Abstract
Mitomycins are quinone-containing antibiotics, widely used as anti-tumor drugs in chemotherapy. Mitomycin-7-O-methyltransferase (MmcR), a key tailoring enzyme involved in the biosynthesis of mitomycin in Streptomyces lavendulae, catalyzes the 7-O-methylation of both C9β- and C9α-configured 7-hydroxymitomycins. We have determined the crystal structures of the MmcR–S-adenosylhomocysteine (SAH) binary complex and MmcR–SAH-Mitomycin A (MMA) ternary complex at resolutions of 1.9 and 2.3 Å, respectively. The study revealed MmcR to adopt a common SAM-dependent O-MTase fold and the presence of a structurally-conserved active site general acid-base pair is consistent with a proton assisted methyltransfer common to most methyltransferases. Given the importance of C7 alkylation to modulate mitomycin redox potential this study may also present a template toward the future engineering of catalysts to generate uniquely bioactive mitomycins.
Keywords: methyltransferase, natural product, biosynthesis, S-adenosyl-L-methionine, cancer, X-ray crystallography
Introduction
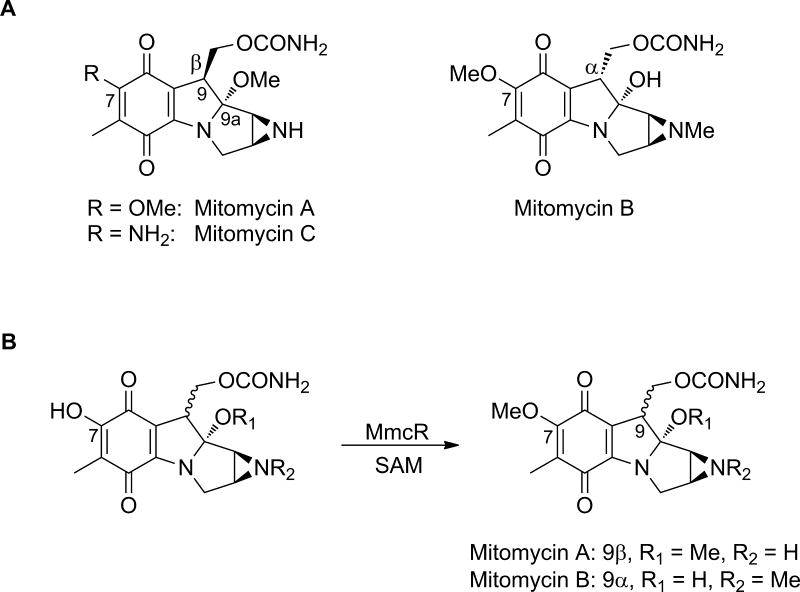
Originally discovered over six decades ago,1–3 the natural product mitomycin C (MMC, Fig. 1A) remains part of the conventional anticancer regimen for solid tumors given its bias toward hypoxia-induced cytotoxicity.4–7 Numerous bioreductants have been implicated in MMC activation via one- or two-electron reduction of the quinone, including NADPH/cytochrome P450 reductase, DT-diaphorase and/or glutathione. Under oxygen-limiting conditions, one-electron reduction predominates, producing a highly reactive quinone methide that alkylates DNA.4,5 However, certain MMC activating agents operate under both anaerobic and aerobic conditions (e.g., DT-diaphorase) and can thereby compromise the hypoxic selectivity of quinone-alkylating agents such as MMC.6 The reactivity, and corresponding cytotoxic selectivity, correlates in part to the redox potential of the quinone ring which, in turn, can be modulated via appended functional groups. Modifications at the C6 and C7 positions, achieved through derivatization of naturally occurring mitomycins, provide further support for this notion.4 Thus, understanding the structural basis for enzymes involved in mitomycin C6 or C7 alkylation may enable the exploitation of these catalysts toward the generation of new and improved mitomycin analogs.
Fig. 1.
(A). Naturally-occurring mitomycins. (B). The reaction catalyzed by MmcR.
Metabolic labeling studies revealed D-glucosamine and 3-amino-5-hydroxybenzoic acid (AHBA) to be the biosynthetic precursors of the mitosane skeleton, the C-10 carbamoyl group to derive from L-arginine or L-citrulline, and the N- and O-methyl moieties to originate from L-methionine.8–11 Based upon annotation of the mitomycin biosynthetic gene cluster from Streptomyces lavendulae NRRL 2564 (producer of mitomycins A-C, Fig. 1A),12 there exist three S-adenosyl-L-methionine (SAM)-dependent methyltransferase-encoding genes in the mitomycin gene cluster.13 Of the corresponding proteins, MmcR shows high sequence similarity to natural product phenolic methyltransferases (MTases) such as bacterial MTases involved in the biosynthesis of calicheamicin, dynemicin, C-1027, fredericamycin,, oxytetracycline, puromycin and plant MTases involved in the biosynthesis of flavonoids. Recent MmcR in vivo and in vitro studies have revealed MmcR to catalyze the efficient methylation of 7-demethylmitomycin A and 7-desmethylmitomycin B (Fig. 1B) over a broader pH range in a metal-independent manner.14 While MmcR was demonstrated to tolerate both C9α- or C9β-configurations in vitro, the enzyme displays a strong preference for C9β-isomer.14 In an effort to further our understanding of the mechanism of MmcR catalysis and the key structural elements for SAM recognition and activation, herein we report the crystal structures of the MmcR–S-adenosylhomocysteine (SAH) binary complex and MmcR–SAH-Mitomycin A (MMA) ternary complex at resolutions of 1.9 and 2.3 Å, respectively. This work reveals MmcR to crystallize as a dimer and adopt a fairly typical MTase structural fold. Given the ability of structurally homologous DNA- and natural product-MTases to utilize non-natural cofactor analogs of SAM to achieve differential alkylation,15–22 this study supports the notion that MmcR (in conjunction with SAM surrogates) may also present an opportunity to further modulate the redox potential of mitomycin.
Materials and Methods
Mitomycin A synthesis
Mitomycin C (109 mg, 0.326 mmol) was suspended in an aqueous solution of NaOH (0.05 M, 8 mL) under Ar at room temperature and was excluded from light. After stirring for 16 h, the sodium mitosane salt (Rf = 0.08, 10% MeOH in CH2Cl2) was cooled to 0 °C and acidified to a pH of 3.9 with 0.1 M H2SO4. The aqueous solution was extracted with EtOAc (4 × 10 mL) then dried with Na2SO4. Evaporation of the solvent yielded the hydroxymitosane as a purple solid (95 mg). Methylation was performed by first dissolving the solid in THF (20 mL) and cooling to 0 °C then slowly adding an ethereal solution of diazomethane (20 mL; prepared by adding 250 mg of N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) to 20 mL of 40% aqueous KOH and 20 mL Et2O). The reaction proceeded for 10 min at 0 °C and was quenched with HOAc (250 μL). After removing the solvent, the residue was purified by column chromatography (SiO2, 5% MeOH in CH2Cl2) providing mitomycin A as an amorphous solid (43 mg, 43%, TLC Rf = 0.51 10% MeOH in CH2Cl2). 1H NMR (acetone-d6, 500 MHz) δ 5.94 (br s, 2 H), 4.72 (dd, J = 10.6, 4.4 Hz, 1 H), 4.30 (t, J = 10.6 Hz, 1 H), 4.02 (s, 3 H), 3.93 (d, J = 12.5 Hz, 1 H), 3.51 (dd, J = 11.0, 4.4 Hz, 1 H), 3.42 (d, J = 12.1 Hz, 1 H), 3.21 (s, 3 H), 2.90 (br s, 1 H), 2.84 (br s, 1 H), 1.79 (s, 3 H); 13C NMR (acetone-d6, 125 MHz) δ 184.3, 179.7, 159.3, 158.4, 153.5, 125.3, 116.0, 108.3, 63.5, 62.5, 51.8, 50.8, 45.9, 38.2, 34.3, 9.2; HRMS (ESI) m/z C16H19N3NaO6 ([M + Na]+) 372.1171, calculated 372.1166.
Protein expression and purification
Recombinant N-His6-MmcR (herein referred to simply as MmcR) was produced in E. coli and affinity purified as previously described.14 The purified enzyme in 20 mM Tris, pH 8, 50mM NaCl was subsequently concentrated to 20 mg/mL, drop frozen in liquid nitrogen, and stored at −80 °C. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA) using BSA as a standard. For the production of selenomethionine (SeMet)-labeled protein, the E. coli methionine auxotroph strain B834 (DE3) was transformed with plasmid N-His6-mmcR14 and the production of the desired (SeMet)-labeled recombinant protein accomplished using auto-induction media.23
Protein crystallization
Crystals of MmcR with SAH were grown by vapor diffusion in hanging drops containing a 1:1 mixture of 1 μl protein and 1 μl crystallization buffer (10% (w/v) polyethylene glycol (PEG) 4000, 15% 2-methyl-2,4-pentadiol (MPD), 100 mM CaCl2, and 100 mM MES (pH 6.0) at 4 °C). Crystals of MmcR in complex with MMA were obtained by soaking the MmcR-SAH crystals in the crystallization solution with 5 mM MMA for 8 hr. Diffraction data were collected from a single crystal that was soaked in crystallization solution containing 25% ethylene glycol, mounted in a cryoloop and flash frozen in a stream of liquid nitrogen.
MmcR structure determination
All diffraction data for SeMet-labeled MmcR-SAH complexes were collected at the General Medicine and Cancer Institute Collaborative Access Team (GM/CA-CAT) 23-ID-D beamline at the Advanced Photon Source of Argonne National Laboratory. Each of the 180 diffraction images for the SeMet-labeled crystals were collected at a crystal-to-detector distance of 270 mm, exposed for 5 s, in a single pass with 1 deg oscillation per frame. Data sets for the MmcR-SAH complex were collected at two wavelengths, 0.97945 and 0.96421 Å. The diffraction images were indexed, integrated and scaled using HKL2000. 24 Diffraction data for SeMet-labeled MmcR-SAH-MMA complexes were collected at the Life Science Collaborative Access Team (LS-CAT) 21-ID-F beamline at the Advanced Photon Source of Argonne National Laboratory. Each of the 120 diffraction images for the SeMet-labeled crystals were collected at the crystal-to-detector distance of 225 mm, exposed for 4 s, in a single pass with 1 deg oscillation per frame and at 0.97872 Å wavelength. The diffraction images were indexed, integrated and scaled using HKL2000. Both complexes crystallized in space group P 21 21 21 with four molecules per asymmetric unit. Unit cell dimensions for MmcR-SAH were a = 88.3 Å, b = 98.9 Å, c = 171.1 Å and for MmcR-SAH-MMA were a = 87.8 Å, b = 98.8 Å, c = 171.0 Å.
Phases for the MmcR-SAH complex were determined using the multiple wavelength anomalous dispersion (MAD) method on SeMet-substituted protein. Initial heavy atom sites were found using the programs PHENIX.HySS25 and SHELXD.26,27 PHENIX.EMMA identified 16 consensus sites. The structure was automatically phased and density modification was performed using autoSHARP.28 Using the initial phase information, a high resolution model of MmcR-SAH was built in ARP/wARP.29 Using the MmcR-SAH structure as a search model, the MmcR-SAH-MMA structure was obtained by molecular replacement using MOLREP.30 The structure was completed via multiple manual interations in COOT31 followed by refinement in REFMAC532 and PHENIX.Refine.33 The quality of all models was assessed using the program MOLPROBITY 34 and PROCHECK.35 The final model of MmcR-SAH contained residues 11–349 in each of the four monomers, 871 waters, 4 molecules of S-adenosyl homocysteine (SAH) and 2-methyl-2,4-pentandiol (MPD). The MmcR-SAH-MMA model contained residues 11-349, 329 water molecules and 4 molecules of SAH and MMA. Topology and parameter files for S-adenosyl homocysteine (SAH) and MPD were obtained from the REFMAC library. Topology and parameter files for MMA were obtained by modifying the MMC structure with SKETCHER.36 All figures are generated using molecular graphics program PyMOL. The binary and ternary complexes have been deposited under accession numbers 3GWZ and 3GXO, respectively.
Results and Discussion
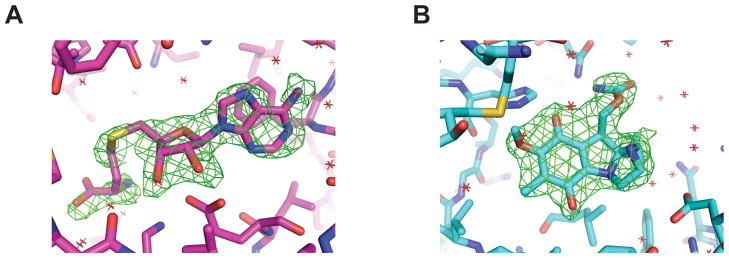
Recombinant MmcR, a 38-kD protein consisting of 349 amino acids, was expressed in Escherichia coli as N-terminal polyhistidine-tagged protein and purified by Ni+2 affinity chromatography. In the MmcR-SAH and MmcR-SAH-MMA binary and ternary complexes, the first 30 residues of chain A, B, C and D (20 residues of which comprised the N-terminal His-tag in each case) were not modeled due to insufficient electron density. The electron density for the rest of the polypeptide chain and the bound ligands is well defined (Fig. 2A and Fig. 2B). The final structure was refined to an Rcryst and Rfree of 17.4% and 21.0% for MmcR-SAH and 20.2% and 25.5% for MmcR-SAH-MMA complex, respectively (Table I).
Fig. 2.
Simulated annealing omit Fo-Fc electron density maps contoured at 3σ. (A) Bound SAH and (B) bound MMA in MmcR-SAH-MMA ternary complex.
Table I.
Crystal parameters, X-ray data collection, phasing, and refinement statistics.
| MmcR-SAH | MmcR-SAH-MitA | ||
|---|---|---|---|
| Crystal parameters | Peak | HRem | |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit-cell parameters (Å, °) | a=88.3, b=98.9, c=171.1 | a=88.5, b = 99.0, c=171.4 | a=87.8, b=98.8, c=171.0 |
| Data collection statistics | |||
| Wavelength (Å) | 0.97945 | 0.96421 | 0.97872 |
| Energy (eV) | 12658.6 | 12858.6 | 12,668 |
| Resolution range (Å) | 50.00–1.91(1.98–1.91) | 100.00-2.00(2.07-2.00) | 50.00–2.30 (2.38–2.30) |
| No. of reflections (measured/unique)a | 220672/115235 | 194218/101748 | 708974/59548 |
| Completeness (%) | 98.6 (87.9) | 98.8(92.2) | 88.8 (70.7) |
| Rmerge b | 0.113 (0.570) | 0.128 (0.903) | 0.092 (0.315) |
| Redundancy | 13.6 (9.2) | 13.3(9.3) | 3.9 (3.3) |
| Mean I/sigma(I) | 11.5 (2.4) | 11.3 (3.9) | |
| Phasing statistics c | |||
| Mean FOM (centric/acentric) | 0.090/0.322 | ||
| Phasing Power (isomorphous/anomalous) | 0.0/0.98 | ||
| Cullis R-factor (isomorphous/anomalous) | 0.0/0.84 | ||
| Refinement and model statistics | |||
| Resolution range | 19.65–1.91 (1.93–1.91) | 43.92–2.30 (2.38–2.30)d | |
| No. of reflections (work/test) | 114971/5763 | 59450/3003 | |
| Rcryst e | 0.174 (0.253) | 0.202 (0.235) | |
| Rfree f | 0.210 (0.333) | 0.255 (0.298) | |
| Rmsd bonds (Å) | 0.006 | 0.007 | |
| Rmsd angles (°) | 1.000 | 1.102 | |
| ESU from Rfree (Å)g | 0.23 | 0.19 | |
| B factor – overall/ waters (Å2)h | 23.4/ 40.4 | 36.5/ 40.9 | |
| No. of protein molecules/all atoms | 4/11428 | 4/10866 | |
| No. of waters | 871 | 329 | |
| Ramachandran Plot by MOLPROBITY (%) | |||
| Favoured regions | 98.7 | 97.6 | |
| Additional allowed regions | 1.2 | 2.4 | |
| Outliers | 0.1 | 0.0 | |
| PBD code | 3GWZ | 3GXO | |
Values in parentheses are for the highest resolution shell.
Rmerge = ΣhΣi|Ii(h) - <I(h)>|/ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
Resolution range for phasing in SHARP was (26.66–3.2) Å.
Resolution range for refinement was cut (38.76–1.90) Å due to low completeness and signal in the remaining resolution shells.
Rcryst = Σh||Fobs|-|Fcalc||/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using ~5.0% of the randomly selected unique reflections that were omitted from structure refinement.
Estimated standard uncertainty based on Rfree.
B-factors from the model refined without TLS.
Overview of the structure
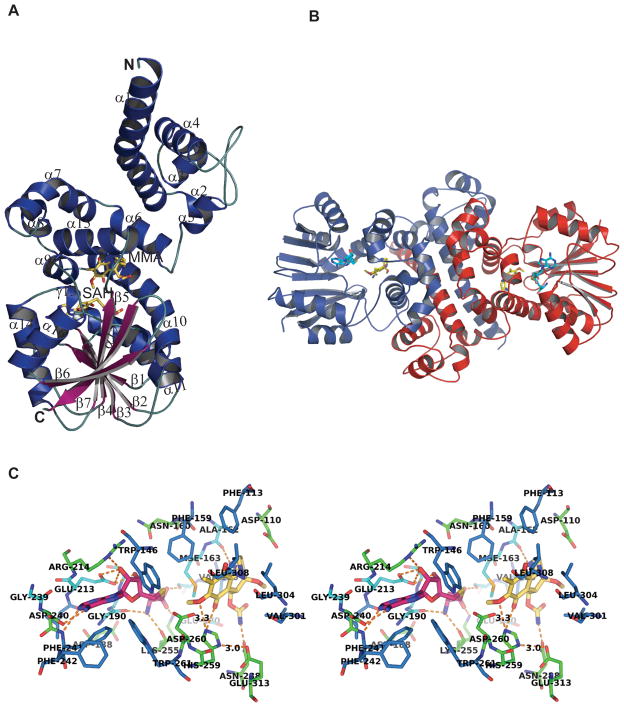
Each subunit of MmcR exhibited a C-terminal catalytic domain and N-terminal dimerization domain. The C-terminal domain contains the conserved DXGXGXG fingerprint needed for cofactor binding. It is folded into a Rossmann fold, comprising a central parallel β-sheet (β1-β7) where all except β7 are parallel to each other, and surrounded by seven alternating α-helices (α10-α16) (Fig. 3A). The sequence of secondary structural elements in this core SAM binding domain is comprised of α10, β1, α11, β2, α12, β3, β4, γ13, α14, β5, α15, α16, β6, and β7. The N-terminal domain involved in dimerization and for substrate binding contains all α-helical secondary structural elements. The secondary elements mainly engaged in dimer formation include the helices, α1, α4, α6, α7 and α15 (Fig. 3B). The interaction area of dimer interface is 3146 Å2 (Fig. 3B) mostly dominated by hydrophobic interactions along with 29 hydrogen bonds and 17 salt bridges. Analysis of crystal packing does not support the formation of a tetramer, as the interaction in the crystal lattice are rather weak. The largest interaction area with an adjacent molecule is only 467 Å2, indicative of crystal contacts rather than a protein-protein interface. This is consistent with biochemical analysis by native sodium dodecyl sulfate/polyacrylamide gel electrophoresis (determined MW 73.2 kDa; calculated MW for the dimer, 75.2 kDa; data not shown). The MmcR-bound product (MMA) is situated at the interface of N- and C-terminal domains (Fig. 3A), and residues from both of these domains are involved in product binding. The fold of MmcR very much resembles previously reported O-MTases such as carminomycin 4-O-methyltransferase (DnrK),37 neocarzinostatin O-methyltransferase (NcsB1),38 chalcone O-methyltransferase (ChOMT),39 isoflavone O-methyltransferase (IOMT),39 calicheamicin O-methyltransferase (CalO1, Chang et. al. unpublished) and caffeic acid/5-hydroxyferulic acid 3/5 O-methyltransferase (COMT).40
Fig. 3.
(A) Overview of the secondary structural elements in the MmcR-SAH-MMA ternary complex (helices are colored blue, strands are colored magenta, ligands SAH and MMA are colored yellow, letters N and C correspond to the N-terminus and C-terminus of the MmcR protein, respectively). (B) Ribbon representation of MmcR-SAH-MMA ternary complex quaternary structure, showing dimer interfaces. In this representation, the two monomers are colored red and blue and ligands SAH and MMA are colored cyan and yellow, respectively. (C) Stereoview of interaction of SAH and MMA in MmcR-SAH-MMA complex. The stick model of SAH and MMA are depicted in magenta and yellow, respectively. The interacting MmcR residues are labeled and illustrated in blue, green and cyan for hydrophobic, charged, and neutral residues, respectively. Polar interactions are depicted by orange hed lines.
The SAM/SAH binding site
In both determined structures, the cofactor product SAH is well defined by electron density (Fig. 2A). MmcR bears a signature motif (DXGXGXG) containing highly conserved residues for the recognition of SAM/SAH and, like all SAM-dependent MTases, contains conserved active site residues to provide a network of hydrogen bonding interactions (Fig. 3C) with the donor and acceptor of the methyl group (see Fig. 4 for sequence alignment with representative homologs). The carboxylate group of SAH forms an electrostatic interaction with side chains of Ser167, Lys255. The terminal amino group participates in a hydrogen bonding network not only in direct interactions with backbone oxygen of Gly190 and Lys255 but also indirectly with backbone nitrogen of Lys255, backbone oxygen of Ile189 and side chain oxygen of Asp188 via water molecules. The adenine ring forms stacking interactions with the side chains of Trp261and Phe241, and Asp240 forms a hydrogen bond to the N6 amino group. The ribose moiety is anchored to MmcR through hydrogen bonds of the O2* and O3* hydroxyl groups to the side chains of Glu213 and Arg214. Lys255 interacts with both the carboxyl group as well as the amino group of SAM/SAH, and the latter also forms a hydrogen bond to the main chain carbonyl oxygen of Gly190. The SAH sulfur atom makes hydrophobic interactions with side chains of Phe159 and Met163. Most of these interactions are globally conserved among the MTase family.
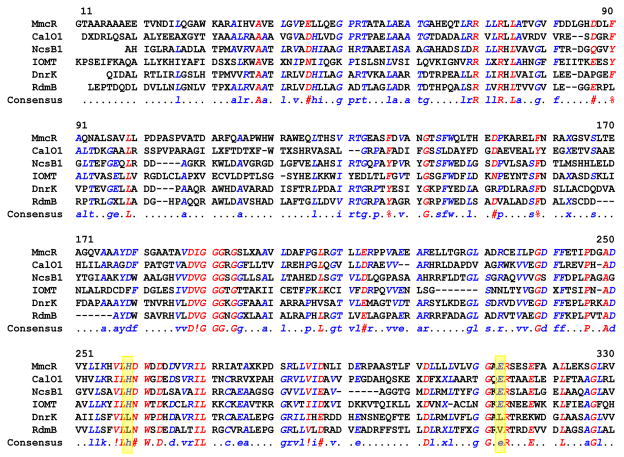
Fig. 4.
Alignment of selected structural homologs of MmcR identified by DALI search: calicheamicin-O-methyltransferase CalO1 (PDB: 3LST); neocarzinostatin-O-methyltransferase NcsB1 (PDB: 3I5U); isoflavone methyltransferase IOMT (PDB: 1FP2); carminomycin-4-O-methyltransferase Dnrk (PDB: 1TW3); aclacinomycin hydroxylase RdmB (PDB:1QZZ)). The alignment was generated by the Multalin-4.1 web-server using default parameters. Residues with sequence consensus above 90% and 50% are colored red and blue, respectively. Yellow boxes highlight the putative catalytic residues.
The substrate binding site
The product, MMA, fits well into the electron density (Fig. 2B) observed at the interface between the N-terminal and C-terminal domain, close to the cofactor binding site (Fig. 3A). The binding of MMA to MmcR is dominated by hydrophobic and van der Waals interactions involving residues Phe113, Phe145, Trp146, Phe159, Ala162, Met163, Val166, Val301, Leu304, Leu305, Leu308, Val309 and Val341 (Fig. 3C). Apart from the van der Waals interactions to the enzyme, MMA is anchored to its binding site also by a few hydrogen bonding interactions. Specifically, MMA N2 forms a hydrogen bond to the main chain oxygen of Ala162, while MMA atoms O7 and O8 interact with the side chain of His259.
Structural comparison of the MmcR-SAH complex and MmcR-SAH-MMA ternary complex
The four subunits of complexes of MmcR-SAH and MmcR-SAH-MMA structures align with a r.m.s. deviation of 0.31 for 8785 structurally corresponding atoms. The dimers of MmcR-SAH and the MmcR-SAH-MMA complex align with r.m.s. deviation of 0.28 for 4237 structurally corresponding atoms. These low values indicate no large-scale changes in tertiary and/or quaternary structure to occur upon substrate binding. The occupancy of the MMA binding site by MPD in MmcR-SAH crystal form (i.e., representing a ‘substrate-bound’ form) may partially explain the high degree of similarity between the MmcR-SAH and the MmcR-SAH-MMA structures. The only notable structural variation occurs within the solvent-exposed loops surrounding the product MMA (residues 100–106, 289–294, 335–342), suggesting subtle loop movements in the context of ligand binding.
Structural homology
A DALI search for MmcR structural homologs in the Protein Data Bank (PDB) database returned several hits. Among those which displayed the highest Z-score were carminomycin 4-O-methyltransferase (DnrK),37 neocarzinostatin O-methyltransferase (NcsB1),38 aclacinomycin hydroxylase (RdmB),41 isoflavone methyltransferase (IOMT)39 and calicheamicin O-methyltransferase (CalO1, Chang et. al. unpublished) with Z-scores 37, 36, 34, 33 and 32 respectively. They share ~25–35% overall sequence identity with MmcR. Highest sequence conservation is found among residues associated with SAM-binding – specifically, the loop that interacts with the homocysteine and ribosyl moieties of the SAH situated between β1 and α11 (which contains the glycine-rich consensus sequence DXGXGXG) and an acidic residue in loop between β3 and β4 (Asp240 in MmcR) which interacts with the exocyclic N6 and ring nitrogen N1 of adenine ring of the cofactor. MmcR gives a r.m.s. deviation of 1.7 Å, 1.9 Å, 2.1 Å, 3.2 Å, and 3.7 Å over 330 Cα, 321 Cα, 323 Cα, 330 Cα and 345 Cα atoms with DnrK, NcsB1, RdmB IOMT, and CalO1 respectively, and like these three MTases, MmcR is dimeric and also shares high structural similarity across the dimerization domain. Similar to most methyltransferases,39 His259 and Glu313 are implicated as general acid/base pair (Fig. 3C and 4) for MmcR catalysis. Specifically, His259 is within range (3.3 Å) (Fig. 3C) to deprotonate the hydroxyl group of 7-desmethyl MMA, while Glu313 is within the hydrogen bonding distance (3.0 Å) (Fig. 3C) of His259 to potentially constrain and orient the catalytic base. Consistent with a putative acid/base-mechanism for MmcR, divalent metals do not influence MmcR activity.14
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants CA84374 and the Laura and Edward Kremers Chair in Natural Products Chemistry (J.S.T.). Analytical support was provided by the School of Pharmacy Analytical Instrumentation Center. GM/CA-CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). LS-CAT has been supported by Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No.W-31-109-ENG-38. We also thank the Center for Eukaryotic Structural Genomics (NIH grant GM074901) for access to crystallographic facilities.
References
- 1.Hata T, Sugawara R. Mitomycin, a new antibiotic from Streptomyces. II. Description of the strain. J Antibiot. 1956;9:147–151. [PubMed] [Google Scholar]
- 2.Hata T, Hoshi T, Kanamori K, Matsumae A, Sano Y, Shima T, Sugawara R. Mitomycin, a new antibiotic from Streptomyces. I. J Antibiot, Series A. 1956;9:141–146. [PubMed] [Google Scholar]
- 3.Wakaki S, Marumo H, Tomioka K, Shimizu G, Kato E, Kamada H, Kudo S, Fujimoto Y. Isolation of new fractions of antitumor mitomycins. Antibiot Chemother. 1958;8:228–240. [PubMed] [Google Scholar]
- 4.Begleiter A. Clinical applications of quinone-containing alkylating agents. Front Biosci. 2000;5:E153–171. doi: 10.2741/begleit. [DOI] [PubMed] [Google Scholar]
- 5.Galm U, Hager MH, van Lanen SG, Ju J, Thorson JS, Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 6.McKeown SR, Coweny RL, Williams KJ. Bioreductive drugs: from concept to clinic. Clinical Oncology (Royal College of Radiologists) 2007;19:427–442. doi: 10.1016/j.clon.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Rockwell S, Dobrucki IT, Kim EY, Marrison ST, Vu VT. Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr Mol Med. 2009;9:442–458. doi: 10.2174/156652409788167087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezanson GS, Vining LC. Studies on the biosynthesis of mitomycin C by Streptomyces verticillatus. Can J Biochem. 1971;49:911–918. doi: 10.1139/o71-131. [DOI] [PubMed] [Google Scholar]
- 9.Anderson MG, Kibby JJ, Rickards RW, Rothschild JM. Biosynthesis of the mitomycin antibiotics from 3-amino-5-hydroxybenzoic acid. J Chem Soc, Chem Commun. 1980;1980:1277–1278. [Google Scholar]
- 10.Hornemann U, Eggert JH. Utilization of the intact carbamoyl group of L-(NH2CO-13C,15N) citrulline in mitomycin biosynthesis by Streptomyces verticillatus. J Antibiot (Tokyo) 1975;10:841–843. doi: 10.7164/antibiotics.28.841. [DOI] [PubMed] [Google Scholar]
- 11.Hornemann Y, Kehrer JP, Nunez CS, Ranieri RL. D-glucosamine and L-citrulline, precursors in mitomycin biosynthesis by Streptomyces verticillatus. J Am Chem Soc. 1974;96:320–322. doi: 10.1021/ja00808a087. [DOI] [PubMed] [Google Scholar]
- 12.Mao Y, Varoglu M, Sherman DH. Molecular characterization and analysis of the biosynthetic gene cluster for the antitumor antibiotic mitomycin C from Streptomyces lavendulae NRRL 2564. Chem Biol. 1999;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- 13.Sitachitta N, Lopanik NB, Mao Y, Sherman DH. Analysis of a parallel branch in the mitomycin biosynthetic pathway involving the mitN-encoded aziridine N-methyltransferase. J Biol Chem. 2007;282:20941–20947. doi: 10.1074/jbc.M702456200. [DOI] [PubMed] [Google Scholar]
- 14.Gruschow S, Chang LC, Mao Y, Sherman DH. Hydroxyquinone O-methylation in mitomycin biosynthesis. JAm Chem Soc. 2007;129:6470–6476. doi: 10.1021/ja0700193. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Weller RL, Thorson JS, Rajski SR. Natural product diversification using a non-natural cofactor analogue of S-adenosyl-L-methionine. J Am Chem Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. RebG- and RebM-catalyzed indolocarbazole diversification. Chembiochem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]
- 17.Singh S, McCoy JG, Zhang C, Bingman CA, Phillips GN, Jr, Thorson JS. Structure and mechanism of the rebeccamycin sugar 4′-O-methyltransferase RebM. J Biol Chem. 2008;283:22628–22636. doi: 10.1074/jbc.M800503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukinavicius G, Lapiene V, Stasevskij Z, Dalhoff C, Weinhold E, Klimasauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) J Am Chem Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- 19.Klimasauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Lüscher B, Weinhold E. Enzymatic site-specific functionalization of protein methyltransferase substrates with alkynes for click labeling. Angew Chem Int Ed. 2010;49:5170–5173. doi: 10.1002/anie.201001240. [DOI] [PubMed] [Google Scholar]
- 21.Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Synthesis of S-adenosyl-L-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nat Protoc. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]
- 22.Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat Chem Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 23.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40:256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 26.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 27.Dall’Antonia F, Baker PJ, Schneider TR. Optimization of selenium substructures as obtained from SHELXD. Acta Crystallogr D Biol Crystallogr. 2003;59:1987–1994. doi: 10.1107/s0907444903017670. [DOI] [PubMed] [Google Scholar]
- 28.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple insomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzym. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 36.Potterton E, Briggs P, Turkenburg M, Dodson EA. Graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 37.Jansson A, Koskiniemi H, Mantsala P, Niemi J, Schneider G. Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products. J Biol Chem. 2004;279:41149–41156. doi: 10.1074/jbc.M407081200. [DOI] [PubMed] [Google Scholar]
- 38.Cooke HA, Guenther EL, Luo Y, Shen B, Bruner SD. Molecular basis of substrate promiscuity for the SAM-dependent O-methyltransferase NcsB1, involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin. Biochemistry. 2009;48:9590–9598. doi: 10.1021/bi901257q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Mol Biol. 2001;8:271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- 40.Palma PN, Rodrigues ML, Archer M, Bonifácio MJ, Loureiro AI, Learmonth DA, Carrondo MA, Soares-da-Silva P. Comparative study of ortho- and meta-nitrated inhibitors of catechol-O-methyltransferase: Interactions with the active site and regioselectivity of O-methylation. Mol Pharmacol. 2006;70:143–153. doi: 10.1124/mol.106.023119. [DOI] [PubMed] [Google Scholar]
- 41.Jansson A, Niemi J, Lindqvist Y, Mantsala P, Schneider G. Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-adenosyl-L-methionine-dependent methyltransferase homolog involved in anthracycline biosynthesis in Streptomyces purpurascens. J Mol Biol. 2003;334:269–280. doi: 10.1016/j.jmb.2003.09.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.