Abstract
To study the importance of intercellular transport for MADS domain transcription factor functioning during floral development, we analyzed the dynamic behavior of fluorescently-tagged MADS domain proteins in transgenic plants by Confocal Laser Scanning Microscopy. These analyses, described in a recent paper in The Plant Journal, provided proof for previous suggestions that the Arabidopsis thaliana C-type protein AGAMOUS has a non-cell-autonomous role in floral meristem integrity. Furthermore, it indicated a possible non-cell-autonomous role for the B-type proteins APETALA3 and PISTILLATA and the E-type protein SEPALLATA3, through lateral intercellular movement in the floral meristem. In this addendum we compare some of the available fluorescent protein-based technologies for the investigation of transcription factor movements and dynamics.
Key words: arabidopsis, MADS, GFP, mEosFP, intercellular transport, non-cell-autonomous function
Only recently it has been realized that intercellular transport of transcription factors through plasmodesmata is a highly dynamic process that can play an important regulatory role in developmental processes.1 Members of the MADS domain transcription factor family are well known for their crucial functions in flower development.2 Previously, the only two known intercellularly transported MADS domain transcription factors were the Antirrhinum majus B-type proteins DEFICIENS (DEF) and GLOBOSA (GLO), which are able to move towards the epidermis in the floral meristem.3 Intriguingly, their respective Arabidopsis thaliana orthologues APETALA3 (AP3) and PISTILLATA (PI) do not seem to have the same transport abilities.4 Moreover, AP3, PI and also the A-type protein APETALA1 (AP1) seem to lack the ability for inward movement in floral meristems.4–6 Nevertheless, it has been suggested that certain Arabidopsis thaliana MADS domain transcription factors do have non-cell-autonomous functions. To investigate this possibility further, we generated transgenic Arabidopsis thaliana plants that express AP3, PI, the C-type protein AGAMOUS (AG) and the E-type protein SEPALLATA3 (SEP3) with a GREEN FLUORESCENT PROTEIN (GFP) tag under the control of an epidermis-specific promoter.7 We analyzed the presence of the GFP-tagged proteins in both epidermal and non-epidermal cell layers by Confocal Laser Scanning Microscopy (CLSM) and in this way demonstrated that only the C-type protein AG can actively move inwards from the epidermal cell layer to the subepidermal cell layer in floral meristems, via secondary plasmodesmata. Additionally, a photobleaching technique demonstrated that all tested MADS domain proteins can diffuse through the epidermal cell layer, presumably in a passive manner through primary plasmodesmata. These observations provide proof for previous suggestions that AG has a non-cell-autonomous role in floral meristem integrity8–10 and also indicate a possible non-cell-autonomous role for AP3, PI and SEP3 during floral development through their lateral intercellular movement. This type of study is possible due to the implementation of various fluorescent protein-based technologies and provides insight into the dynamical behavior of transcription factor molecules during plant development. There is a strong need for this type of study, because our knowledge about the role of protein transport in developmental processes is limited. Below, possibilities and pitfalls of a few commonly used technologies are discussed.
FRAP and FLIP on a Larger Scale
In our study, we made use of the phenomenon that the GFP fluorophore can irreversibly lose its fluorescent ability when it is photochemically altered by high intensity 488 nm laser light. This photobleaching allows the tracking of fluorescently-tagged protein movement by monitoring the recovery of fluorescence in a photobleached area, commonly called Fluorescence Recovery After Photobleaching (FRAP).11,12 Alternatively, photobleaching can be used to track the movement of photobleached fluorescently-tagged proteins by monitoring the decrease in fluorescence in non-bleached areas, known as Fluorescence Loss In Photobleaching (FLIP). These FRAP and FLIP methods are generally used to investigate intracellular movement of molecules and to show continuity or transport between subcellular compartments in the cell. However, we applied photobleaching on a larger scale to investigate the movement of proteins between epidermal cells that specifically express GFP-tagged MADS domain proteins. In contrast to the usage of endogenous regulatory sequences, which in general results in expression of the proteins in all three clonal cell layers,13 the two-dimensional aspect of the fluorescent epidermal cell layer greatly simplified monitoring of the fluorescent recovery in bleached areas. Furthermore, it would also be feasible to demonstrate possible compartmentalization of developing tissues into symplasmic subdomains, by following the spread of photobleached fluorescently-tagged proteins while repeatedly photobleaching a few cells. Symplasmic subdomains are clusters of interconnected cells that are fully or partially isolated from intercellular transport from surrounding tissues.14–16
Photoconvertible and Photoactivatable Fluorescent Tags
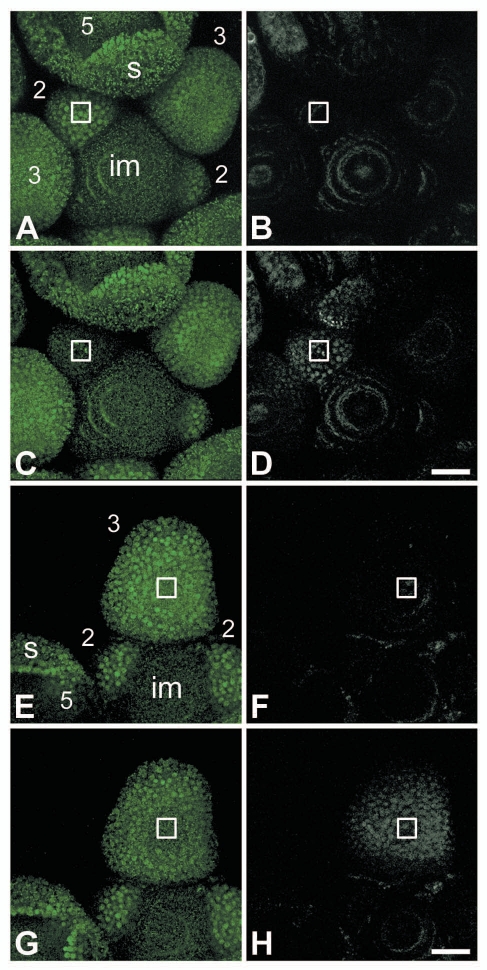
As an alternative for the above described photobleaching techniques, it is nowadays also possible to visualize behavior of a subset of tagged proteins by changing the color of the fluorescent tag.17 For instance, the photoconvertible monomeric EosFP (mEosFP) tag can be switched from green to red fluorescence by irreversibly changing the structure of the fluorophore with laser light of around 400 nm.18 We created transgenic Arabidopsis thaliana plants that express selected MADS box genes with this photoconvertible tag under the control of their own promoter, to study the intercellular movement of MADS domain proteins in their native environment. Unfortunately, there seemed to be extensive scattering of the photoconverting 405 nm laser light far away from the selected photoconversion area, as we observed unwanted photoconversion of mEosFP-tagged proteins in remote areas (Fig. 1). Changing the embedding medium for the samples from 0.8% agar to water did not change this scattering phenomenon. Thus, the system for photoconversion of a targeted area lacked the necessary precision to properly study intercellular protein transport over short and medium distances in inflorescences. Although the mEosFP tag and other photoconvertible tags have been used successfully in studies on the dynamics of subcellular components,17,19,20 in our set-up the curved and cell-dense inflorescence tissue might easily diffract the short wavelength photoconverting laser light.21 Whether this scattering of laser light occurs less with 488 nm laser light or whether it is just less noticeable in the photobleaching experiments is unclear at this moment. Similar problems with scattering may occur with photoactivatable fluorescent proteins, such as photoactivatable-GFP and DRONPA,17,22,23 which are frequently used to track the subcellular distribution of proteins and could also be used to track intercellular movements of proteins. These photoactivatable fluorophores do not change the color of their fluorescence when irradiated with 400–405 nm laser light, but are switched on by it.
Figure 1.
Photoconversion of AP1:mEosFP with 405 nm laser light. Confocal microscopy of mEosFP-tagged MAD S domain transcription factor APETA LA1 (AP1:mEosFP) in a wild-type Arabidopsis thaliana inflorescence before and after photoconversion of the mEosFP tag with 405 nm laser light at 100% power (A–D) and 25% power (E–H). The nuclei filled with unchanged, green AP1:mEosFP are visible as bright green dots, while the green autofluorescence is visible as small green speckles. Nuclei filled with the photoconverted red version of AP1:mEosFP are visible as bright white dots, while the preconversion scans show the presence of red autofluorescence in white. AP1:mEosFP is not present in the inflorescence meristem (im) and starts to appear from floral bud stage 1 onwards. In a stage 5 floral bud especially the tips of the sepals (s) show high levels of AP1:mEosFP. (A–D) Photoconversion was performed in a target area (indicated by the square) in a stage 2 floral bud by scanning one z-section 30 times with 405 nm laser light at 100% power. Localisations of green AP1:mEosFP before (A) and after photoconversion (C); showing overall reduced green fluorescence in the targeted floral bud after photoconversion. Localizations of red AP1:mEosFP before (B) and after photoconversion (D); demonstrating that not only the targeted floral bud was photoconverted, but also the adjacent floral buds. (E–H) Photoconversion was performed in a target area (indicated) in a stage 3 floral bud by scanning one z-section 30 times with 405 nm laser light at 25% power. Localizations of green AP1:mEosFP before (E) and after photoconversion (G); showing reduced green fluorescence in the middle of the targeted floral bud. Localizations of red AP1:mEosFP before (F) and after photoconversion (H); demonstrating that almost the whole targeted floral bud contains photoconverted AP1:mEosFP. Scale bars are 25 µm.
Conclusions
Ideally, the dynamic behavior of MADS domain transcription factors should be studied under native conditions. However, our first attempts to do this with photoconvertible mEosFP-tagged MADS domain proteins expressed under the control of endogenous regulatory sequences were unsuccessful. Nevertheless, our recent paper in The Plant Journal resolved some of the mysteries of the dynamic behavior of MADS domain transcription factors in the floral meristem, by monitoring the whereabouts of GFP-tagged proteins with CLSM after specifically expressing them in the epidermal cell layer and by applying a photobleaching technique. What the biological relevance of the observed movements is and what implications they have for the establishment of MADS domain protein expression patterns, remain to be studied.
Acknowledgements
We are grateful to Jörg Wiedenmann for providing the mEosFP vector. This project was cofinanced by the Centre for BioSystems Genomics (CBSG), which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12949
Supplementary Material
References
- 1.Lucas WJ, Ham LK, Kim JY. Plasmodesmata—bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Causier B, Schwarz-Sommer Z, Davies B. Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol. 2010;21:73–79. doi: 10.1016/j.semcdb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Perbal MC, Haughn G, Saedler H, Schwarz-Sommer Z. Non-cell-autonomous function of the Antirrhinum floral homeotic proteins DEFICIENS and GLOBOSA is exerted by their polar cell-to-cell trafficking. Development. 1996;122:3433–3441. doi: 10.1242/dev.122.11.3433. [DOI] [PubMed] [Google Scholar]
- 4.Jenik PD, Irish VF. The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development. 2001;128:13–23. doi: 10.1242/dev.128.1.13. [DOI] [PubMed] [Google Scholar]
- 5.Sessions A, Yanofsky MF, Weigel D. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science. 2000;289:779–781. doi: 10.1126/science.289.5480.779. [DOI] [PubMed] [Google Scholar]
- 6.Wu XL, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, et al. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 7.Urbanus SL, Martinelli AP, Dinh QD, Aizza LCB, Dornelas MC, Angenent GC, et al. Intercellular transport of epidermis-expressed MADS domain transcription factors and their effect on plant morphology and floral transition. Plant J. 2010;63:60–72. doi: 10.1111/j.1365-313X.2010.04221.x. [DOI] [PubMed] [Google Scholar]
- 8.Sieburth LE, Drews GN, Meyerowitz EM. Non-autonomy of AGAMOUS function in flower development: use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development. 1998;125:4303–4312. doi: 10.1242/dev.125.21.4303. [DOI] [PubMed] [Google Scholar]
- 9.Jenik PD, Irish VF. Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development. 2000;127:1267–1276. doi: 10.1242/dev.127.6.1267. [DOI] [PubMed] [Google Scholar]
- 10.Cartolano M, Efremova N, Kuckenberg M, Raman S, Schwarz-Sommer Z. Enhanced AGAMOUS expression in the centre of the Arabidopsis flower causes ectopic expression over its outer expression boundaries. Planta. 2009;230:857–862. doi: 10.1007/s00425-009-0966-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin JS, Kenworthy AK. Photobleaching approaches to investigate diffusional mobility and trafficking of Ras in living cells. Methods. 2005;37:154–164. doi: 10.1016/j.ymeth.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Urbanus SL, de Folter S, Shchennikova AV, Kaufmann K, Immink RGH, Angenent GC. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009:9. doi: 10.1186/1471-2229-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gisel A, Barella S, Hempel FD, Zambryski PC. Temporal and spatial regulation of symplastic trafficking during development in Arabidopsis thaliana spices. Development. 1999;126:1879–1889. doi: 10.1242/dev.126.9.1879. [DOI] [PubMed] [Google Scholar]
- 15.Rinne PLH, van der Schoot C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1998;125:1477–1485. doi: 10.1242/dev.125.8.1477. [DOI] [PubMed] [Google Scholar]
- 16.Kim I, Kobayashi K, Cho E, Zambryski PC. Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2005;102:11945–11950. doi: 10.1073/pnas.0505622102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman S, Oparka KJ, Roberts AG. New tools for in vivo fluorescence tagging. Curr Opin Plant Biol. 2005;8:565–573. doi: 10.1016/j.pbi.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinclair AM, Trobacher CP, Mathur N, Greenwood JS, Mathur J. Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 2009;59:231–242. doi: 10.1111/j.1365-313X.2009.03863.x. [DOI] [PubMed] [Google Scholar]
- 20.Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 21.Dobrucki JW, Feret D, Noatynska A. Scattering of exciting light by live cells in fluorescence Confocal imaging: Phototoxic effects and relevance for FRAP studies. Biophys J. 2007;93:1778–1786. doi: 10.1529/biophysj.106.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runions J, Brach T, Kuhner S, Hawes C. Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot. 2006;57:43–50. doi: 10.1093/jxb/eri289. [DOI] [PubMed] [Google Scholar]
- 23.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.