Abstract
Our recent study revealed the involvement of the Arabidopsis histone deacetylase HDA6 in modulating ABA and salt stress responses. In this report, we further investigated the role of HDA19 in ABA and salt stress responses. The Arabidopsis HDA19 T-DNA insertion mutant, hda19-1, displayed a phenotype that was hypersensitive to ABA and salt stress. Compared with wild-type plants, the expression of ABA responsive genes, ABI1, ABI2, KAT1, KAT2 and RD29B, was decreased in hda19-1 plants when treated with ABA. Our study indicates that HDA6 and HDA19 may play a redundant role in modulating seed germination and salt stress response, as well as ABA- and salt stress-induced gene expression in Arabidopsis.
Key words: histone deacetylases, HDA6, HDA19, abscisic acid, salt stress
In eukaryotic cells, gene activity is controlled not only by DNA sequences but also by epigenetic marks that can be transmitted to a cell progeny during mitosis or meiosis.1 Epigenetic changes involve modification of DNA activity by methylation, histone modification or chromatin remodelling without alteration of the nucleotide sequence. Among the multiple epigenetic mechanisms that modify chromatin to regulate gene expression, histone modifications including acetylation play a major role. Histone acetylation levels are determined by the action of histone acetyltransferases and histone deacetylases (HDAs). The involvement of HDAs in plant responses to environmental stresses has recently been reported. Thus, mutations in HDA6, an Arabidopsis RPD3-type HDA encoding gene, affect jasmonate response and senescence.2 Another Arabidopsis RPD3-type HDA, HDA19, has also been implicated in plant responses to adverse environmental conditions.3,4 Additional results have confirmed that histone acetylation has a decisive function in regulating plant responses to abiotic stresses such as low temperatures, drought and salt stress.5–8 In addition, Arabdiopsis HDA19 has been implicated to interact with the transcription factor AtERF7 to regulate gene expression in response to abiotic stresses.9
In our recent study, we investigated the effects of abscisic acid (ABA) and salt stress on the histone acetylation and methylation of abiotic stress response genes.8 We found that both ABA and salt stress can induce histone H3K9K14 acetylation and H3K4 trimethylation but decrease H3K9 dimethylation of some ABA and abiotic stress responsive genes. In addition, we found that the HDA6 mutant axe1-5 and HDA6 RNA-interference (HDA6-RNAi) plants displayed a phenotype that is more sensitive to ABA and salt stress. Compared with wild-type plants, the expression of ABA and abiotic stress responsive genes was decreased in axe1-5 and HDA6-RNAi plants. In this report, we further investigated the role of HDA19 in ABA and salt stress response. Our study indicates that HDA6 and HDA19 may play a redundant role in modulating seed germination and salt stress response, as well as ABA- and salt stress-induced gene expression in Arabidopsis.
hda19-1 Plants were Hypersensitive to ABA and NaCl
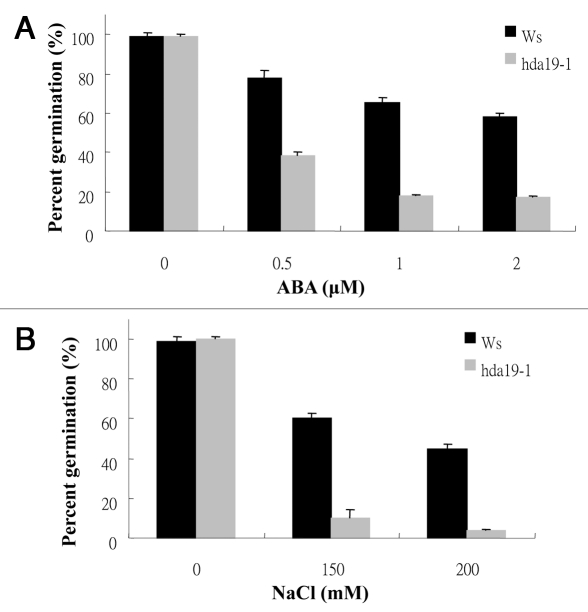
To investigate the function of HDA19 in ABA and abiotic stress response, the sensitivity of hda19-1 mutants to ABA and salt was analyzed. hda19-1 (also called hd1-1) is a T-DNA insertion mutant which carries a T-DNA inserted in the second exon of HDA19 and it is in Ws background.10 As shown in Figure 1A, hda19-1 seeds displayed hypersensitivity to ABA compared with wild-type. At 2 µM ABA, the percent germination of wild-type seeds was 60%, whereas the percent germination of hda19-1 seeds was about 20% only. When treated with NaCl, it was found that hda19-1 seeds also displayed lower percent germination compared with wild-type (Fig. 1B). At 200 mM NaCl, the percent germination of wild-type seeds was about 50%, whereas the percent germination of hda19-1 seeds was about 5% only.
Figure 1.
Seed germination rates of hda19-1 mutants treated with ABA and NaCl. (A) Germination rates of two-day-old Ws wild-type and hda19-1 seeds treated with ABA. (B) Germination rates of two-day-old Ws wild-type and hda19-1 seeds treated with NaCl. Each data point represents the mean of a single experiment performed in triplicate (n ≥ 100).
hda19-1 Plants Displayed Decreased Expression of ABA and Abiotic Stress Responsive Genes
The expression of ABA responsive genes, such as ABA INSENSITIVE 1 (ABI1), ABI2, 3-KETO-ACYL-COA THIOLASE 1 (KAT1), KAT2 and RESPONSIVE TO DESSICATION 29B (RD29B) was determined in hda19-1 plants. Without ABA treatment, hda19-1 and wild-types displayed similar expression patterns of ABI1, ABI2, RD29B, KAT1 and KAT2 (Fig. 2). With ABA treatment, hda19-1 displayed a lower expression of ABA-responsive genes compared with wild-type, suggesting that HDA19 is required for full induction of ABA-responsive genes. hda19-1 mutants displayed a phenotype that is similar to had6 mutants in ABA and salt response as well as gene expression.8
Figure 2.
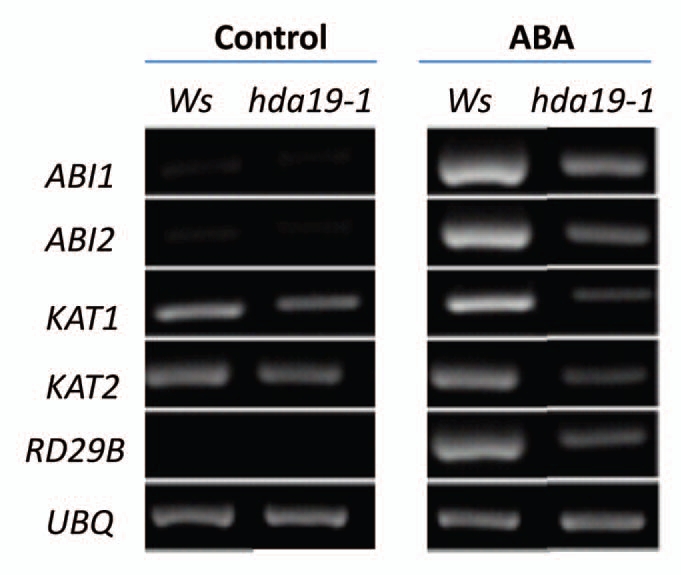
RT-PCR analysis of ABA responsive gene expression in hda19-1 mutants. Two-week-old Ws wild-type and hda19-1 plants were treated with 100 µM of ABA for 3 h. Total RNA for RT-PCR analysis was isolated from leaf tissues. Ubiquitin (UBQ) was shown as an internal control.
A Mode of HDA6 and HDA19 Function in the Regulation of Gene Expression in ABA and Abiotic Stress Response
A recent study suggested that HDA19 may act in a protein complex of AtERF7, a transcription repressor,9 to regulate abiotic stress response genes. AtERF7 is an EAR-motif-containing transcriptional repressor that is involved in regulating ABA and drought stress responses in Arabidopsis. It was found that the AtERF7 interacts with the Arabidopsis homologue of a human global corepressor, AtSin3,11 which in turn may interact with HDA19. It was proposed that AtERF7, AtSin3 and HDA19 can form a transcriptional repressor complex to regulate ABA and drought response in Arabidopsis.9,12 Reduced expression of AtERF7 and AtSin3 by RNA-interference caused ABA hypersensitivity including root development repression and delayed germination, a phenotype similar to hda6 and hda19 mutants. The similarity in the ABA and salt response as well as gene expression among hda6 and hda19 mutants suggests that HDA6 and HDA19 may play a redundant role in ABA and abiotic stress response. In addition to AtERF7, another EAR-motif-containing transcriptional repressor, AtERF4, was also found to be involved in regulating ABA and abiotic stress responses in Arabidopsis.13 HDA6 and HDA19 may therefore form a similar transcriptional complex with AtERF4 and AtERF7 to regulate gene expression in Arabidopsis (Fig. 3). AtERF4 and AtERF7 may repress gene expression through chromatin modification mediated by histone deacetylation. The activity of AtERF4 and AtERF7 can also be affected by the protein kinase PKS3.9
Figure 3.
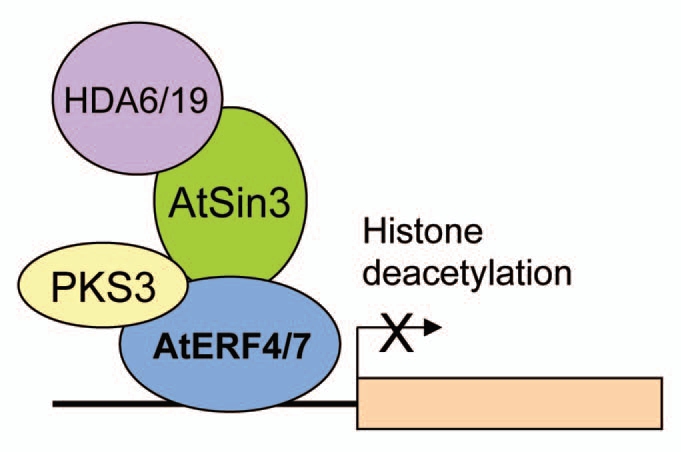
A mode of HDA 6 and HDA 19 function in the regulation of gene expression in ABA and abiotic stress response. Arabidopsis AtERF4 and AtERF7 bind to the promoters of the target genes and recruits AtSin3, a co-repressor, which in turn may interact with histone deacetylases, HDA6 and HDA19. AtERF4 and AtERF7 may therefore repress gene expression through chromatin modification mediated by histone deacetylation. The activity of AtERF4 and AtERF7 can also be affected by the protein kinase PKS3.
Acknowledgements
This work is supported by grants from National Science Council of Taiwan (96-2311-B-002-013-MY2, 97-2311-B-002-004-MY3 and 99-2321-B-002-027-MY3) and National Taiwan University (97R0066-36).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/13168
References
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 3.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benhamed M, Bertrand C, Servet C, Zhou DX. Arabidopsis GCN5, HD1 and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell. 2006;18:2893–2903. doi: 10.1105/tpc.106.043489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, et al. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Jeong JC, Zhu Y, Sokolchik I, Miyazaki S, Zhu JK, et al. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc Natl Acad Sci USA. 2008;105:4945–4950. doi: 10.1073/pnas.0801029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Luo M, Wang Y, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic Acid and drought stress responses. Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian L, Wang J, Fong MP, Chen M, Cao H, Gelvin SB, et al. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics. 2003;165:399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 12.Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]