Abstract
The eukaryotic cell cycle is a process controlled by protein assemblies, of which the key subunits are serine-threonine cyclin-dependent kinases (CDKs). Timely association and dissociation of these assemblies ensure that the cell division program is executed correctly. The challenge to unravel the rules of the plant cell cycle results from the multiplicity of the process-regulating genes that emerged through genome duplications during the evolution of flowering plants. Despite the increasing knowledge on the plant cell cycle control, little is known about the composition of the different CDK-Cyclin complexes and their spatiotemporal occurrence. The binary interactions of the previously annotated 58 Arabidopsis thaliana core cell cycle proteins were tested in two high-throughput protein-protein interaction (PPI) assays: the bimolecular fluorescence complementation (BiFC) and the yeast two-hybrid. The resulting PPI network was integrated with available cycle phase-dependent gene expression data and subcellular localization information, revealing distinct cell cycle clusters acting at different cell division stages. Additionally, the BiFC assay revealed that three D-type cyclins, CYCD4;1, CYCD4;2 and CYCD5;1, form active kinase complexes with CDKA;1 and CDKB1;1 in vivo because they induce cell divisions in differentiated tobacco (Nicotiana benthamiana) epidermal cells. We demonstrate that these complexes promote cell proliferation in Arabidopsis and we discuss their putative mode of action in plant development.
Key words: Arabidopsis, cell cycle, protein-protein interaction, cell division, meristem
Cell division is an evolutionarily conserved process controlled by the alternating activity of CDKs.1,2 Proper timing and localization of the CDK activity depends on the association with their regulatory subunits, cyclins.3 This activity is further regulated by the phosphorylation status of the CDKs and by the binding of inhibitors and scaffold proteins.4 Oscillatory expression of cyclins determines the cell cycle phase substrate specificity of CDKs5–11 and cyclins present in the G1 phase regulate the CDK-dependent G1-to-S transition, while the G2/M cyclins are indispensable for progression through mitosis.
A plethora of information is available on the cell cycle PPI network in yeast,12 but corresponding experimental data on the plant cell cycle are scarce. Therefore, a systematic approach to functionally characterize plant cell cycle proteins is of particular interest. A PPI analysis was carried out for 58 Arabidopsis core cell cycle regulators to identify all physical connections among these proteins, potentially occurring during a plant cell division.13 Two PPI assays, the BiFC and yeast two-hybrid, were combined with information obtained from DNA microarrays on the transcription periodicity of Arabidopsis genes during the cell cycle. Merging of the physical PPI data with transcriptional expression peaks allowed the identification of plant cell cycle phase-specific modules. Moreover, novel regulatory relations were determined based on the transcriptional correlation among the interacting protein couples, revealing distinct cell cycle subnetworks that operate at different cell division stages.
Ectopic Expression of CDKA/B-CYCD4/5 Protein Complexes Induces Cell Division in Tobacco Epidermis
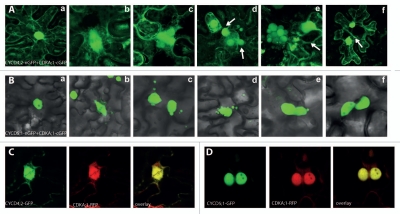
In plants, as in other eukaryotes, the CDK-CYCD complexes control the G1-to-S transition by their ability to act on the retinoblastoma protein (RB) mediated by the presence of the LxCxE amino acid motif in cyclins.14 Phosphorylation of RB by CDK-CYCD causes the dissociation of the RB-bound E2F/DP transcription factors and thereby enables the transcription of genes required for the S-phase progression.14–16 The BiFC screen provided information for the direct PPIs among the 58 Arabidopsis core cell cycle proteins in tobacco epidermal cell.13 From the identified 341 cell cycle complexes, six CDK-CYC couples, CDKA;1-CYCD4;1 CDKA;1-CYCD4;2; CDKA;1-CYCD5;1, CDKB1;1-CYCD4;1, CDKB;1-CYCD4;2 and CDKB;1-CYCD5;1 induced ectopic cell divisions in differentiated epidermal cells of tobacco. As the phenotypes of the respective CDKA;1 and CDKB1;1 combinations were identical, only the CDKA;1-CYCD4/5 complexes are shown (Fig. 1). In nondividing epidermal cells of tobacco, the fluorescent CDKA/B-CYCD4 complex localized to the nuclei and cytoplasm (Fig. 1Aa), whereas during cell division the complex was associated with the spindle zone (Fig. 1Ab). The ectopic cell divisions resulted in either two daughter cells due to completed cytokinesis (Fig. 1Ac), multiple nuclei due to karyokinesis without subsequent cytokinesis, or enlarged daughter nuclei due to endomitosis (Fig. 1Ad and Ae). Some cells divided multiple times, as inferred from the emerged cross walls (Fig. 1Ad and Af). In contrast, the CDKA/B-CYCD5;1 complexes were exclusively nuclear (Fig. 1Ba), but the fluorescent complexes were associated with the spindle as well (Fig. 1Bb). In these cells, divisions gave rise to cells with either multiple differently sized nuclei or one main nucleus and several satellite nuclei (micronuclei) (Fig. 1Bc and Bd). In some cases, the satellite nuclei were connected with the main nuclei via nuclear bridges (Fig. 1Be and Bf), indicative of improper karyokinesis or reformation of a nuclear membrane around improperly separated or lagging chromosomes.17,18 We can speculate that the CDKA/B-CYCD5;1 complexes initiated division, but failed to maintain the kinase activity required for further mitotic events. To exclude the possibility that the ectopic cell divisions resulted from the stabilization of CDK-CYCD complexes by the irreversible association of the two green fluorescent protein (GFP) fragments, all CYCD and CDK proteins were coexpressed in the tobacco epidermis after fusion to a full-length GFP or red fluorescent protein (RFP). In all combinations tested, the two proteins co-localized and induced ectopic cell divisions, exemplified by the coexpression of the CYCD4;2-GFP and CDKA;1-RFP fusions (Fig. 1C) and CYCD5;1-GFP and CDKA;1-RFP (Fig. 1D). In addition, with the exception of the CYCD4;1-GFP fusion (with a very low frequency), none of the single proteins was able to induce cell divisions when transiently expressed in the tobacco epidermis.
Figure 1.
Cell division in epidermal cells of tobacco (Nicotiana benthamiana) induced by ectopic expression of CDK-CYCD complexes. (A) Transient expression and detection of interaction between CYCD 4;2-nGFP and CDKA;1-cGFP. (a) Localization of the CDKA;1-CYCD 4;2 complex to the nucleus and to the cytoplasm in non-dividing epidermal cells. (b) In dividing epidermal cells, association of the CDKA;1-CYCD 4;2 complex with the spindle. (c–f) After division, frequent formation of two daughter nuclei, multiple, or enlarged nuclei was observed in the epidermal cells. (B) Transient expression and detection of interaction between CYCD 5;1-nGFP and CDKA;1-cGFP. (a) Exclusive localization of the complex to the nuclei in non-dividing epidermal cells. (b) Localization of the complex to the spindle in dividing cells. (c–f) After cell division, satellite micronuclei around the main nucleus were observed. (C) Transient expression of CYCD 4;2-GFP and CDKA;1-RFP, when the two proteins co-localize and ectopic cell divisions take place. (D) Transient expression of CYCD 5;1-GFP and CDKA;1-RFP; two daughter nuclei are close to the recent division site.
The CDKA-CYCD4 Complexes Promote Cell Proliferation in Arabidopsis
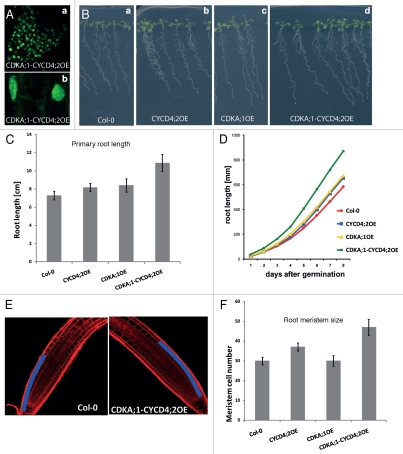
We investigated whether the CDK-CYCD4 complexes would induce ectopic cell divisions when stably overproduced in Arabidopsis. Because the two D4-type cyclins function redundantly in Arabidopsis19 and no differences between CDKB1;1 and CDKA;1 complexes had been observed, only the CDKA;1-CYCD4;2 transgenic lines are described. Based on the fluorescence signal, the CDKA;1-CYCD4;2 complex was detected in most plant tissues. In Arabidopsis, similarly to tobacco epidermis cells, the CDKA;1-CYCD4;2 complex was nuclear and slightly cytoplasmic (Fig. 2Aa and Ab). One representative homozygous transgenic line overexpressing a single T-DNA from both the CYCD4;2-nGFP and CDKA;1-cGFP constructs, designated CDKA;1-CYCD4;2OE, was phenotypically analyzed. In parallel, plants overexpressing the CDKA;1-GFP (CDKA;1OE), CYCD4;2-GFP (CYCD4;2OE) constructs and the wild-type Columbia-0 (Col-0) were used as controls. The roots of ten-day-old seedlings expressing the CDKA;1-CYCD4;2 complex were 45% longer than those of the Col-0, while the roots of the lines overexpressing the single CDKA;1 and CYCD4;2 proteins were only slightly (12% and 15%) longer (Fig. 2B and C). The expression of the CDKA;1-CYCD4;2 complex also enhanced the root growth rate more than the CDKA;1 and CYCD4;2 proteins expressed alone (Fig. 2D). To assess whether the increase in the root length in the CDKA;1-CYCD4;2OE line is caused by promotion of cell division, we measured the root meristem size as the number of cortex cells in a file extending from the quiescent center to the first elongated cell five days after germination, when a fixed number of meristematic cells is established (Fig. 2E and F).20 While the wild-type roots and the single CDKA;1OE line had on average 30 meristematic cells in the cortex file of the proximal meristem, the CYCD4;2OE and the CDKA;1-CYCD4;2OE lines had on average 37 and 47 cells, respectively (Fig. 2F). Therefore, the increase in the root length and enhanced growth rate in the lines expressing CYCD4;2 and CDKA;1-CYCD4;2, positively correlated with enhanced meristematic activity in these organs.
Figure 2.
Phenotypic analysis of the root meristem of Arabidopsis plants stably overexpressing the CDKA;1-CYCD4;2 complex. (A) Localization of the CDKA;1-CYCD4;2 complex in Arabidopsis roots. (B) Root growth of CDKA;1-CYCD4;2OE line compared to that of CDKA;1OE, CYCD4;2OE and the wild type. (C) Measurement of the root length. (D) Root growth rate. (E) Longitudinal view of the Arabidopsis root meristem of wild-type and CDKA;1-CYCD4;2-overexpressing plants in which cortex cell files in the meristem are marked in blue. (F) Number of root meristem cells of five-day-old seedlings.
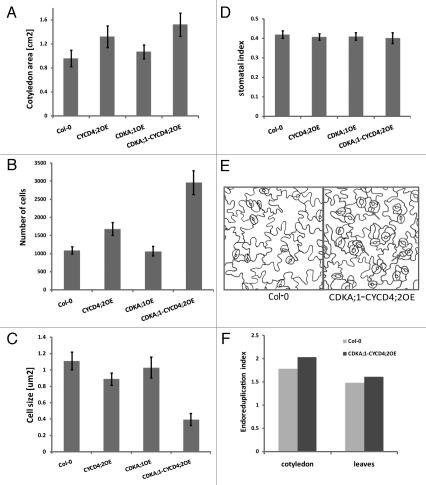
Previously, the overexpression of D4-type cyclins in Arabidopsis had been reported to result in no distinct leaf phenotype, although in the CYCD4;2OE lines the number of epidermal cells had increased and their size decreased.19 In contrast, in our experiments, in plants overexpressing either the CYCD4;2 or the CDKA;1-CYCD4;2 complex, the cotyledons were 35% and 58% larger than those of the wild type and the CDKA;1OE line, respectively (Fig. 3A). To determine whether cell division or cell expansion contributed to the differences in organ size, we analyzed the epidermal cells of cotyledons of CYCD4;2OE, CDKA;1OE, CDKA;1-CYCD4;2OE and wild-type plants by quantitative microscopy.21 The abaxial epidermis of 21-day-old Arabidopsis cotyledons was examined with regard to cell number, cell size and stomatal index (Fig. 3A–E). The total epidermal cell number in the cotyledons of the CDKA;1-CYCD4;2OE line was much higher than that of the wild-type and of the lines overexpressing the single CYCD4;2 or CDKA;1 proteins (Fig. 3B), the average size of the epidermal cells was drastically reduced in the cotyledons of CDKA;1-CYCD4;2OE plants (Fig. 3C). Additionally, the stomatal index remained the same in all analyzed plant lines (Fig. 3D), although some undifferentiated stomata we observed in the CDKA;1-CYCD4;2OE line (Fig. 3E).
Figure 3.
Phenotypes of cotyledon epidermal cells of 21-day-old Arabidopsis plants grown in vitro and overexpressing the CDKA;1-CYCD4;2 complex, CDKA;1 and CYCD4;2. (A) Cotyledon area. (B) Average number of epidermal cells. (C) Average size of epidermal cells. (D) Stomatal index. (E) Drawing-tube images of the abaxial epidermis of cotyledons. (F) Endoreduplication index presented as the mean number of endoreduplication cycles per nucleus.
A positive correlation between ploidy level and cell size was observed in wild-type epidermal cells and trichomes, suggesting that ploidy levels could determine cell size.22,23 Thus, we anticipated that the smaller epidermal cell size in the CDKA;1-CYCD4;2OE line would correlate with lower ploidy levels. Surprisingly, the ploidy levels in cotyledons of the CDKA;1-CYCD4;2OE transgenic line were characterized with more endoreduplication events in these organs than in the wild type (Fig. 3F). In agreement with previous reports, the ploidy level of the CYCD4;2OE line was not affected19 (data not shown).
Because CYCD4;1 and CYCD4;2 control cell division in the stomatal lineage in hypocotyls,19 we analyzed the hypocotyls of the CDKA;1-CYCD4;2OE line and found, consistently, that the number of stomata had increased by 55% in seven-day-old seedlings overexpressing CYCD4;2 and by 150% in the seedlings of the CDKA;1-CYCD4;2OE line (Fig. 4A). In the hypocotyls of the CDKA;1-CYCD4;2OE transgenic plants, more and smaller cells were observed in the non-protruding files and some non-differentiated stomatal precursors (Fig. 4B). Taken together, the D4-type cyclins positively influenced the root meristem size and increased the number of epidermal cells in cotyledons and hypocotyls. However, a co-expression of CYCD4;2 and CDKA;1 proteins enhanced the phenotype, indicating that the CDKA;1-CYCD4;2 complex promotes proliferation, probably through enhancing both G1-to-S and G2-to-M transitions.
Figure 4.

Cell divisions in hypocotyls of 7-day-old seedlings induced by overexpression of CYCD4;2 and CDKA;1-CYCD4;2. (A) Number of stomata in hypocotyls. (B) Hypocotyl epidermis of the wild-type and CDKA;1-CYCD4;2OE plants.
Conclusions
We used a transient BiFC assay in tobacco epidermal cells to analyze the binary cell cycle PPIs as a fast, flexible and reproducible approach for a high-level expression of proteins of interest.13 Although the BiFC assay is very sensitive, PPIs associated with particular cell cycle phases can be missed in non-dividing and differentiated tobacco epidermal cells. Nevertheless, in addition to the identification of 341 binary PPIs (of which 293 interactions had not been reported before), tobacco epidermal cells accounted for the discovery of novel and functional complexes between D4/D5-type cyclins and CDKA and CDKB proteins. D4-type cyclins have previously been proposed to function as positive regulators of cell division in different tissues of Arabidopsis and to form active kinase complexes with CDKA;1.19,24 However, binding of CYCD4 to CDKB2;1 proteins had been shown only in vitro19 and the functionality of these complexes had not been demonstrated in Arabidopsis. Our in vivo data support a function for the CDKB1;1 and D4-type cyclin complexes at either G2-to-M transition or during the S phase, when the expression of the CDKB1;1 is regulated by the G1/S-specific E2F transcription factors.21 The differences in the reported overexpression phenotypes between CYCD4 (this study) and CYCD3,25 proteins illustrates the functional diversity and complexity of the plant D-type cyclins. Similarly to CYCD3;1-overexpressing plants,25 we observed more and smaller epidermal cells in Arabidopsis cotyledons overproducing the CDKA;1-CYCD4;2 complex. Interestingly, the CDKA;-CYCD4;2OE plants showed an increase in ploidy levels, in contrast to the CYCD3;1 overexpression phenotype. Increase in endoreduplication has been observed in plants overexpressing E2F/DP transcription factors,26 which are known to act downstream of the CDK-CYCD-dependent phosphorylation events.
Acknowledgements
We thank Martine De Cock for help with the manuscript preparation. J.B. was indebted to the European Union-Human Resources and Mobility for an Early Stage Training (Grant MEST-CT-2004-514632) for a predoctoral fellowship.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13037
References
- 1.Krylov DM, Nasmyth K, Koonin EV. Evolution of eukaryotic cell cycle regulation: stepwise addition of regulatory kinases and late advent of the CDKs. Curr Biol. 2003;13:173–177. doi: 10.1016/s0960-9822(03)00008-3. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 3.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, et al. Mechanism of CDK activation revealed by the structure of a cyclinA—CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 4.Dewitte W, Murray JAH. The plant cell cycle. Annu Rev Plant Biol. 2003;54:235–264. doi: 10.1146/annurev.arplant.54.031902.134836. [DOI] [PubMed] [Google Scholar]
- 5.Peeper DS, Parker LL, Ewen ME, Toebes M, Hall FL, Xu M, et al. A- and B-type cyclins differentially modulate substrate specificity of cyclin—cdk complexes. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin W., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Robles AI, Martinez LA, Liu F, Gimenez-Conti IB, Conti CJ. Expression of G1 cyclins, cyclin-dependent kinases and cyclin-dependent kinase inhibitors in androgen-induced prostate proliferation in castrated rats. Cell Growth Differ. 1996;7:1571–1578. [PubMed] [Google Scholar]
- 8.Dynlacht BD. Regulation of transcription by proteins that control the cell cycle. Nature. 1997;389:149–152. doi: 10.1038/38225. [DOI] [PubMed] [Google Scholar]
- 9.Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams PD, Li X, Sellers WR, Baker KB, Leng X, Harper JW, et al. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol Cell Biol. 1999;19:1068–1080. doi: 10.1128/mcb.19.2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts JM. Evolving ideas about cyclins. Cell. 1999;98:129–132. doi: 10.1016/s0092-8674(00)81007-7. [DOI] [PubMed] [Google Scholar]
- 12.de Lichtenberg U, Jensen LJ, Brunak S, Bork P. Dynamic complex formation during the yeast cell cycle. Science. 2005;307:724–727. doi: 10.1126/science.1105103. [DOI] [PubMed] [Google Scholar]
- 13.Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, et al. Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell. 2010;22:1264–1280. doi: 10.1105/tpc.109.073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakenfull EA, Riou-Khamlichi C, Murray JAH. Plant D-type cyclins and the control of G1 progression. Phil Trans R Soc Lond B. 2002;357:749–760. doi: 10.1098/rstb.2002.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC. G1 to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol. 2002;5:480–486. doi: 10.1016/s1369-5266(02)00301-1. [DOI] [PubMed] [Google Scholar]
- 16.Rossi V, Varotto S. Insights into the G1/S transition in plants. Planta. 2002;215:345–356. doi: 10.1007/s00425-002-0780-y. [DOI] [PubMed] [Google Scholar]
- 17.Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–515. doi: 10.1242/jcs.115.3.507. [DOI] [PubMed] [Google Scholar]
- 18.Gisselsson D. Classification of chromosome segregation errors in cancer. Chromosoma. 2008;117:511–519. doi: 10.1007/s00412-008-0169-1. [DOI] [PubMed] [Google Scholar]
- 19.Kono A, Umeda-Hara C, Adachi S, Nagata N, Konomi M, Nakagawa T, et al. The Arabidopsis D-type cyclin CYCD4 controls cell division in the stomatal lineage of the hypocotyl epidermis. Plant Cell. 2007;19:1265–1277. doi: 10.1105/tpc.106.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dello Ioio R, Scaglia Linhares F, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 21.Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, et al. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melaragno JE, Mehrotra B, Coleman AW. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folkers U, Berger J, Hülskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124:3779–3786. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- 24.Nieuwland J, Maughan S, Dewitte W, Scofield S, Sanz L, Murray JAH. The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proc Natl Acad Sci USA. 2009;106:22528–22533. doi: 10.1073/pnas.0906354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewitte W, Riou-Khamlichi C, Scofield S, Healy JMS, Jacqmard A, Kilby NJ, et al. Altered cell cycle distribution, hyperplasia and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, et al. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]