Abstract
Sulfur element plays a pivotal role in plant growth and development. Recently, we have demonstrated that miR395 is crucial for the sulfate homeostasis through regulating the sulfate uptake, transport and assimilation in Arabidopsis thaliana. miR395 controls the sulfate concentration in the shoot by targeting three ATP sulfurylase genes (APS), which encode the first enzymes catalyzing sulfate activation in sulfur assimilation pathway. Furthermore, miR395 also regulates the transport of sulfate between leaves. Under sulfate starvation conditions, upregulated miR395 represses the expression of SULTR2;1, which then confined the transport of sulfate from mature to young leaves. Of note, transcript expression analysis suggested that, unlike APS1 and APS4 mRNA, APS3 and shoot SULTR2;1 is in accordance with miR395 in response to sulfate deprivation. We proposed that the differential regulation of targets by miR395 may be required for adaptation to the sulfate deficiency environment. In addition, our results revealed that there is reciprocal regulation between SULTR2;1 and APS genes through miR395.
Key words: sulfate, miR395, APS1, APS3, APS4, SULTR2;1, sulfate transport, sulfate assimilation
MicroRNAs (miRNAs) are a class of noncoding small RNAs, which post-transcriptionally regulate target mRNAs by cleavage or/and translation repression.1,2 Several plant miRNAs have been identified to be involved in nutrients response, such as nitrogen, phosphor or sulfur.3,4 Recent research suggested that miR395 is inducible by sulfate deprivation, and it targets two families of genes, ATP Sulfurylases and SULTR2;1, both of which function in sulfate metabolism pathway.5–7 Our latest research revealed how miR395 functions in sulfate metabolism by regulating its target genes.5 In miR395 overexpressing transgenic plant that exhibits sulfur deficiency symptoms, three APS genes (APS1, APS3, APS4) are repressed, which suppresses the activation of sulfate and then results in the over-accumulation of sulfate in the shoot. Meanwhile, the SULTR2;1 transcripts are down-regulated, which then disrupts the transport of sulfate from mature leaves to young. Additionally, SULTR1;1 and SULTR1;2 are significantly upregulated, which contribute to the influx of sulfate from soil. In spite of the sulfate over-accumulation in the shoot, the root sulfate content is lower than that of wild-type plants, which can be attributed to the induced SULTR4;1 and SULTR4;2, the products of which serve for the efflux of sulfate from root vacuoles. These evidences suggest that miR395 regulates the homeostasis of sulfate in Arabidopsis thaliana.
Although all target genes are repressed in the miR395 overexpressing transgenic plants, they display varied responses to sulfate starvation in wild-type plants.5 Under sulfate depletion, SULTR2;1 is induced in roots and repressed in shoots, respectively. Kawashima et al.6 revealed that the spatially overlapping expression of miR395 and SULTR2;1 led to the negative correlation between them in shoots, and the different tissue-specific expression allowed the positive correlation between them in roots. As expected, APS1 and APS4 transcripts decreased in response to sulfate deficiency. The expression of APS3 gene was induced by sulfate starvation, although its repressor miR395 was upregulated. We suggested that APS3 might avoid the cleavage by miR395 in a manner similar to SULTR2;1 and serve to activate sulfate for assimilation under sulfate starvation condition when APS1 and APS4 were repressed. Therefore, to further investigate the accurate tissue-specific expression of these APS genes and their specific functions is required for understanding the mechanism underling sulfate assimilation.
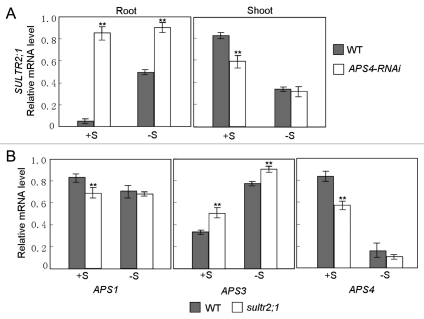
In addition to the regulation of target genes by miR395, we also found that the lack of APS4 gene results in the enhanced expression of miR395.5 This indicates that miR395 and APS4 are connected by a negative feedback loop. Our previous results also showed that APS1 and APS3 were suppressed in the APS4-silienced plants.5 Depending on these evidences, we proposed that APS4 regulated the expression of APS1 and APS3 by miR395. To investigate whether SULTR2;1 was also affected by APS4, we detected SULTR2;1 transcript levels in the root and shoot of APS4-silienced plants (Fig. 1A). As expected, SULTR2;1 was downregulated in the shoot, which can be attributed to the induction of miR395. However, SULTR2;1 was significantly induced in the root, suggesting that it responded to the sulfate starvation resulting from the lack of APS4. This is consistent with our previous conclusion that SULTR2;1 is dually regulated by sulfate deficiency. According to these evidences, we draw the conclusion that APS4 genes affected the expression of SULTR2;1. It is necessary to make sure whether SULTR2;1 can affect the expression of APS genes. We analyzed the expression of the three APS genes in the shoot of sultr2;1 mutant (Fig. 1B). Under sulfate sufficiency conditions, APS1 and APS4, but not APS3, were downregulated in sultr2;1 mutant, which was similar to their expression patterns in wild type under sulfate deficiency, although the fold-changes were not equal totally. Unlike wild-type plants, the sulfate concentration of young leaves was lower than that of mature leaves in sultr2;1 mutants. We proposed that the sulfate deficiency of young leaves accounted for the expression patterns of APS genes in sultr2;1 mutant. In conclusion, there are cross-talks between sulfate transport and assimilation pathways by miR395 targeting SULTR2;1 and APS genes.
Figure 1.
Real-time PCR analysis of miR395 target genes. (A) The expression of SULTR2;1 in the root and shoot of APS4-RNAi plants. (B) The expression of APS1, APS3 and APS4 in the shoot of sultr2;1 mutant plants. (A and B) Plants were grown for 10 days on MS medium with 1,500 lM sulfate (+S) or MS medium without sulfate (−S). RNA was isolated from roots and shoots, respectively. The error bars represent SD from triplicate samples. Student's t-test indicated that the values marked by two asterisks are significantly different from the corresponding wild-type value (p < 0.01; n = 3).
Recently, several miRNAs have been identified that they are involved in the feedback regulation of their target.8–10 Our results suggested that miR395 targets can feedback regulate the expression of miR395, which then controls the expression of other target genes. The complicate regulation of sulfate metabolism pathway may enable plants to survive in sulfate fluctuation environment.
Acknowledgements
We thank the Arabidopsis Biological Resource Center for the pools of T-DNA insertion mutants. This research was supported by the Science Foundation of Ministry of Agriculture of the People's Republic of China (2009ZX08009-066B), the National Natural Science Foundation of China (90408022), the National High Technology Research and Development Program of China (863 Program) (2006AA02Z129).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12608
References
- 1.Brodersen P, Sakvarelidze-Achard L, Brunun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 2.Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, et al. Biochemical evidence for translational repression by Arabidopsis MicroRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-Mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang G, Yang FX, Yu DQ. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 2010;62:1046–1057. doi: 10.1111/j.1365-313X.2010.04216.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, et al. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 7.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 8.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 9.Vaucheret H, Mallory AC, Bartel DP. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]