Abstract
Cost reduction in plant breeding and conservation programs depends largely on correctly defining the minimal sample size required for the trustworthy assessment of intra- and inter-cultivar genetic variation. White clover, an important pasture legume, was chosen for studying this aspect. In clonal plants, such as the aforementioned, an appropriate sampling scheme eliminates the redundant analysis of identical genotypes. The aim was to define an optimal sampling strategy, i.e., the minimum sample size and appropriate sampling scheme for white clover cultivars, by using AFLP data (283 loci) from three popular types. A grid-based sampling scheme, with an interplant distance of at least 40 cm, was sufficient to avoid any excess in replicates. Simulations revealed that the number of samples substantially influenced genetic diversity parameters. When using less than 15 per cultivar, the expected heterozygosity (He) and Shannon diversity index (I) were greatly underestimated, whereas with 20, more than 95% of total intra-cultivar genetic variation was covered. Based on AMOVA, a 20-cultivar sample was apparently sufficient to accurately quantify individual genetic structuring. The recommended sampling strategy facilitates the efficient characterization of diversity in white clover, for both conservation and exploitation.
Keywords: sampling strategy, white clover, AFLP, genetic diversity parameters
Introduction
The characterization of genetic variation, and its partitioning within and among populations, is important for plant breeding and conservation research focused on the management of genetic resources (Herrmann et al., 2005; Nybom and Bartish, 2000). Prior to the mid 1960’s, genetic variation was estimated by using morphological and physiological characters (phenotypic characterization). However, with this approach, and apart from the bias arising from environmental variability, estimates of diversity were based on a limited set of loci. Subsequently, molecular marker techniques have come to offer an attractive alternative to such a limited scope (Kölliker et al., 2001), through facilitating the rapid estimation of genome-wide intra- and inter-cultivar genetic variation at the DNA level. Molecular marker techniques are therefore preferred to phenotypic characterization if they are reasonably cost-effective.
Among the variety of molecular marker techniques available, the amplified fragment length polymorphism (AFLP) technique, first described by Vos et al. (1995), has proved to be the most suitable for assessing intra-species diversity (Bonin et al., 2007). It has been successfully applied for determining genetic diversity in a multitude of legume forages including red clover (Trifolium pratense L.; Herrmann et al., 2005), alfalfa (Medicago sativa L.; Segovia-Lerma et al., 2003), bird’s foot trefoil (Lotus corniculatus L.; Sardaro et al., 2008), sulla (Hedysarium coronarium L.; Marghali et al., 2005), and white clover (Trifolium repens L.; Kölliker et al., 2001). This technique requires no sequence knowledge for developing species-specific primers, such as SSR (simple sequence repeat) markers (Peakall et al., 1998). Nevertheless, the cost of using AFLP genetic markers depends greatly on the number of samples to be analyzed.
White clover (2n = 4x = 32) is one of the most important pasture legumes in temperate climates, worldwide. As its gametophytic self-incompatibility system requires outbreeding (Williams, 1987) to thus maintain high levels of diversity in natural (ecotype) and synthetic (cultivar) populations (Gustine and Sanderson, 2001), apparently there is the need for a relatively large number of samples to detect the prevailing genetic variation. An optimal sampling strategy is further complicated by lateral clonal spread via stolons (Brink et al., 1999; Chapman, 1983). As both clonal propagation and recruitment from seeds are the rule, white clover populations are comprised of many genetically different individuals (genets), each consisting of several identical individuals (ramets) (Welham et al., 2002). The effectiveness of a sampling scheme thus depends on avoiding the redundant genotyping of clones. One way to avoid this is to place the samples sufficiently apart in a grid-based sampling approach.
Numerous parameters have been developed to better understand and estimate inter- and intra-population genetic diversity. Genetic differentiation (Φst) is a widely used parameter for investigating diversity at the inter-population level. As to the intra- level, diversity parameters can be classified into two groups.
The first group consists of parameters based on allele richness. One popular parameter based on allele richness is the mean number of alleles per locus (A), estimated by the Ewens sampling formula (Ewens, 1972). This formula was based on an ideal population with a random matting population of constant size, without either migration or selection. As natural populations rarely comply with formula assumptions, accuracy in this case was later questioned. Several attempts were then made to develop novel approaches to standardize estimation of A, such as rarefaction (Petit et al., 1998), repeat random subsampling (Leberg, 2002), the Bayesian method (Belkhir et al., 2006) and non-linear regression (Bashalkhanov et al., 2009). The use of A, when estimating genetic diversity, is restricted to studies with codominant genetic markers. The Shannon index (I) is another diversity parameter based on allele richness, and which is extensively used in genetic diversity studies relying on dominant genetic markers, as AFLPs (Ward and Jasieniuk, 2009). According to its mathematical formula, the Shannon index (I) is associated with the number of alleles and their frequency. This index gives the same weight to both the number and frequency of alleles, without emphasizing which are common or rare (Zhao et al., 2006). One reason for its popular use is that it does not rely on Hardy-Weinberg equilibrium. This makes the Shannon index comparable across studies (Bussell, 1999). The second group are heterozygosity-based diversity parameters, including expected heterozygosity (He), which is the most commonly used parameter for estimating intra-population genetic diversity. As with the Shannon index (I), the expected heterozygosity (He) formula is associated with the number and frequency of alleles. Nevertheless, alleles with high frequencies (common alleles) contribute more to He values than those with low ones (rare alleles) (Zhao et al., 2006).
An inappropriate sampling strategy can severely bias genetic diversity parameters, whereas excessive sampling inflates costs (Suzuki et al., 2004). Even so, studies of white clover genetic diversity have been inapt at explicitly optimizing sample sizes or verifying sampling effectiveness (Kölliker et al., 2001; Gustine and Elwinger, 2003; Van Treuren et al., 2005; Bortolini et al., 2006; Rizza et al., 2007). Herein, an investigation was undertaken of the impact of both sample size and the sampling scheme, on assessing genetic diversity within and among white clover cultivars. Three commercially available clover-cultivars were grown in experimental plots mimicking the prevailing conditions in pastures. Several samples per cultivar were collected and genotyped using genome-wide AFLP markers. The main aim was to infer the minimal sample size required to accurately assess genetic variation within and among cultivars, based on widely used parameters, viz., the Shannon index (I), expected heterozygosity (He) and genetic differentiation (Φst). In addition, we tested whether the applied grid-based sampling scheme avoided the inclusion of clones.
Materials and Methods
Plant material and sampling scheme
Three varieties of white clover, diverse in phenotypic characteristics and origin, were used. These were the Aberherald (medium-leaf type) from the UK, the Riesling (large-leaf type) from the Netherlands, and the Rivendel (small-leaf type) from Denmark. In 2005, seeds of each variety were sown in an experimental plot, together with perennial ryegrass (0,5 g of white clover seeds and 2,5 g ryegrass seeds per square meter), at the Institute for Agriculture and Fisheries Research (ILVO), Merelbeke, Belgium. In 2008, 45 samples per plot were collected and subdivided into a 0.4 x 0.4 m grid pattern. Sampling was carried out at the center of each square, to so maintain fixed distances (40 cm) between sampling points in both dimensions. The spatial coordinates of each collected sample were recorded. The sampling distance itself was set at 40 cm based on previous reports of clone sizes in wild clover populations, as well as visual assessment of the field studied. In addition to the 45 field samples, leaf samples were collected from 30 greenhouse-sown seedlings of each cultivar. Up to that time, the 75 samples of each variety used to investigate appropriate sample size was more than the norm in studies of genetic diversity in white clover (Kölliker et al., 2001; Gustine and Elwinger, 2003; Van Treuren et al., 2005; Bortolini et al., 2006; Rizza et al., 2007).
AFLP analyses
Prior to DNA extraction, leaf material was freeze-dried for 48 h,and then homogenized and ground (Tissue Lyser, QIAGEN) to a fine powder. Total DNA was extracted from 20 mg of freeze-dried leaf material by applying the modified Cetyltrimethylammonium Bromide (CTAB) method, as described by Doyle and Doyle (1990). DNA concentration was estimated with NanoDrop ND-1000 spectrophotometer software v. 3.0.1 (NanoDrop Technologies). AFLP analysis was according to Vos et al. (1995), with minor modifications. The enzymes EcoRI and MseI were used for DNA digestion. Each individual plant was fingerprinted with six primer combinations (Table 1). Fragment separation and detection were with an ABI Prism 3130xl capillary sequencer. GeneScan 500 Rox-labelled size standard (Perkin Elmer) was loaded into each sample. Fluorescent AFLP patterns between 50 and 500 bp were scored using a version 4.0 Genemapper (Applied Biosystems).
Table 1.
AFLP primer combinations used for selective amplification and the number of polymorphic loci.
| Primer combinations | Number of polymorphic loci |
|---|---|
| E1+ACA / M2+CAG | 51 |
| E+AGG / M+CTT | 61 |
| E+ACC / M+CAT | 57 |
| E+ACT / M+CAT | 47 |
| E+AAC / M+CAT | 45 |
| E+AAC / M+CTG | 22 |
E indicates EcoRI primer.
M indicates MseI primer.
Data analysis
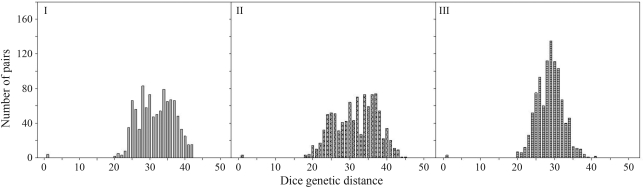
The extent of clonality among the collected samples was quantified, in order to evaluate the effectiveness of the applied sampling scheme. A histogram of pairwise Dice genetic similarity coefficients was calculated to account for minor, but potential, differences within genets, due to somatic mutation and scoring errors (Meirmans and Van Tienderen, 2004). Based on the bimodal distribution of this histogram, a threshold was set up, as outlined by Meirmans and Van Tienderen (2004), as a means of assigning individuals to genets (Figure 1).
Figure 1.
Frequency distribution of the number of pairwise genetic distances among 45 individuals of the white clover varieties Aberherald (I), Rivendel (II) and Riesling (III). Pairwise distances were calculated using GENOTYPE/GENODIVE software.
The evolution of intra- and inter-population genetic diversity parameters over an increasing sample size was investigated, in order to determine the minimal sampling size. Hence, a computer simulation method was employed to generate randomly selected subsets with 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70 and 74 individuals, drawn from the total sample (n = 75).
Next, for each sample size, intra- and inter-cultivar genetic diversity parameters were calculated for each of 50 randomly drawn subsamples per sample size using GenAlEx v.6 (Peakall and Smouse, 2006). Intra-cultivar genetic diversity was calculated as the expected heterozygosity (He) and the Shannon diversity index (I), the two most widely used genetic-diversity parameters (Suzuki et al., 2004). The analysis of molecular variance (AMOVA) was applied for estimating the Φst parameter representing the proportion of genetic variation among the three cultivars. Significance of Φst was tested using 9999 random permutations.
Finally, the intra-cultivar genetic diversity estimate for a given sample-size was divided by the value for the total sample of 75 individuals, and then multiplied by 100. Thus, mean values were converted into index percentages of total variation (Zhu et al., 2007). The inter-cultivar genetic diversity parameter (Φst) and the index percentages of the intra-cultivar genetic diversity parameters (He and I), were then regressed against sample sizes, by the following formula: ln(Y) = b0 + (b1/t), where b0 and b1 are parameters, t sample size, and Y index percentages or Φst.
Results
The six EcoRI/MseI primer pair combination generated 283 clearly identifiable loci. The number of AFLP polymorphic bands detected by each primer pair, across three cultivars, ranged from 22 to 61 (Table 1). Very few samples were assigned to the same genet upon applying a Dice similarity threshold of 0.97, and all belonged to neighboring grids (intersample distance 40 cm) (Figure 1). The largest genet consisted of 4 samples and was found in the Aberherald plot. Only 3 samples in the Rivendel plot and 2 in the Riesling plot were replicates. As replication could thus be considered negligible, replicates were not removed from the dataset in subsequent analysis.
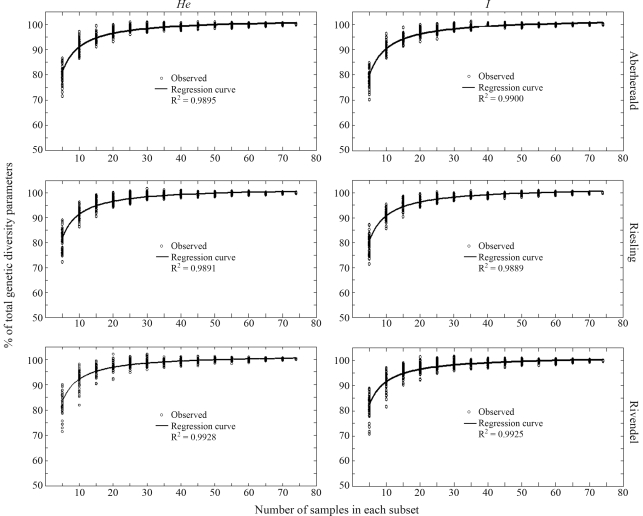
Intra-cultivar genetic diversity was comparable for the three cultivars. The mean expected heterozygosity (He) per cultivar for total sample size (n = 75) was 0.319, 0.289 and 0.272 for Aberherald, Rivendel and Riesling, respectively, and the mean Shannon diversity index (I) 0.487, 0.459 and 0.421, respectively. Regression of He and I across different sample sizes (from 5 to 74), showed that sample size had a profound impact on estimating intra-cultivar genetic diversity (Figure 2). As regards the first three sample sizes (5, 10 and 15 individuals), the curve ascended steeply and then flattened out at around 20 samples for both He and I. A sample size of 20 individuals accounted for 97%, 96% and 97% for He, and 97%, 96% and 97% for I, of the total genetic diversity measured in all the 75 samples of Aberherald, Rivendel and Riesling white clover cultivars, respectively. A sample size of 30 came extremely close to total genetic diversity, with 98.8%, 98.5% and 98.8% for He, and 98.7%, 98.6% and 98.5% for I.
Figure 2.
The influence of sample size on intra-cultivar genetic diversity parameters. Dots represent either the amount of expected heterozygosity (He) or the Shannon diversity index (I) of total variation. The regression formula applied was ln(Y) = b0 + (b1/t), where b0 and b1 are parameters, t represents sample size, and Y refers to the index of percentage of each sample. Samples in various subsets were subtracted 50 times from the total sample.
By AMOVA based on all 75 samples per cultivar, most genetic variation was shown to be among varieties (78.8%), whereas 21.2% was among cultivars (Table 2).
Table 2-.
Analysis of molecular variance (AMOVA) in three white clover varieties. P values were obtained by 9,999 permutations.
| Source | df | SS | MS | Est. Var. | %D |
|---|---|---|---|---|---|
| Variance among varieties | 2 | 2259.74 | 1129.87 | 14.35 | 21.2 * |
| Variance within varieties | 222 | 11851.04 | 53.38 | 53.38 | 78.8 |
| Total | 224 | 14110.78 | 67.74 | 100 |
Statistics include: df, degrees of freedom; SS, sum of squares; MS, mean squares; Est. var., estimated variance; and %D, distribution of total variance.
p = 0.0001.
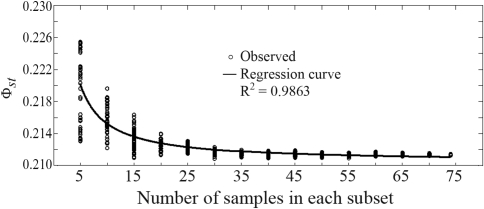
The overall and highly significant (p = 0.0001) genetic differentiation index (Φst) was 0.212. Resampling analysis of Φst showed an overestimation of Φst when using 5 to 15 samples (Figure 3). Although Φst remained stable at 20 samples, variance among subsamples diminished still further.
Figure 3.
The influence of sample size on estimated Φst among cultivars. Dots show the calculated genetic differentiation index (Φst) of each subset. Samples in various subsets were drawn 50 times from a total of 75 individuals. The regression formula applied was ln(Y) = b0 + (b1/t), where b0 and b1 are regression parameters, t the sample size and Y the genetic differentiation index (Φ st) of each subset.
Discussion
The main aim was to present an optimal sampling strategy for studying genetic diversity in white clover cultivars. Hence, 75 samples from three cultivars, mutually different in phenotypic characteristics and geographical origins, were genotyped at a large number of AFLP loci (n = 283). These proved to be sufficient for the sure assessment of population genetic parameters (Mohapatra et al., 2009).
Influence of the applied sampling scheme
The grid-based sampling method has already been successfully applied when investigating genetic diversity in white clover. However, the reported width of clonal patches varied from several centimeters to several meters (Harberd, 1963; Cahn and Harper, 1976; Gustine and Elwinger, 2003). As an adequate sampling interval was unavailable, the determination of a grid-sampling scheme, with suitable spatial intervals, became necessary to avoid collecting spatially correlated samples and to maximize the information obtained. On using a grid-based sampling approach with a 40 cm intersample distance, very few clonal replicates were encountered, after growing clover plants for three years in plots mimicking common clover-pasture conditions (Figure 1). In other words, patch-sizes were less than 40 cm. Although an intersample distance of 40 cm was found suitable for studying diversity, this should be further investigated before recommending it. Further investigation including census sampling (harvesting all individuals within the frame) or using a higher density sampling grid are indicated. In natural populations, very old and large genets can occur (Harberd, 1963). Differences in clone-size are also likely to be magnified by environmental heterogeneity (Vandepitte et al., 2009), which is less prominent in pastures.
The influence of sample size
Based on the lack of considerable change in genetic diversity beyond 30 samples (Figures 2 and 3), a total sample-size of 75 individuals should adequately represent the real AFLP variation within and among the cultivars studied. It is very unlikely that genetic diversity would once again increase after reaching a plateau. Furthermore, the intra-genetic diversity parameters obtained, as well as AMOVA results, are in accordance with reports of higher levels of variation within than among clover cultivars (Kölliker et al., 2001). The noted intra-cultivar variation of 78% was close to the 84% reported by Kölliker et al. (2001). This result is commonly attributed to obligate outcrossing (Hamrick and Godt, 1996), and the breeding system of white clover (Annicchiarico and Piano, 1995).
Sample size was important when estimating both intra- and inter-genetic parameters (Figures 2 and 3). In subsets of less than 20 samples per cultivar, inter-genetic diversity (Φst) was overestimated and intra-genetic diversity (He and I) was severely underestimated. In subsets of 20 or more individuals, the intra-genetic diversity parameters obtained (He and I) covered, on an average, more than 95% of the total genetic diversity, the threshold proposed by Sedcole (1977). Furthermore, the influence of sample-size did not greatly differ among the three cultivars and the two intra-cultivar genetic diversity parameters used (Figure 3).
Such patterns of biased discordance in small sample-sizes have also been observed in previous studies (Isabel et al., 1995; Wu et al., 1999). On using simulations, Berg and Hamrick (1995) showed that, on reducing sample sizes to below 20 per population, estimates of inter-genetic diversity (Φst) became increasingly inflated. They also forewarned that this may cause severe misinterpretation, based on the observation that population genetic parameters calculated using 15 individuals or less, often generated higher estimates than when using larger sample sizes. Likewise, a sample-size of 20 was found to be adequate for studying the level of genetic diversity within and among cultivars of open-pollinating species, this including red clover (Trifolium pretense L.; Kongkiatngam et al., 1995) and tall fescue (Festuca arundinacea Schreb.; Xu et al., 1994). In accordance with these studies, we recommend a sample size between 20 and 30 samples to quantify the genetic diversity within and among white clover cultivars. Although the use of 20 samples maximizes cost-effectiveness, the use of 30 attains utmost precision, whereby its adequacy, for example, in detecting minor differences at the level of intra-cultivar genetic variation.
The study of genetic diversity in genetic resources could provide significant information as to its potential for breeding purposes (Singh et al., 2006). The proposed sampling strategy (sampling scheme and sample-size) should facilitate the efficient management and exploitation of white clover germplasm in breeding and conservation programs.
Acknowledgments
The authors would like to thank Sabine Van Glabeke and Nancy Mergan for their outstanding technical assistance. We would also like to thank Miriam Levenson for the English language editing.
Footnotes
Associate Editor: Dario Grattapaglia
References
- Annicchiarico P, Piano E. Variation within and among Ladino white clover ecotypes for agronomic traits. Euphytica. 1995;86:135–142. [Google Scholar]
- Bashalkhanov S, Pandey M, Rajora OP. A simple method for estimating genetic diversity in large populations from finite sample sizes. BMC Genetics. 2009;10:e84. doi: 10.1186/1471-2156-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Dawson KJ, Bonhomme F. A comparison of rarefaction and Bayesian methods for predicting the allelic richness of future samples on the basis of currently available samples. J Hered. 2006;97:483–492. doi: 10.1093/jhered/esl030. [DOI] [PubMed] [Google Scholar]
- Berg EE, Hamrick JL. Fine-scale genetic structure of a Turkey oak forest. Evolution. 1995;49:110–120. doi: 10.1111/j.1558-5646.1995.tb05963.x. [DOI] [PubMed] [Google Scholar]
- Bonin A, Ehrich D, Manel S. Statistical analysis of amplified fragment length polymorphism data: A toolbox for molecular ecologists and evolutionists. Mol Ecol. 2007;16:3737–3758. doi: 10.1111/j.1365-294X.2007.03435.x. [DOI] [PubMed] [Google Scholar]
- Bortolini F, Dall’Agnol M, Schifino-Wittmann MT. Molecular characterization of the USDA white clover (Trifolium repens L.) core collection by RAPD markers. Genet Resour Crop Evol. 2006;53:1081–1087. [Google Scholar]
- Brink GE, Pederson GA, Alison MW, Ball DM, Bouton JH, Rawls RC, Stuedemann JA, Venuto BC. Growth of white clover ecotypes, cultivars, and germplasms in the southeastern USA. Crop Sci. 1999;39:1809–1814. [Google Scholar]
- Bussell JD. The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliaceae) Mol Ecol. 1999;8:775–789. [Google Scholar]
- Cahn MG, Harper JL. The biology of leaf mark polymorphism in Trifolium repens L.1. Distribution of phenotypes at a local scale. Heredity. 1976;37:309–325. [Google Scholar]
- Chapman DF. Growth and Demography of Trifolium repens Stolons in Grazed Hill Pastures. J Appl Ecol. 1983;20:597–608. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–15. [Google Scholar]
- Ewens WJ. Sampling theory of selectively neutral alleles. Theor Pop Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Gustine DL, Elwinger GF. Spatiotemporal genetic structure within white clover populations in grazed swards. Crop Sci. 2003;43:337–344. [Google Scholar]
- Gustine DL, Sanderson MA. Molecular analysis of white clover population structure in grazed swards during two growing seasons. Crop Sci. 2001;41:1143–1149. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Phil Trans R Soc Lond Ser B Biol Sci. 1996;351:1291–1298. [Google Scholar]
- Harberd DJ. Observations on natural clones of Trifolium repens L. New Phytol. 1963;62:198–204. [Google Scholar]
- Herrmann D, Boller B, Widmer F, Kölliker R. Optimization of bulked AFLP analysis and its application for exploring diversity of natural and cultivated populations of red clover. Genome. 2005;48:474–486. doi: 10.1139/g05-011. [DOI] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Bousquet J. Complete congruence between gene diversity estimates derived from genotypic data at enzyme and random amplified polymorphic DNA loci in black spruce. Proc Natl Acad Sci USA. 1995;92:6369–6373. doi: 10.1073/pnas.92.14.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölliker R, Jones ES, Jahufer MZZ, Forster JW. Bulked AFLP analysis for the assessment of genetic diversity in white clover (Trifolium repens L.) Euphytica. 2001;121:305–315. [Google Scholar]
- Kongkiatngam P, Waterway MJ, Fortin MG, Coulman BE. Genetic variation within and between two cultivars of red clover (Trifolium pratense L.) - Comparisons of morphological, isozyme, and RAPD markers. Euphytica. 1995;84:237–246. [Google Scholar]
- Leberg PL. Estimating allelic richness: Effects of sample size and bottlenecks. Mol Ecol. 2002;11:2445–2449. doi: 10.1046/j.1365-294x.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Marghali S, Panaud O, Lamy F, Ghariani S, Sarr A, Marrakchi M, Trifi-Farah N. Exploration of intra- and inter-population genetic diversity in Hedysarum coronarium L. by AFLP markers. Genet Resour Crop Evol. 2005;52:277–284. [Google Scholar]
- Meirmans PG, Van Tienderen PH. GENOTYPE and GENODIVE: Two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- Mohapatra KP, Sehgal RN, Sharma RK, Mohapatra T. Genetic analysis and conservation of endangered medicinal tree species Taxus wallichiana in the Himalayan region. New Forests. 2009;37:109–121. [Google Scholar]
- Nybom H, Bartish IV. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst. 2000;3:93–114. [Google Scholar]
- Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A. Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: Implications for the transferability of SSRs in plants. Mol Biol Evol. 1998;15:1275–1287. doi: 10.1093/oxfordjournals.molbev.a025856. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv Biol. 1998;12:844–855. [Google Scholar]
- Rizza MD, Real D, Reyno R, Porro V, Burgueño J, Errico E, Quesenberry KH. Genetic diversity and DNA content of three South American and three Eurasiatic Trifolium species. Genet Mol Biol. 2007;30:1118–1124. [Google Scholar]
- Sardaro MLS, Atallah M, Tavakol E, Russi L, Porceddu E. Diversity for AFLP and SSR in natural populations of Lotus corniculatus L. from Italy. Crop Sci. 2008;48:1080–1089. [Google Scholar]
- Sedcole JR. Number of plants necessary to recover a trait. Crop Sci. 1977;17:667–668. [Google Scholar]
- Segovia-Lerma A, Cantrell RG, Conway JM, Ray IM. AFLP-based assessment of genetic diversity among nine alfalfa germplasms using bulk DNA templates. Genome. 2003;46:51–58. doi: 10.1139/g02-100. [DOI] [PubMed] [Google Scholar]
- Singh M, Chabane K, Valkoun J, Blake T. Optimum sample size for estimating gene diversity in wild wheat using AFLP markers. Genet Resour Crop Evol. 2006;53:23–33. [Google Scholar]
- Suzuki JI, Herben T, Maki M. An under-appreciated difficulty: Sampling of plant populations for analysis using molecular markers. Evol Ecol. 2004;18:625–646. [Google Scholar]
- Van Treuren R, Bas N, Goossens PJ, Jansen J, Van Soest LJM. Genetic diversity in perennial ryegrass and white clover among old Dutch grasslands as compared to cultivars and nature reserves. Mol Ecol. 2005;14:39–52. doi: 10.1111/j.1365-294X.2004.02391.x. [DOI] [PubMed] [Google Scholar]
- Vandepitte K, Roldan-Ruiz I, Leus L, Jacquemyn H, Honnay O. Canopy closure shapes clonal diversity and fine-scale genetic structure in the dioecious understorey perennial Mercurialis perennis. J Ecol. 2009;97:404–414. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Jasieniuk M. Sampling weedy and invasive plant populations for genetic diversity analysis. Weed Sci. 2009;57:593–602. [Google Scholar]
- Welham CVJ, Turkington R, Sayre C. Morphological plasticity of white clover (Trifolium repens L.) in response to spatial and temporal resource heterogeneity. Oecologia. 2002;130:231–238. doi: 10.1007/s004420100791. [DOI] [PubMed] [Google Scholar]
- Williams WM. Genetics and breeding. In: Baker MJ, Williams WM, editors. White Clover. CAB International; Wallingford: 1987. pp. 343–419. [Google Scholar]
- Wu J, Krutovskii KV, Strauss SH. Nuclear DNA diversity, population differentiation, and phylogenetic relationships in the California closed-cone pines based on RAPD and allozyme markers. Genome. 1999;42:893–908. [Google Scholar]
- Xu WW, Sleper DA, Krause GF. Genetic diversity of tall fescue germplasm based on RFLPs. Crop Sci. 1994;34:246–252. [Google Scholar]
- Zhao R, Cheng Z, Lu WF, Lu BR. Estimating genetic diversity and sampling strategy for a wild soybean (Glycine soja) population based on different molecular markers. Chin Sci Bull. 2006;51:1219–1227. [Google Scholar]
- Zhu W, Zhou T, Zhong M, Lu B. Sampling strategy for wild soybean (Glycine soja) populations based on their genetic diversity and fine-scale spatial genetic structure. Frontiers Biol China. 2007;2:397–402. [Google Scholar]