Abstract
Effectors are pathogen-encoded proteins that are thought to facilitate infection by manipulation of host cells. Evidence showing that the effectors of some eukaryotic plant pathogens are able to interact directly with cytoplasmic host proteins indicates that translocation of these proteins into host cells is an important part of infection. Recently, we showed that the flax rust effectors AvrM and AvrL567 are able to internalize into plant cells in the absence of the pathogen. Further, N-terminal sequences that were sufficient for uptake were identified for both these proteins. In light of the possibility that the internalization of fungal and oomycete effectors may require binding to specific phospholipids, the lipid binding activities of AvrM and AvrL567 mutants with different abilities to enter cells were tested. While AvrL567 was not found to bind to phospholipids, AvrM bound strongly to phosphatidyl inositol, phosphatidyl inositol monophosphates and phosphatidyl serine. However, a fragment of AvrM sufficient to direct uptake of a fusion protein into plant cells did not bind to these phospholipids. Thus, our results do not support the role of specific binding of AvrM and AvrL567 to phospholipids for uptake into the plant cytoplasm.
Key words: effector, uptake, translocation, lipid binding, phosphatidylinositol
Eukaryotic pathogens, including fungi and oomycetes, secrete effector proteins that are subsequently delivered into host cells to facilitate infection.1–3 Many oomycete effector proteins contain an N-terminal RxLR motif required for delivery into host cells4,5 and are able to enter plant cells in the absence of the pathogen.6 However, the mechanism of RxLR protein uptake into cells has yet to be determined. In our recent study,7 the AvrM and AvrL567 effectors from the biotrophic flax rust fungus were also shown to enter plant cells in a pathogen-independent process. Further, through deletion analysis, minimum regions of each protein required for uptake were identified.7 Unlike the oomycete RxLR effectors, no motif was found to be strongly conserved between the uptake regions of these two effectors or in other rust effectors.7
A number of proteins with the ability to cross membranes are known and include the HIV Tat protein8,9 and the penetratin homeodomain of the transcription factor Antennapedia.10,11 Through deletion studies it was shown that uptake of these proteins is mediated by small regions of each protein, less than 30 amino acids in size, known as cell penetrating peptides (CPPs). These sequences can also mediate uptake of attached cargoes, including low molecular weight drugs,12 proteins,13 nucleic acids14 and peptides.15 CPPs have diverse sequences and structures and studies on their uptake show that internalization can occur by different mechanisms—possibly even simultaneously—including clathrin-dependent endocytosis,16 lipid raft-mediated macropinocytosis,17 clathrin- and caveolin-independent endocytosis18 and direct membrane penetration.19 In the case of the amphipathic penetratin peptide, membrane penetration may occur by integration into phospholipid membranes, with two critical tryptophan residues penetrating into the lipid bilayer and positively charged side chains associating non-specifically with negatively charged phospholipids, disrupting the membrane and possibly facilitating the formation of small vesicles or inverted micelles within the membrane before release into the cytoplasm.19–23 On the other hand, for endocytic transport, penetratin may associate with negatively charged membrane-associated components, such as the glycosaminoglycan heparan sulfate prior to uptake.16,24,25 Recently, it has been proposed that the RxLR uptake motif of plant pathogenic oomycete effectors and potential uptake regions of rust fungal effectors, bind specifically to phosphatidylinositol phosphate (PtdInsP) head groups, which may act as receptors initiating effector endocytosis into cells.26 Here, we present data on the lipid binding activities of AvrM and AvrL567 in the context of their internalization into plant cells.
Lipid Binding Specificities of AvrM and AvrL567
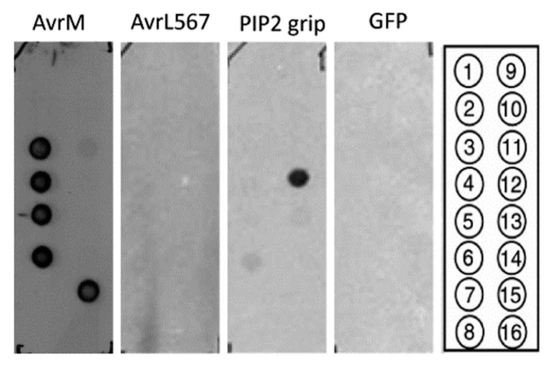
Based on primary sequence analysis, no lipid binding domains were identified in AvrM or AvrL567. The majority of lipid binding domains lack primary sequence similarities or motifs,27 although they are often characterized by the presence of important positively charged residues. Therefore, the binding activity of both proteins to a range of lipids found in biological membranes was experimentally tested. A lipid blot consisting of 100 pmol amounts of different phospholipids immobilized onto a nitrocellulose membrane was probed with 0.25 µg/mL (6.5 nM) His-tagged AvrM or 0.5 µg/mL (17.5 nM) His-tagged AvrL567 and detected with polyclonal antibodies raised against purified AvrM7 and AvrL567, respectively. His-tagged AvrM lacking the signal peptide was found to bind to PtdIns, PtdIns(3)P, PtdIns(4)P, PtdIns(5)P and phosphatidylserine, with binding being strongest for PtdIns(3)P and PtdIns(5)P. In contrast, no signal was detected when blots were incubated with AvrL567 (Fig. 1). When the concentration of AvrL567 was increased ten-fold (175 nM), a weak binding to PtdIns(3)P and PtdIns(5) P was also detected (data not shown). At 0.5 µg/mL, the PIP2 Grip protein, consisting of the pleckstrin homology domain of phospholipase C fused to GST, demonstrated specific binding towards PtdIns(4,5)P2 when detected using an anti-GST antibody28 and the negative control, His-tagged GFP, demonstrated no binding when probed with anti-GFP as expected.
Figure 1.
Binding of His-tagged AvrM and AvrL567 to lipids. PIP strips from Echelon Biosciences were blocked with 4% non-fat milk/PBS (0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride, pH 7.4), incubated with purified proteins in blocking buffer overnight at 4°C and washed three times for 10 min each in PBST [PBS + 0.1% (v/v) Tween-20]. The PIP2 grip protein (Echelon Biosciences) was used as a positive control and GFP was used as a negative control. Bound AvrM, AvrL567, PIP2 grip and GFP were detected with anti-AvrM,7 anti-AvrL567, anti-GST28 and anti-GFP (Roche Diagnostics Ltd., Mannheim, Germany) antibodies, respectively. Blots were rinsed, incubated with anti-rabbbit IgG-peroxidase conjugate (A9169, Sigma) and bound proteins were detected using the Supersignal West Pico Chemiluminescent Substrate (Pierce Chemical). All proteins were used at 0.5 µg/mL with the exception of AvrM which was used at 0.25 µg/mL. Compounds spotted on the membrane: 1, lysophosphatidic acid; 2, lysophosphatidylcholine; 3, PtdIns; 4, PtdIns(3)P; 5, PtdIns(4)P; 6, PtdIns(5)P; 7, phosphatidyl ethanolamine; 8, phosphatidyl choline; 9, sphingosine-1-phosphate; 10, PtdIns(3,4)P2; 11, PtdIns(3,5)P2; 12, PtdIns(4,5)P2; 13, PtdIns(3,4,5)P3; 14, phosphatidic acid; 15, phosphatidyl serine; 16, blank. Each spot consists of 100 pmol of immobilized lipid.
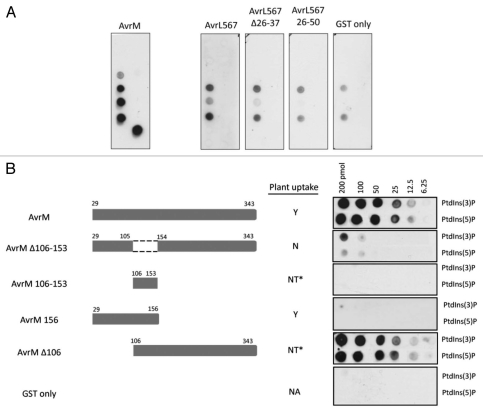
The above experiments used different antisera to detect protein bound to lipid blots. To allow direct comparison of lipid binding of both effector proteins and detection of lipid binding of truncated fragments of these effectors, proteins were expressed as N-terminal GST fusions and binding was detected using anti-GST antibodies. At 6.5 nM, the AvrM-GST fusion showed similar phospholipid binding to that of the wild-type protein (Fig. 2A). Similarly, AvrL567-GST at 175 nM was found to have the same binding pattern as protein lacking the GST tag. However, at 175 nM, the negative control of unmodified GST showed a similar binding pattern to that of AvrL567, indicating that AvrL567 binding was close to background (Fig. 2A).
Figure 2.
Binding of GST-tagged AvrM and AvrL567 mutants to lipids. (A) GST-tagged AvrM (6.5 nM) and AvrL567 mutants (175 nM) were affinity purified, used to probe PIP strips spotted with various lipids as for the His-tagged proteins and detected with anti-GST antibodies. GST at 175 nM was used as a negative control. (B) GST-tagged AvrM uptake mutants binding PtdIns(3)P and PtdIns(5)P. Serial dilutions (200, 100, 50, 25, 12.5, 6.25 pmol) of each phospholipid were spotted onto nitrocellulose membranes and incubated with purified-GST fusion proteins. Membranes were washed and bound protein detected with anti-GST. Data of uptake into plants was previously assessed.7 Y, Uptake detected; N, no uptake detected; NT, not tested; NA, not applicable; *The similar AvrM fragments, AvrM106-156 and AvrM−35-106, were shown to internalize into host cells.
Lipid Binding Activities of AvrM and AvrL567 Uptake Mutants
To explore the possibility that lipid binding might be required for effector uptake into plant cells, GST-fusions to uptake mutants identified in our earlier study7 were analyzed for lipid binding activity. In the case of AvrL567, both a C-terminal fragment lacking amino acids 26–37, which are required for cell internalization,7 and an N-terminal fragment consisting of residues 26–50 also showed no binding to phospholipids above background (Fig. 2A). As we previously showed that a protein fusion consisting of amino acids 1–50 (including the signal peptide) was sufficient for uptake,7 this result is not consistent with a role for specific phospholipid binding in entry of AvrL567 into cells.
Because AvrM showed its strongest binding towards PtdIns(3)P and PtdIns(5)P, we tested several AvrM truncations for binding to dilution series of these lipids on nitrocellulose membranes (Fig. 2B). Deletion of the N-terminal amino acids up to position 106, did not affect binding to PtdIns(3)P and PtdIns(5)P, while deletion of residues 106–153, which are required for uptake, substantially reduced but did not completely abolish lipid binding. Thus this region may be required for lipid binding, either directly or through effects on overall protein folding. Indeed this fusion protein expressed very poorly in E. coli, suggesting possible folding defects. However, no lipid binding activity was detected for a GST fusion containing AvrM residues 106–153, indicating that this region is not sufficient for PtdInsP binding. Similarly, residues 29–156 of AvrM, which are sufficient for uptake in planta, also did not show lipid binding. This indicates that the uptake region is not sufficient for AvrM binding to specific phospholipids, which therefore seems to also require the C-terminal effector domain.
Together, these results do not support the conclusion that specific binding to particular phospholipid headgroups is required for pathogen-independent internalization of AvrM and AvrL567 into host cells. This does not rule out a role for direct membrane association of these uptake peptides. Interestingly, the AvrM uptake region shows similarity to the penetratin peptide in that it contains a pattern of alternating hydrophobic and charged residues, so it may also integrate into the lipid bilayer via formation of an amphipathic structure. Conserved hydrophobic positions in the AvrL567 uptake region may function similarly, but it is also possible that different rust effectors adopt different uptake mechanisms.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13013
References
- 1.Hogenhout SA, Van der Hoorn RAL, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant-Microbe Interact. 2009;22:115–122. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- 2.Stergiopoulos I, De Wit PJGM. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 3.Panstruga R, Dodds PN. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehmany AP, Gordon A, Rose LE, Allen RL, Armstrong MR, Whisson SC, et al. Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell. 2005;17:1839–1850. doi: 10.1105/tpc.105.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–158. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 6.Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rafiqi M, Gan P, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, et al. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell. 2010;22:2017–2032. doi: 10.1105/tpc.109.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel AD, Pabo CO. Cellular uptake of the Tat protein from the human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 9.Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 10.Joliot A, Pernelle C, Deagostini-Bazin H, Prochiantz A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc Natl Acad Sci USA. 1991;88:1864–1868. doi: 10.1073/pnas.88.5.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorén PEG, Persson D, Karlsson M, Nordén B. The Antennapedia peptide penetratin translocates across lipid bilayers—the first direct observation. FEBS Lett. 2000;482:265–268. doi: 10.1016/s0014-5793(00)02072-x. [DOI] [PubMed] [Google Scholar]
- 12.Rothbard JB, Garlington S, Lin Q, Kirschberg T, Kreider E, McGrane PL, et al. Conjugation of arginine oligomers to cyclosporin A facilitates topical delivery and inhibition of inflammation. Nat Med. 2000;6:1253–1257. doi: 10.1038/81359. [DOI] [PubMed] [Google Scholar]
- 13.Nagahara H, Vocero-Akbani AM, Snyder EL, Ho A, Latham DG, Lissy NA, et al. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 14.Simmons CG, Pitts AE, Mayfield LD, Shay JW, Corey DR. Synthesis and membrane permeability of DNA-peptide conjugates. Bioorg Med Chem Lett. 1997;7:3001–3006. [Google Scholar]
- 15.Shibagaki N, Udey MC. Dendritic cells transduced with protein antigens induce cytotoxic lymphocytes and elicit antitumor immunity. J Immunol. 2002;168:2393–2401. doi: 10.4049/jimmunol.168.5.2393. [DOI] [PubMed] [Google Scholar]
- 16.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J Biol Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 17.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 18.Ter-Avetisyan G, Tünnemann G, Nowak D, Nitschke M, Herrmann A, Drab M, et al. Cell entry of arginine-rich peptides is independent of endocytosis. J Biol Chem. 2009;284:3370–3378. doi: 10.1074/jbc.M805550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joliot A, Prochiantz A. Transduction peptides: from technology to physiology. Nat Cell Biol. 2004;6:189–196. doi: 10.1038/ncb0304-189. [DOI] [PubMed] [Google Scholar]
- 20.Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 21.Drin G, Cottin S, Blanc E, Rees AR, Temsamani J. Studies on the internalization mechanism of cationic cell-penetrating peptides. J Biol Chem. 2003;278:31192–31201. doi: 10.1074/jbc.M303938200. [DOI] [PubMed] [Google Scholar]
- 22.Zorko M, Langel Ü. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Del Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Yesylevskyy S, Marrink S-J, Mark AE. Alternative mechanisms for the interaction of the cell-penetrating peptides penetratin and the TAT peptide with lipid bilayers. Biophys J. 2009;97:40–49. doi: 10.1016/j.bpj.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goncalves E, Kitas E, Seelig J. Binding of oligoarginine to membrane lipids and heparan sulfate: structural and thermodynamic characterization of a cell-penetrating peptide. Biochemistry. 2005;44:2692–2702. doi: 10.1021/bi048046i. [DOI] [PubMed] [Google Scholar]
- 25.Jiao C-Y, Delaroche D, Burlina F, Alves ID, Chassaing G, Sagan S. Translocation and endocytosis for cell-penetrating peptide internalization. J Biol Chem. 2009;284:33957–33965. doi: 10.1074/jbc.M109.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.BM Tyler, SD Kale, B Gu, DGS Capelluto, D Dou, FD Arredondo, et al. Blocking entry of rust effectors into host cells; Plant & Animal Genomes XVIII Conference; 2010. p. 284. [Google Scholar]
- 27.Catimel B, Schieber C, Condron M, Patsiouras H, Connolly L, Catimel J, et al. The PI(3,5)P2 and PI(4,5)P2 interactomes. J Proteome Res. 2008;7:5295–5313. doi: 10.1021/pr800540h. [DOI] [PubMed] [Google Scholar]
- 28.Upadhyaya NM, Yang M, Kositratana W, Ghosh A, Waterhouse PM. Molecular analysis of rice ragged stunt oryzavirus segment 9 and sequence conservation among isolates from Thailand and India. Arch Virol. 1995;140:1945–1956. doi: 10.1007/BF01322684. [DOI] [PubMed] [Google Scholar]