the Alambari river was a natural triploid and had two extra chromosomes, resulting in 2n = 77. The remarkable karyotypic similarity among the specimens of C. cf. zebra suggests a close evolutionary relationship. On the other hand, the distinct patterns of 5S rDNA distribution may be the result of gene flow constraints during their evolutionary history.
Keywords: Characidium, heterochromatin, 18S rDNA, 5S rDNA, natural triploidy
Introduction
Characidium is the richest genus in number of species within the subfamily Characidiinae (Crenuchidae). It comprises about 50 valid species widespread throughout the Neotropical region, from Panama to Argentina (Buckup, 2003). The phylogenetic status of this assemblage has been controversial since the first description of Characidiinae, which has already been regarded as a member of Hemiodontidae, a subfamily of Characidae or an independent family (Buckup, 1998). Studies based on morphological data using cladistic methods indicated that Characidiinae along with Crenuchinae formed the family Crenuchidae (Buckup, 1998).
Characidium zebra represents a dispersed and polymorphic species, often misidentified as C. fasciatum (Buckup and Reis, 1997). This species is characterized by the presence of a completely scaled isthmus. The presence of several plesiomorphic morphological traits led to the conclusion that this species was primitive in relation to its congeners, occupying a basal position in the phylogeny of Characidium (Buckup, 1993). The type locality of C. zebra is in the Amazon drainage, but there are no studies defining its actual range (Buckup and Reis, 1997). Due to its phylogenetic position, the cytogenetic analysis of species related to C. zebra may represent an interesting starting point for understanding events related to the karyotypic evolution within the genus.
The objective of this work was to study the karyotypes of five samples of C. cf. zebra collected in three major Brazilian river basins (Tietê, Paranapanema and Paraguay) in order to contribute to the understanding of the processes involved in the karyotypic differentiation of Characidium species.
Material and Methods
Specimens of Characidium cf. zebra from five collection sites along distinct hydrographic basins in Brazil were analyzed. The specimens were fixed in 10% formaldehyde, stored in 70% ethanol for identification, and deposited in the fish collection of the Laboratório de Biologia e Genética de Peixes (LBP) at UNESP, Botucatu, São Paulo, Brazil (Table 1).
Table 1-.
Specimens of Characidium cf. zebra analyzed.
| LBP | Sample Localities | F | M | Coordinates |
|---|---|---|---|---|
| 8704 | Paraitinga river – Salesópolis – SP | 12 | 7 | S 23°30’40 W 45°51’32 |
| 8714 | Alambari river – Botucatu – SP | 25 | 10 | S 22°56’06 W 48°19’18 |
| 6727 | Novo river – Avaré – SP | 10 | 4 | S 23°01’26 W 48°49’32 |
| 8715 | Araquá river – Botucatu – SP | 8 | 5 | S 22°47’13 W 48°28’89 |
| 8533 | Juba river – Nova Fernandópolis – MT | 6 | 5 | S 14°58’23 W 57°44’40 |
LBP: deposit number at the fish collection of the Laboratório de Biologia e Genética de Peixes, Instituto de Biociências, UNESP. F: females and M: males; SP – São Paulo: MT – Mato Grosso.
Chromosome preparations were obtained from kidney and gill tissue following the technique described by Foresti et al. (1981). Nucleolar organizer regions (NORs) were evidenced by silver nitrate staining according to Howell and Black (1980) and C-banding was performed based on Sumner (1972).
Molecular cytogenetic analyses comprised the identification of GC- and AT-rich regions by fluorochrome staining with chromomycin A3 (Schweizer, 1976) and DAPI (Schweizer et al., 1978), respectively. Mapping of both 18S and 5S rDNA sites on metaphase chromosomes was performed by fluorescent in situ hybridization (FISH) according to Pinkel et al. (1986), with slight modifications. The probes used were a 18S rDNA sequence obtained after PCR from the nuclear DNA of Prochilodus argenteus (Hatanaka and Galetti Jr, 2004) using the primers NS1 5’-GTAGTCATATGCTTGTCTC-3’ and NS8 5’-TCCGCAGGTTCACCTACGGA-3’ (White et al., 1990) and a 5S rDNA sequence amplified using the primers A (5’- TACGCCCGATCTCGTCCGATC-3’) and B (5’-GCTGGTATGGCCGTAGC-3’) of Leporinus elongatus (Martins and Galetti Jr, 1999). The 18S rDNA probe was labeled with 14-dATP biotin by nick translation, according to the manufacturer’s instructions (Bionick Labeling System – Invitrogen). Signal detection and amplification were performed with fluorescein-conjugated avidin (FITC) and biotinylated anti-avidin antibodies. The 5S rDNA probe was labeled with digoxigenin-11-dUTP (Roche Applied Science) by PCR and the hybridization signal was detected with anti-digoxigenin conjugated with rhodamine.
Chromosomes were counterstained with DAPI or propidium iodide (in the case of the triploid specimen) and analyzed under an epifluorescence photomicroscope (Olympus BX61). Images were captured using Image Pro Plus, 6.0 software (MediaCybernetics). The chromosomes were morphologically classified according to their arms ratio, as established by Levan et al. (1964), and were arranged in decreasing size.
Results
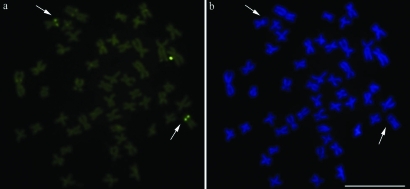
The five samples of Characidium cf. zebra presented a constant diploid number of 2n = 50 and a karyotype composed of 32 metacentric and 18 submetacentric chromosomes. Constitutive heterochromatin was identified only at the centromeric regions of most chromosomes in individuals of both sexes. Pair 23, presented additional interstitial heterochromatic blocks on its long arms in some metaphases, which colocalized with the NORs (Figure 1). No sex chromosome heteromorphism was identified. The Ag-NORs, evidenced after silver nitrate staining (Figure 1, inset), CMA3 staining (Figure 2a) and FISH with a 18S rDNA probe (Figure 3d), were located at an interstitial position in the long arm of pair 23. Chromomycin A3 staining allowed the demonstration that the NORs were located in GC-rich segments in all the populations studied.
Figure 1-.
Karyotype present in the five populations of Characidium cf. zebra analyzed after Giemsa staining (a) and C-banding (b). In the inset, the NOR-bearing chromosomes. Bar = 10 μm.
Figure 2-.
Metaphases of Characidium cf. zebra after chromomycin A3 (a) and DAPI (b) staining. The arrows indicate GC-rich (a) and AT-poor (b) sites. Bar = 10 μm.
Figure 3-.
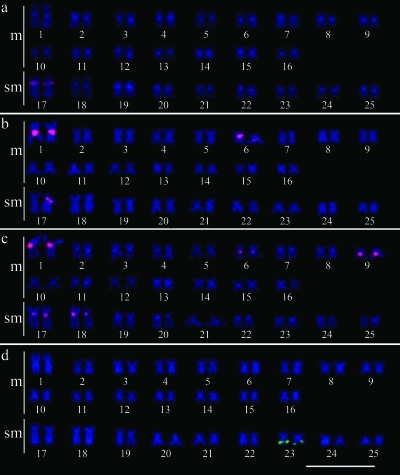
Karyotype of Characidium cf. zebra showing the 5S rDNA on pair 17, typical of the populations from the Alambari, Novo and Araquá rivers (a); in pairs 1, 6 and one homologue of pair 17, from the Paraitinguinha river (b) and in pairs 1, 6, 9, 17 and 18 from the Juba river (c). In (d), the karyotype present in the five populations of Characidium cf. zebra with the 18S rDNA on pair 23. Bar = 10 μm.
FISH with the 5S rDNA probe showed that the specimens from the Paraitinguinha river presented 5S rRNA genes at interstitial positions of five chromosomes (pairs 1 and 6 and one homologue of pair 17) (Figure 3b). In the individuals from the Novo, Alambari and Araquá rivers (Figure 3a), this ribosomal sequences were located at an interstitial position on pair 17. The 5S rDNA sites were localized at interstitial regions of five chromosome pairs (1, 6, 9, 17 and 18) in the samples from the Juba river (Figure 3c).
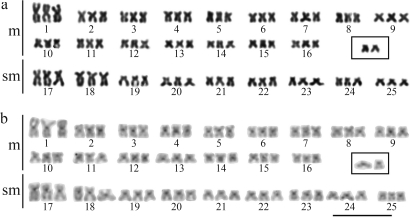
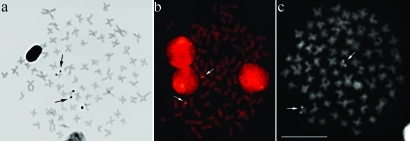
All the cells of one specimen collected in the Alambari river presented 77 chromosomes, characterizing a case of natural triploidy (Figure 4). This individual also carried two extra chromosomes (Figure 4a, inset). C-banding revealed heterochromatic blocks only in the centromeric regions of all chromosomes of the normal set (Figure 4b) in both diploid and triploid specimens. The extra chromosomes were almost completely euchromatic (Figure 4b, inset), except for their centromeric regions. Although the specimen was a triploid, its NORs, identified by silver nitrate staining, CMA3, and FISH with the 18S rDNA probe, were restricted to two chromosomes (Figure 5).
Figure 4-.
Karyotype of a triploid specimen of Characidium cf. zebra from the Alambari river after Giemsa staining (a) and C-banding (b). In the inset, the supernumerary chromosomes. Bar = 10 μm.
Figure 5-.
Metaphases of a triploid specimen of Characidium cf. zebra showing NORs after: silver nitrate staining (a), FISH with a 18S rDNA probe (b) and CMA3 staining (c). Note that there are only two NOR-bearing chromosomes. Bar = 10 μm.
Discussion
Cytogenetic studies in five samples of Characidium cf. zebra showed a constant 2n = 50 with similar karyotypes comprising metacentric and submetacentric chromosomes. The largest chromosome pair (pair 1) in this karyotype is remarkably larger than the second pair (pair 2) in all the individuals, thus representing an important marker for the species. The largest chromosomes in species such as Characidium cf. gomesi (Centofante et al., 2001; Maistro et al., 1998, Pansonato-Alves et al., 2010b), C. lanei (Noleto et al., 2009, Pansonato-Alves et al., 2010a) and C. schubarti (Pansonato-Alves et al., 2010a) are those from the 17th submetacentric pair. The presence of the marker chromosome pair 1, the similar distribution pattern of heterochromatin and of the NORs suggest that the studied samples of C. cf. zebra are closely related.
Based on the phylogenetic position of C. zebra proposed by Buckup (1993), the absence of differentiated sex chromosomes in this species would represent a plesiomorphic trait within the genus, whereas the presence of heteromorphic sex chromosomes in some representatives of the genus should be regarded as an apomorphy (Centofante et al., 2003). According to Centofante et al. (2003), the occurrence of a ZW sex chromosome system with a heterochromatic W chromosome is restricted to a few species closely related to C. gomesi. However, among the ten Characidium species cytogenetically studied (Miyazawa and Galetti Jr, 1994; Maistro et al., 1998; Centofante et al., 2001, 2003 and Pansonato-Alves et al., 2010a,b), only C. cf. zebra and C. lagosantensis lack heteromorphic sex chromosomes. This suggests that the presence of differentiated sex chromosomes is the most common condition of the character among the species of the genus.
The NOR-bearing chromosome pair 23 was identical in the five studied samples of Characidium cf. zebra. The region corresponding to the NOR was faintly stained by DAPI, reinforcing its AT-poor nature (Figure 2b). NORs have been shown to be usually stained by GC-specific fluorochromes, such as chromomycin A3 and mithramycin in fish (Pendás et al., 1993; Vicari et al., 2005) and amphibians (Schmid, 1982). However, some heterochromatic segments unrelated to NORs may also brightly fluoresce after GC-specific fluorochrome staining (Artoni et al., 1999).
FISH with 5S rDNA probes in metaphase chromosomes of Characidium revealed a peculiar distribution of these sequences. Vicari et al. (2008) mapped the 5S rRNA genes to an interstitial position in a single metacentric pair of a C. cf. gomesi population from the Tibagi river. Pansonato-Alves et al. (2010b) located the same genes at an interstitial position in the 25th submetacentric pair in specimens from two populations of C. cf. gomesi from the Paranapanema river basin and at an interstitial position in pairs 20 and 25 in individuals from the Tietê river basin. In the present study, the populations of C. cf. zebra from the Alambari, Araquá and Novo rivers had 5S rDNA sites at an interstitial position on pair 17. The 5S rDNA clusters were located in several chromosomes in populations from the Paraitinguinha (in pairs 1, 6 and one homologue of pair 17) and Juba rivers (in pairs 1, 23, 6, 9, 17 and 18). These findings indicate that, in spite of a close relationship among the studied samples, specific modifications have occurred on the basic genome and restrictions to gene flow during their evolutionary history may have determined the currently observed patterns. Furthermore, the 5S rDNA probes produced signals at centromeric heterochromatic regions in the populations from the Juba and Paraitinguinha rivers. This suggests that, some of them may correspond to pseudogenes representing inactive 5S rDNA-like sequences, as proposed for Centropyge aurantonotus (Affonso and Galetti Jr, 2005).The 5S rDNA and 18S rDNA sequences were located in distinct chromosome positions. This is in agreement with the most frequent pattern found in vertebrates, in which the two ribosomal gene families follow independent evolutionary pathways (Lucchini et al., 1993; Suzuki et al., 1996).
The presence of multiple 5S rDNA sites at interstitial regions of several chromosomes, a derived situation as compared to the plesiomorphic condition of a single interstitial site, may be due to structural rearrangements in the interphase nucleus (Schweizer and Loidl, 1987). This hypothesis contradicts the suggestion that an interstitial position would protect 5S rDNA sites (Martins and Galetti Jr, 2001), as chromosomal domains and heterochromatin dispersion could play a major role in the distribution of these genes.
The occurrence of natural triploidy in a single specimen of C. cf. zebra from the Alambari river may be related to the geography and climate of the Botucatu region, where other natural triploids have already been reported (Maistro et al., 1994). In fish, external fecundation and completion of the second mitotic division only take place after spawning. Environmental temperature changes may facilitate the retention of the second polar body and lead to the formation of natural triploids (Cuellar and Uyeno, 1972). Triploid fish are artificially produced by several techniques, such as thermal or pressure shocks and chemical treatment after fertilization, resulting in a large number of phenotypically normal polyploids (Purdom, 1983). In these procedures, the second meiotic division is disrupted, inhibiting anaphase II (Thorgaard et al., 1981). The outcome is a triploid fish bearing three chromosome sets: two maternal and one paternal, as an unreduced diploid oocyte is fertilized by a normal haploid spermatozoon. The same features were observed in natural triploids of the genus Cyprinus (Vasilev et al., 1975) and Salmo (Thorgaard and Allen Jr, 1987).
The occurrence of natural triploidy in Characidium cf. zebra seems to result from a process similar to that reported for C. gomesi (Centofante et al., 2001), although the former species lacks heteromorphic sex chromosomes. However, in addition to the 75 chromosomes of the triploid specimen, two supranumerary chromosomes were observed, resulting in a 2n = 77 (Figure 4). Cases of triploidy associated with B chromosomes have already been reported in Astyanax scabripinnis (Fauaz et al., 1994; Maistro et al., 1994) and Curimata modesta (Venere and Galetti Jr, 1985). In these studies, the extra chromosomes were easily identified by C-banding due to their heterochromatic nature. All the chromosomes of the triploid C. cf. zebra specimen presented centromeric heterochromatin, hindering the precise identification of the extra elements by C-banding (Figure 4). Nevertheless, two acro/subtelocentric chromosomes could be detected, contrasting to the meta- and submetacentric chromosomes commonly reported in Characidium karyotypes.
An extra element was also found in a single diploid C. cf. zebra from the Passa Cinco river, Ipeúna, SP by Miyazawa and Galetti Jr (1994). That chromosome was identified as an euchromatic acrocentric B chromosome, similar to those observed in our triploid specimen. There were no B chromosomes in the other 35 diploid specimens of C. cf. zebra from the Alambari river. This may indicate that the occurrence of extra chromosomes constitute a unique condition present in the triploid specimen and that they are a consequence of the same event that led to the formation of the triploid fish.
Conspicuous nucleolar organizer regions were located in only two chromosomes of the triploid specimen after silver nitrate and CMA3 staining and FISH with a 18S rDNA probe. This may indicate a complete functional inactivation of the NOR in the third NOR-bearing homologue, as already reported in a Cyprinidae (Mayr et al., 1986). However, the lack of CMA3 and 18S rDNA signals in the third chromosome of our specimen suggests that other events, such as unequal crossovers, may be responsible for the loss or significant deletion of rDNA sequences in this chromosome.
The remarkable karyotypic similarities in the five populations of C. cf. zebra studied reinforces the possibility of a close relationship among them. Nevertheless, the distinct pattern of 5S rDNA distribution in the samples from the Paraitinguinha and Juba rivers most likely derive from constraints in gene flow during their evolutionary history. This, in turn, may indicate a tendency of fixation of allopatric differentiation mechanisms among populations isolated in headwaters of different hydrographic systems, which is related to the lifestyle of the species.
Footnotes
Associate Editor: Yatiyo Yonenaga-Yassuda
References
- Affonso PRAM, Galetti PM., Jr Chromosomal diversification of reef fishes from genus Centropyge (Perciformes, Pomacanthidae) Genetica. 2005;123:227–233. doi: 10.1007/s10709-004-3214-x. [DOI] [PubMed] [Google Scholar]
- Artoni RF, Molina WF, Bertollo LAC, Galetti PM., Jr Heterochromatin analysis in the fish species Liposarcus anisitsi (Siluriformes) and Leporinus elongatus (Characiformes) Genet Mol Biol. 1999;22:39–44. [Google Scholar]
- Buckup PA. The monophyly of the Characidiinae, a Neotropical group of characiform fishes (Teleostei, Ostariophysi) Zool J Linn Soc. 1993;108:224–245. [Google Scholar]
- Buckup PA. Relationships of the Characidiinae and phylogeny of characiform fishes (Teleostei, Ostariophysi) In: Malabarba LR, Reis RE, Vari RP, Lucena CAS, Lucena ZMS, editors. Phylogeny and Classification of Neotropical Fishes Edipucrs, Porto Alegre. 1998. pp. 123–144. [Google Scholar]
- Buckup PA. Family Crenuchidae (South American Darters) In: Reis RE, Kullander ESO, Ferraris CJ Jr, editors. Check List of the Freshwater Fishes of South and Central America. 2nd edition. Edipucrs; Porto Alegre: 2003. pp. 87–95. [Google Scholar]
- Buckup PA, Reis RE. Characidiin genus Characidium (Teleostei, Characiformes) in southern Brazil, with description of three new species. Copeia. 1997;3:531–548. [Google Scholar]
- Centofante L, Bertollo LAC, Moreira-Filho O. Comparative cytogenetics among sympatric species of Characidium (Pisces, Characiformes). Diversity analysis with the description of a ZW sex chromosome system and natural triploidy. Caryologia. 2001;54:253–260. [Google Scholar]
- Centofante L, Bertollo LAC, Buckup PA, Moreira-Filho O. Chromosomal divergence and maintenance of sympatric Characidium fish species (Crenuchidae, Characidiinae) Hereditas. 2003;138:213–218. doi: 10.1034/j.1601-5223.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Cuellar O, Uyeno T. Triploidy in rainbow trout. Cytogenetics. 1972;11:508–515. doi: 10.1159/000130217. [DOI] [PubMed] [Google Scholar]
- Fauaz G, Vicente VE, Moreira-Filho O. Natural triploidy and B chromosomes in the Neotropical fish genus Astyanax (Characidae) Braz J Genet. 1994;17:157–163. [Google Scholar]
- Foresti F, Almeida-Toledo LF, Toledo-Filho AS. Polymorphic nature of nucleolus organizer regions in fishes. Cytogenet Cell Genet. 1981;31:137–144. doi: 10.1159/000131639. [DOI] [PubMed] [Google Scholar]
- Howell WM, Black DA. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia. 1980;36:1014–1015. doi: 10.1007/BF01953855. [DOI] [PubMed] [Google Scholar]
- Hatanaka T, Galetti PM., Jr Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae) Genetica. 2004;122:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Lucchini S, Nardi I, Barsacchi G, Batistoni R, Andronico F. Molecular cytogenetics of the ribosomal (18S + 28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome. 1993;36:762–773. doi: 10.1139/g93-101. [DOI] [PubMed] [Google Scholar]
- Martins C, Galetti PM., Jr Chromosomal localization of 5s rDNA genes in Leporinus fish (Anostomidae, Characiformes) Chromosome Res. 1999;7:363–367. doi: 10.1023/a:1009216030316. [DOI] [PubMed] [Google Scholar]
- Martins C, Galetti PM., Jr Two rDNA arrays in Neotropical fish species: Is it a general rule for fishes? Genetica. 2001;111:439–446. doi: 10.1023/a:1013799516717. [DOI] [PubMed] [Google Scholar]
- Maistro EL, Dias AL, Foresti F, Oliveira C, Moreira O. Natural triploidy in Astyanax scabripinnis (Pisces, Characidae) and simultaneous occurrence of macro B-chromosomes. Caryologia. 1994;47:233–239. [Google Scholar]
- Maistro EL, Mata EP, Oliveira C, Foresti F. Unusual occurrence of a ZZ/ZW sex-chromosome system and supernumerary chromosomes in Characidium cf. fasciatum (Pisces, Characiformes, Characidiinae) Genetica. 1998;104:1–7. doi: 10.1023/A:1003242020259. [DOI] [PubMed] [Google Scholar]
- Mayr B, Rab P, Kalat M. NORs and counterstain-enhanced fluorescence studies in Cyprinidae of different ploidy level. Genetica. 1986;69:111–118. doi: 10.1007/BF00263267. [DOI] [PubMed] [Google Scholar]
- Miyazawa CS, Galetti PM., Jr First cytogenetical studies in Characidium species (Pisces, Characiformes, Characidiinae) Cytologia. 1994;59:73–79. [Google Scholar]
- Noleto RB, Amorin AP, Vicari MR, Artoni RF, Cestari MM. An unusual ZZ/ZW sex chromosome system in Characidium fishes (Crenuchidae, Characiformes) with the presence of rDNA sites. J Fish Biol. 2009;75:448–453. doi: 10.1111/j.1095-8649.2009.02342.x. [DOI] [PubMed] [Google Scholar]
- Pansonato-Alves JC, Paiva LRS, Oliveira C, Foresti F. Interspecific chromosomal divergences in the genus Characidium (Teleostei, Characiformes, Crenuchidae) Neotrop Ichthyol. 2010a;8:77–86. [Google Scholar]
- Pansonato-Alves JC, Vicari MR, Oliveira C, Foresti F. Chromosomal diversification in samples of Characidium cf. gomesi (Teleostei, Crenuchidae) J Fish Biol. 2010b doi: 10.1111/j.1095-8649.2010.02847.x. epub. [DOI] [PubMed] [Google Scholar]
- Pendás AM, Morán P, Garcia-Vásquez G. Ribosomal RNA genes are interspersed throughout a heterochromatic chromosome arm in Atlantic salmon. Cytogenet Cell Genet. 1993;63:128–130. doi: 10.1159/000133517. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdom CE. Genetic engineering by the manipulation of chromosomes. Aquaculture. 1983;33:287–300. [Google Scholar]
- Schmid M. Chromosome banding in Amphibia. VII. Analysis of the structure and variability of NORs in Anura. Chromosoma. 1982;87:327–344. [Google Scholar]
- Schweizer D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 1976;58:307–324. doi: 10.1007/BF00292840. [DOI] [PubMed] [Google Scholar]
- Schweizer D, Loidl J. A model for heterochromatin dispersion and the evolution of C band patterns. Chromosome Today. 1987;9:61–74. [Google Scholar]
- Schweizer D, Ambros P, Andrle M. Modification of DAPI banding on human chromosomes by prestaining with a DNA-binding oligopeptide antibiotic, Distamycin A. Exp Cell Res. 1978;111:327–332. doi: 10.1016/0014-4827(78)90177-5. [DOI] [PubMed] [Google Scholar]
- Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–306. doi: 10.1016/0014-4827(72)90558-7. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Sakurai S, Matsuda Y. Rat rDNA spacer sequences and chromosomal assignment of the genes to the extreme terminal region of chromosome 19. Cytogenet Cell Genet. 1996;72:1–4. doi: 10.1159/000134149. [DOI] [PubMed] [Google Scholar]
- Thorgaard GH, Allen SK., Jr . Chromosome manipulation and markers in fishery management. In: Ryman N, Utter F, editors. Population Genetic and Fishery Management. University of Washington Press; Seattle: 1987. pp. 319–331. [Google Scholar]
- Thorgaard GH, Jazmin ME, Stier AR. Polyploidy induced by heat shock in rainbow trout. Trans Am Fish Soc. 1981;110:546–550. [Google Scholar]
- Vasilev VP, Makeeva AP, Ryabov IN. On the triploidy of remote hybrids of carp (Cyprinus carpio) with other representative Cyprinidae. Genetika. 1975;11:49–56. [PubMed] [Google Scholar]
- Venere PC, Galetti PM., Jr Natural triploidy and chromosome B in the fish Curimata modesta (Curimatidae, Characiformes) Rev Bras Genet. 1985;7:681–687. [Google Scholar]
- Vicari MR, Artoni RF, Bertollo LAC. Comparative cytogenetics of Hoplias malabaricus (Pisces, Erythrinidae): A population analysis in adjacent hydrographic basins. Genet Mol Biol. 2005;28:103–110. [Google Scholar]
- Vicari MR, Artoni RF, Moreira-Filho O, Bertollo LAC. Diversification of a ZZ/ZW sex chromosome system in Characidium fish (Crenuchidae, Characiformes) Genetica. 2008;134:311–317. doi: 10.1007/s10709-007-9238-2. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: PCR Protocols: A Guide to Methods and Applications. Academic Press Inc; London/New York: 1990. pp. 312–315. [Google Scholar]