Abstract
In a recent publication we analyzed the global effects triggered by IAA overproduction in S. meliloti RD64 under free-living conditions by comparing the gene expression pattern of wild type 1021 with that of RD64 and 1021 treated with IAA and other four chemically or functionally related molecules. Among the genes differentially expressed in RD64 and IAA-treated 1021 cells we found two genes of pho operon, phoT and phoC. Based on this finding we examined the mechanisms for mineral P solubilization in RD64 and the potential ability of this strain to improve Medicago growth under P-starved conditions. Here, we further analyze the expression profiles obtained in microarray analysis and evaluate the specificity and the extent of overlap between all treatments. Venn diagrams indicated that IAA- and 2,4-D-regulated genes were closely related. Furthermore, most differentially expressed genes from pSymA were induced in 1021 cells treated with 2,4-D, ICA, IND and Trp as compared to the untreated 1021 cells. RT-PCR analysis was employed to analyze the differential expression patterns of nitrogen fixation genes under free-living and symbiotic conditions. Under symbiotic condition, the relative expression levels of nif and fix genes were significantly induced in Mt- RD64 plants and in Mt-1021 plants treated with IAA and 2,4-D whereas they were unchanged or repressed in Mt-1021 plants treated with the other selected compounds when compared to the untreated Mt-1021 plants.
Key words: 2,4-D; IAA; Medicago truncatula; nitrogen-fixation; Sinorhizobium meliloti
We have previously shown that IAA triggered the upregulation of a central backbone of metabolism such as TCA cycle and the accumulation of PHB granules in free-living Rhizobium.1 Under symbiotic conditions increased acetylene reduction and plant or seed dry weight production were observed for plants nodulated by IAA-overproducing strains.1,2 More recently we showed that IAA led to an improvement of stress responses both in free-living and symbiotic conditions. It is known that plants develop a plethora of physiological, developmental and biochemical changes to deal with environmental stress conditions.3–6 These changes require the activation of biochemical pathways that probably act additively and synergistically and depend largely on efficient nitrogen fixation in the root nodules, a sensitive target for abiotic stresses.7,8 In this addendum, we comment our recent published data and report that Mt-RD64 plants exhibited enhanced expression of nitrogen fixation genes. The treatment of Mt-1021 plants with exogenous IAA led to similar upregulation. We speculate that this positive alteration might be of agronomic advantage: it could improve the adaptation of these plants to stressful environments as we have found for the salt-stress and P-starvation.9,14
Changes in Gene Expression Patterns Under Free-Living Condition
Microarray analysis was previously performed to compare the transcript profile of wild-type S. meliloti 1021 with that of RD64 and 1021 treated with IAA and other four chemically or functionally related molecules such as indole (Ind), tryptophan (Trp), indole-3-carboxylic acid (ICA) and 2,4-dichlorophenoxyacetic acid (2,4-D).9 In order to derive useful biological knowledge from the datasets obtained in this analysis a more detailed classification of genes differentially expressed was performed.
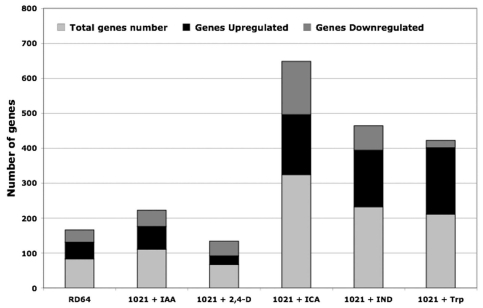
The transcripts of 324, 232 and 211 genes were significantly up- or down-regulated after ICA-, IND- and Trp-treatment, respectively, whereas 112 and 67 genes were differentially expressed in 1021 cells treated with IAA and 2,4-D, respectively (Fig. 1). The number of genes regulated in RD64 cells (126) was closer to that found for the latter samples. This finding suggests that a common response mechanism is activated after IAA- and 2,4-D-treatment and that this mechanism differs from that induced by ICA-, IND- and Trp-treatment.
Figure 1.
Number of genes significantly up or downregulated in RD64 and in 1021 cells treated for 3 hours with 0.5 mM Ind, Trp, ICA, 2,4-D or IAA than in untreated 1021 cells. Numbers shown are for genes where p ≤ 0.05 and M ≥ 0.7 (upregulated) or M ≤ 0.7 (downregulated). The M value refers to the log2 of the ratio of intensities of each spots in the two channels. Microarray analysis is carried out as previously reported.7
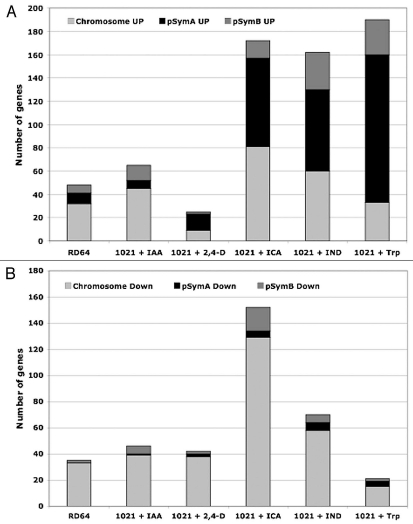
The evaluation of replicon location showed that the upregulated genes had a plasmid location for 1021 + 2,4-D, 1021 + ICA, 1021 + IND and 1021 + Trp (Fig. 2A), since more than 50% of all predicted protein-encoding genes were located on the megaplasmid pSymA, which is known to contain many genes specifically involved in symbiosis.10 Indeed, among the genes significantly upregulated in these cells many nif and fix genes were found.9 For 1021 + IAA and RD64 cells the upregulated genes were mainly mapped on chromosome (Fig. 2A). On the other hand, the majority of downregulated genes were located on the chromosome for all samples (Fig. 2B).
Figure 2.
Replicon assignment of genes induced (A) or repressed (B) in RD 64 and in 1021 cells treated for 3 hours with 0.5 mM Ind, Trp, ICA, 2,4-D or IAA than in untreated 1021 cells.
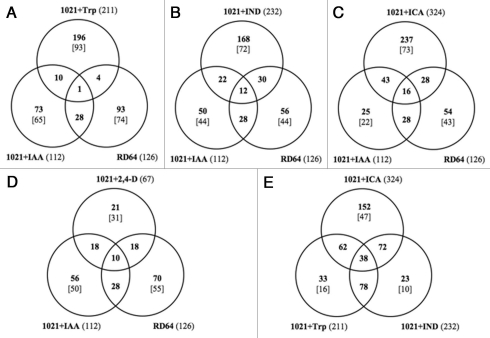
To examine the specificity and the overlapping of the treatments examined we compared the expression profiles of 1021 + IAA and RD64 cells with those of 1021 + 2,4-D, 1021 + ICA, 1021+IND and 1021 + Trp cells by using Venn diagrams and classified the genes up or downregulated into groups (Fig. 3).11 This analysis led to identification of 38 genes that were differentially expressed in both 1021 + IAA and RD64 cells (Fig. 3D). Moreover, 69% of 2,4-D-regulated genes were also regulated in these cells, thus indicating that similar response mechanisms were activated after treatment with these two auxin molecules (Fig. 3D). When similar comparisons were made with 1021+ICA (Fig. 3C), 1021 + IND (Fig. 3B) and 1021+Trp (Fig. 3A) we found that a high percentage of genes were differentially expressed solely in these cells. This result was particularly evident in 1021 + Trp cells for which only 1% of genes were also expressed in 1021 + IAA and RD64 cells (Fig. 3A). By contrast, the analysis of differences and cross-talk of gene expression among 1021 + ICA, 1021 + IND and 1021 + Trp cells revealed that the percentage of genes solely regulated in each treatments was very low, with 90% of genes expressed in 1021 + IND cells also expressed in 1021 + ICA and 1021 + Trp cells (Fig. 3E), suggesting a common regulatory mechanism involved in the response of 1021 cells to chemically related molecules such as ICA, IND and Trp. These results also indicated the existence of greater cross-talk between IAA- and 2,4-D-response processes than between IAA- and ICA-, IND- or Trp-response processes.
Figure 3.
Venn diagram showing the classification of s. meliloti genes significantly up or down regulated in RD64 and in 1021 cells treated for three hours with 0.5 mM Ind, Trp, ICA, 2,4-D or IAA than in untreated 1021 cells on the basis of microarray analyses. Numbers in parentheses represent the total genes with p ≤ 0.05 and M ≥ 0.7 (upregulated) or M ≤ 0.7 (downregulated). The M value description is as reported in Figure 1. In total, there were 112 IAA-regulated genes, 126 RD64-regulated genes, 67 2,4-D-regulated genes, 211 Trp-regulated genes, 232 IND-regulated genes and 324 ICA-regulated genes. Numbers in bracket represent the percentages of genes differentially expressed exclusively in each samples.
Quantitative Real-Time PCR Analysis Under Free-Living and Symbiotic Conditions
Microarrays results indicated that under free-living conditions the majority of genes differentially regulated in 1021 + 2,4-D, 1021 + ICA, 1021 + IND and 1021 + Trp were located on pSymA, which contains genes necessary for nodulation and nitrogen fixation. Therefore, RT-PCR analysis was employed to evaluate the relative expression levels of two nif genes (nifA and nifK) and two fix genes (fixL and fixK2) under free-living conditions and in symbiosis with Medicago plants.12
Under free-living conditions, the expression of nifA, fixL and fixK2 genes was induced in 1021 + 2,4-D, 1021 + ICA, 1021 + IND and 1021 + Trp cells when compared to untreated 1021 cells (Table 1). The highest induction level was observed for nifA gene, the specific activator of nitrogen fixation genes.13 These findings were consistent with the upregulation of genes located on pSymA and involved in nodulation and nitrogen fixation observed in microarray analysis.7 For 1021 + IAA and RD64 cells nifK and fixK2 genes were downregulated, whereas nifA and fixL genes were induced or unaffected.
Table 1.
Quantitative RT-PCR analysis of nif e fix genes in free-living S. meliloti cells
| Sample | Relative levela | |||
| nifA | nifK | fixL | fixK2 | |
| RD64 | 2.6 ± 0.3 | 0.43 ± 0 | 2.3 ± 0.2 | 0.27 ± 0.03 |
| 1021 + IAA | 2.3 ± 0.3 | 0.53 ± 0.06 | 0.93 ± 0.1 | 0.45 ± 0.07 |
| 1021 + 2,4-D | 22.5 ± 0.6 | 0.81 ± 0.05 | 1.60 ± 0.1 | 5.7 ± 0.7 |
| 1021 + IND | 25.6 ± 1.3 | 0.29 ± 0.3 | 1.10 ± 0.05 | 3.0 ± 0 |
| 1021 + Trp | 8.3 ± 0.7 | 0.35 ± 0.03 | 1.4 ± 0.1 | 3.7 ± 0 |
| 1021 + ICA | 37.0 ± 4.0 | 0.85 ± 0.07 | 2.1 ± 0.2 | 4.4 ± 0.2 |
Relative gene expression levels from comparative CT method. The relative expression level was >1 for genes more highly expressed in RD 64 cells and in 1021 cells treated for 3 h with 0.5 mM Ind, Trp, ICA, 2,4-D or IAA than in untreated 1021 cells. The values reported in the Table are the means ± SD of at least three biological replicates conducted at different times. All averages differ significantly according to the Tukey's test (p < 0.01). Total RNA isolation, cDNA synthesis and quantitative PCR were performed as previously described.9 Specific primer pairs, designed using the Primer3 software, are shown. nifA: 5′-CCT TGC AAG AGC ATT CCT TC-3′ and 5′-TCT TTG ACC TGG CGA GAG TT-3′; nifK: 5′-GGA GGT CAT TGG TGA CGA CT-3′ and 5′-TTG ATC GAG CCA GGG TTT AC-3′; fixL: 5′-AAA AGC GCA TCA TCG GTA TC-3′ and 5′-TTT CGC CCA TCT CAT TTA GG-3′; fixK2: 5′-AAG CCA AAC CAC AGT CCA TC-3′ and 5′-CAT CTG AAA GGA GGC GGT AG-3′.
During symbiosis the expression levels of these genes were completely different except for Mt-1021 plants treated with 2,4-D (Table 2). All the selected genes were upregulated in Mt-RD64 plants and in Mt-1021 plants treated with IAA and 2,4-D as compared to the untreated Mt-1021 plants. By contrast, for Mt-1021 plants treated with ICA, IND and Trp the expression of nifA, nifK and fixL genes was unaffected, whereas fixK2 gene was downregulated as compared to the untreated Mt-1021 plants.
Table 2.
Quantitative RT-PCR analysis of nif e fix genes in S. meliloti cells during symbiosis
| Sample | Relative levela | |||
| nifA | nifK | fixL | fixK2 | |
| Mt-RD64 | 1.9 ± 0.2 | 2.3 ± 0.2 | 3.2 ± 0 | 2.3 ± 0.3 |
| Mt-1021 + IAA | 2.1 ± 0.2 | 2.8 ± 0.4 | 3.0 ± 0.2 | 1.5 ± 0 |
| Mt-1021 + 2,4-D | 1.9 ± 0.3 | 2.0 ± 0.2 | 2.7 ± 0.1 | 1.6 ± 0.1 |
| Mt-1021 + IND | 1.2 ± 0.2 | 1.0 ± 0.3 | 1.2 ± 0.2 | 0.66 ± 0 |
| Mt-1021 + Trp | 1.2 ± 0.2 | 1.1 ± 0.2 | 0.93 ± 0.09 | 0.66 ± 0.06 |
| Mt-1021 + ICA | 0.85 ± 0.05 | 1.0 ± 0.2 | 1.6 ± 0.2 | 0.47 ± 0.05 |
Relative gene expression levels from comparative CT method. The relative expression level was >1 for genes more highly expressed in nodules of Mt-RD64 plants or in nodules of Mt-1021 plants treated for 4 hours with 0.5 mM Ind, Trp, ICA, 2,4-D or IAA than in untreated Mt-1021 plants. The values reported in the table are the means ± SD of at least three biological replicates conducted at different times. All averages differ significantly according to the Tukey's test (p < 0.01). Experimental procedures were as described in Table 1.
Final Remarks
Our previous investigations provided evidence that overexpression of IAA in S. meliloti 1021 played a positive role in the adaptation to different environmental conditions, including P starvation, both in free-living bacteria and in nodulated Medicago plants.7,14 We also reported that plants nodulated by the IAA overproducing strain exhibited improved nitrogen-fixing ability, both under normal and salt stress conditions.12 Here we suggest that this finding was connected to the upregulation of genes that control nitrogen fixation such as nifA, fixL and fixK2 (Table 2). We also speculate that the effects observed under symbiotic conditions were specifically due to the hormonal activity of IAA, since the synthetic auxin 2,4-D led to similar effects that were not revealed in the case of structurally related molecules.
Acknowledgements
We thank A. Sollo and C. Lepore for technical assistance. This work was supported by the CNR Department Molecular Design project: “Sviluppo delle esportazioni di prodotti agroalimentari del Mezzogiorno” supported by MIUR, program MIUR-CNR, and by the European Union, INCO-DEV SONGLINES grant, project ICA4-CT-2001-10059 and by the project SALVE for Biodiversity, PSR Campania 2007–2013.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13068
References
- 1.Imperlini E, Bianco C, Lonardo E, Camerini S, Cermola M, Moschetti G, et al. Effect of indole-3-acetic acid on Sinorhizobium meliloti survival and on symbiotic nitrogen fixation and stem dry weight production. Appl Microbiol Biotechnol. 2009;83:727–738. doi: 10.1007/s00253-009-1974-z. [DOI] [PubMed] [Google Scholar]
- 2.Camerini S, Senatore B, Lonardo E, Imperlini E, Bianco C, Moschetti G, et al. Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch Microbiol. 2008;190:67–77. doi: 10.1007/s00203-008-0365-7. [DOI] [PubMed] [Google Scholar]
- 3.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 5.Tesfaye M, Liu J, Allan DL, Vance CP. Genomic and genetic control of phosphate stress in legumes. Plant Physiol. 2007;144:594–603. doi: 10.1104/pp.107.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J. The transcriptional control of plant responses to phosphate limitation. J Exp Bot. 2004;55:285–293. doi: 10.1093/jxb/erh009. [DOI] [PubMed] [Google Scholar]
- 7.Zahran HH. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev. 1999;63:968–989. doi: 10.1128/mmbr.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clement M, Lambert A, Herouart D, Boncompagni E. Identification of new upregulated genes under drought stress in soybean nodules. Gene. 2008;426:15–22. doi: 10.1016/j.gene.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Bianco C, Defez R. Improvement of phosphate solubilization and Medicago plant yield by an indole-3-acetic acid-overproducing strani of Sinorhizobium meliloti. Appl Env Microbiol. 2010;76:4626–4632. doi: 10.1128/AEM.02756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, Barloy-Hubler F, et al. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc Natl Acad Sci USA. 2001;98:9883–9888. doi: 10.1073/pnas.161294798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kestler HA, Muller A, Gress TM, Buchholz M. Generalized Venn diagrams: a new method of visualizing complex genetic set relations. Bioinformatics. 2005;21:1592–1595. doi: 10.1093/bioinformatics/bti169. [DOI] [PubMed] [Google Scholar]
- 12.Fisher HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 14.Bianco C, Defez R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid over-producing Sinorhizobium meliloti strain. J Exp Bot. 2009;60:3097–3107. doi: 10.1093/jxb/erp140. [DOI] [PubMed] [Google Scholar]