Abstract
We have determined the structure of Bacteroides thetaiotaomicron TetX2 at 2.8 Å resolution, and shown that it is a class A flavin dependent oxidoreductase. TetX2 has broad activity against a range of tetracyclines including one of the most recent tetracyclines, tigecycline (Tygacil®). Comparison of TetX2 with that of the weakly homologous Pseudomonas fluorescens para-hydroxybenzoate hydroxylase (PHBH) (21% identity) shows substantial differences among residues at the substrate binding site although FAD is positioned in a similar conformation between the two enzymes and is poised to carry out catalysis.
Keywords: TetX2, oxidoreductase, X-ray crystallography, antibiotic resistance
Introduction
Tetracyclines are broad-spectrum antibiotics that are commonly used against both Gram-positive and Gram-negative bacteria as well as in agriculture as growth promoters1. Tetracyclines inhibit protein synthesis by non-covalently binding to the ribosome and blocking the binding of aminoacyl-tRNA to the ribosomal acceptor site2. Since the discovery of tetracyclines in 1940s, bacterial adaptation has resulted in the diminished effectiveness of these antibiotics. Tetracycline resistance mechanisms have been classified into three groups based on their mechanism of resistance: efflux, ribosomal protection, and more recently discovered, enzymatic modification1,3. TetX is a tetracycline degrading enzyme that was originally isolated from Bacteroides transposons Tn4351 and Tn44003. Continued use of tetracycline derivatives in clinical and non-clinical environments suggest that TetX and other novel resistance strategies will continue to arise with important consequences to human health.
Previous in vitro studies have shown that TetX hydrolyzes a broad range of tetracyclines including the front line drug, Tygacil® (tigecycline)4,5. TetX is a 44 kDa flavin adenine dinucleotide (FAD) containing monooxygenase that requires NADPH, Mg2+ and O2 for enzymatic activity4. TetX2, an ortholog of TetX with 99.8% sequence identity to TetX, was originally isolated from transposon CTnDOT in Bacteroides thetaoiotaomicron6. TetX2 crystallized in space group P21 with four molecules in the asymmetric unit. Based upon its modest sequence similarity (21%) to Pseudomonas fluorescens FAD-hydroxylase, para-hydroxybenzoate hydroxylase (PHBH) and enzymatic activity, TetX2 was initially classified as a potential class A monooxygenase. Our structure, shows TetX2 to have a tightly bound FAD and as expected for a class A monooxygenase lacks an independent NADPH binding domain7. Based on comparative analysis of the TetX2 and PHBH structures, we have identified a putative substrate binding pocket for TetX2. A comparison of PHBH and TetX2 residues within the substrate binding pocket shows little conservation and is consistent with different roles and substrates in vivo.
Materials and Methods
Expression and Purification of Recombinant Proteins
An expression vector, pET-28b(+) containing Bacteroides thetaiotaomicron tet(X2) (residues 11–388) was a generous gift from Dr. G. D. Wright (McMaster University, Canada). TetX2 was expressed and purified according to protocol modified from Yang et al. (2004)4. Tet(X2) containing vector was transformed into E. coli BL21 (DE3) Star strain (Stratagene). Bacteria were cultured in a shaking incubator at 37°C in 2 × YT media supplemented with 50 μg/ml kanamycin. Seleno-methionine labeled TetX2 (SeMet-TetX2) was expressed according to the methionine inhibition pathway method8. The cell culture was grown in M9 Minimal media supplemented with 50 μg/ml of kanamycin. Prior to induction of protein expression 50 mg/L of L-selenomethionine, 100 mg/L of leucine, isoleucine, phenylalanine, and 50 mg/L of threonine, lysine, valine were added to the media. Expression of TetX2 was induced with isopropyl β-D-1-thiogalactopyranoside to 0.5 mM at an optical density of ~0.8 at 600 nm and incubated at 16°C overnight. Cells were harvested by centrifugation at 8000 × g for 10 min, and the pellets were stored at −80°C. Frozen cells were thawed on ice and resuspended in lysis buffer consisting of 20 mM Tris pH 8, 500 mM NaCl, 0.2 mM phenylmethanesulfonyl fluoride (PMSF) and 5 mM β-mercaptoethanol (BME) and lysed by sonication. Cell lysate was centrifuged at 15,000 × g for 30 min and the soluble fraction was applied on a Ni-His Bind Chromatography column. TetX2 was eluted with 500 mM imidazole containing buffer. An equimolar amount of exogenous FAD was added to the fractions containing TetX2 after each purification step. Pooled fractions were dialysed against 20 mM Tris pH 8, 100 mM NaCl, 0.2 mM PMSF and 5 mM BME overnight. The N-terminal His6-tag was removed by digestion of TetX2 protein with Thrombin for 24 h at 4°C (Novagen). The TetX2 digest was analyzed on a 15% SDS-PAGE gel and re-applied to a Ni-His Bind Chromatography column. Fractions with cleaved His6-tag were pooled and dialyzed against 20 mM Tris pH 8, 100 mM NaCl, 0.2 mM PMSF, 5 mM BME, and 1 mM EDTA, overnight. The protein sample was loaded onto a HiTrap Q-XL Sepharose anion exchange chromatography column equilibrated in 20 mM Tris pH 8, 200 mM NaCl, 0.2 mM PMSF, 5 mM BME, and 1 mM EDTA. The protein was eluted with linear gradient of 0–1 M NaCl. Finally, the TetX2 containing fractions were pooled, concentrated and loaded onto HiLoad16/60 Superdex-200 column (GE Healthcare).
Crystallization and Data Collection
Single TetX2 crystals were obtained after 3 days from 2.3 M ammonium sulfate, 0.1 M CHES pH 8.6, and 0.1 M potassium formate at 4°C using hanging-drop vapor diffusion method. SeMet-TetX2 crystals were obtained under similar crystallization conditions using the sitting-drop vapor diffusion method.
Multiple-wavelength anomalous dispersion (MAD) data were collected at Advanced Light Source (ALS) beamline 4.2.2 using a NOIR-1 MBC detector. SeMet-TetX2 diffraction data was processed using HKL20009 (Table I). SeMet-TetX2 crystals diffracted to 2.8 Å resolution. The protein crystallized in space group P21 with cell dimensions a = 87.65 Å, b = 67.41 Å, c = 152.35 Å and angles α = γ = 90.0° and β = 101.68°.
Table I.
Summary of Data Collection and Refinement Statistics
| Data Collection | SeMet TetX2 |
|---|---|
| Wavelength (Å) | 0.97889 |
| Resolution (Å)a | 50 – 2.80 (2.85 - 2.80) |
| Space group | P21 |
| Unit Cell (Å) | a = 87.70, b = 67.33, c = 153.79 α = γ = 90.0°, β = 100.31° |
| Total number of reflectionsa | 132098 |
| Unique reflections | 39866 |
| Average redundancya | 3.3 (3.2) |
| Completeness (%)a | 92.0 (86.7) |
| Rmerge (%)a,b | 11.8 (28.6) |
| Output <I/sigI>a | 16 (3.9) |
| Refinement | |
| Rwork (%)c | 24.00 |
| Rfree (%)d | 28.24 |
| r.m.s.d.e from ideality | |
| Bonds (Å) | 0.009 |
| Angles (°) | 1.255 |
| Average B-factor (Å2) | 25.05 |
| Ramachandranf | |
| favored (%) | 92.1 |
| additional allowed (%) | 7.9 |
| disallowed (%) | 0 |
| PDB accession number | 3P9U |
Values for the last shell are in parentheses.
Rmerge Σ|I − <I>|/ΣI, where I is measured intensity for reflections with indices of hkl.
Rwork Σ |Fo − Fc|/Σ |Fo| for all data with Fo > 2 σ (Fo) excluding data to calculate Rfree.
Rfree Σ |Fo − Fc|/Σ |Fo| for all data with Fo > 2 σ (Fo) excluded from refinement.
Root mean square deviation.
Calculated by using MolProbity
Structure Determination and Refinement
The initial structure determination by molecular replacement (MR) was performed using BALBES10. The best solution was obtained when PhzS from Psedomonas auerugosa (PDB ID: 3c96) was used as a search model (21% sequence identity). The solution from MR suggested four molecules in the asymmetric unit. The initial model was submitted to phenix.autobuild and phenix.refine for automatic building and structure refinement11. Diffraction data collected at the peak wavelength 0.97889 Å were used for structure determination by Single-wavelength Anomalous Dispersion (SAD) using phenix.autosol11. The partial model obtained by MR was used to search for Se sites. Automated model building by phenix.autobuild11 resulted in successful placement of ~55–60% of the protein structure with the initial R-factor = 45% and R-free = 47.5%. The model was further built manually in COOT12 and refined by phenix.refine. The initial refinement strategy included rigid-body, positional and group ADP refinement with non-crystallographic symmetry (NCS) restraints. FAD was fit manually into unoccupied density found in all 4 molecules. Additionally, TLS refinement was carried out in phenix.refine11. The diffraction was strongly anisotropic and an anisotropy correction using the Diffraction Anisotropy Server was applied to the SAD data set that resulted in a 2% drop in R-factor and R-free after refinement13. Structure coordinates were deposited in the Protein Data Bank under accession number 3P9U.
Results and Discussion
The crystal structure of TetX2 shows it is a class A flavin oxidoreductase
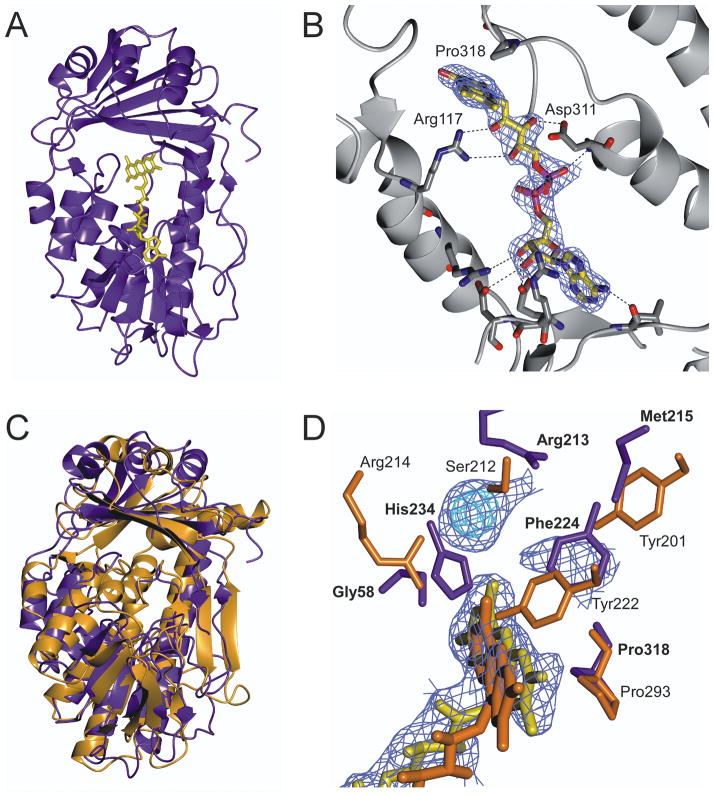
The final model consists of 4 molecules (A, B, C, D) with r.m.s.d. = 0.1 Å between copies, 84 water molecules and R-factor and R-free of 24.0 and 28.2 % respectively. Electron density for residues 11, 248–249 and 6 C-terminal residues was weak and could not be modeled. TetX2 shows a very low overall sequence identity with other FAD-dependant hydroxylases (Fig. 1), but shares several conserved motifs associated with formation of an FAD binding pocket and similar overall topology to the class A family of oxidoreductases. TetX2 is a monomer comprised of two domains binding one FAD molecule (Fig. 2A). FAD is stabilized in the active site through a network of hydrogen interactions with residues from both N - and C - terminal domains (Fig. 2B). FAD is in an extended conformation with adenosine positioned close to the Rossmann fold and the isoalloxazine ring extending toward the smaller domain near the putative substrate binding site. The larger (N-terminal) domain contains a Rossmann fold (βαβ-fold) that includes the highly conserved GXGXXG motif required for binding the adenine moiety of flavin14. The flavin binding pocket also contains a highly conserved GD motif (residues 310–311) found in most FAD-dependent oxidoreductases15. The residues in the GD motif form a specific pocket for the flavin molecule through the formation of hydrogen bond interactions between the Oδ of Asp311 and the O3′ of ribose, and OP2 of the pyrophosphate of the FAD to the backbone amide of Asp311. In TetX2, a third DG motif involved in the binding the pyrophosphate moiety of FAD16, that is found in most FAD – hydroxylases, has an Asn168 in place of Asp resulting in a slightly altered FAD binding pocket. In addition, Arg117 forms highly conserved hydrogen bonds to the ribityl chain of FAD that may orient the flavin toward the substrate in the TetX2 active site17. TetX2 lacks an independent NADPH binding domain which is a characteristic of class A monooxygenases and is consistent with the hypothesis that NADPH is immediately released upon FAD reduction 16.
Fig. 1. TetX2 shares common sequence motifs with other FAD-dependant monooxygenases.
Multiple sequence alignment of TetX2 from Bacteroides thetaiotaomicron (TetX2), p-hydroxybenzoate hydroxylase (PHBH) from Pseudomonas fluorescence, phzS from Pseudomonas aeruginosa (PHZS), and salicylate hydroxylase (SAL) from Pseudomonas putida shows that TetX2 shares conserved sequence motifs, GXGXXG, DG, modified GD, with other FAD-monooxygenases. Sequence were aligned with program CLASTALW and represented by BOXSHADE 19.
Fig. 2. Crystal structure of wild-type TetX2 from Bacteroides thetaiotaomicron, a FAD-dependent monooxygenase determined at 2.8 Å.
A) Ribbon representation of crystal structure of TetX2 with bound FAD (yellow). B) FAD binding is coordinated through a number of hydrogen interactions with residues surrounding the FAD-binding pocket. C) Superposition of structures of TetX2 (blue) and PHBH (orange) results in 4.1 Å r.m.s.d. The overall topology of TetX2 is similar to PHBH and other FAD-dependent hydroxylases, forming two domains with one FAD molecule and of the absence an independent NADHP binding domain. D) Overlay of active sites of TetX2 (blue residues) and PHBH (orange residues) with strong unknown density shown in blue and cyan. Residues surrounding the active site are not well conserved with the exception of Pro318 (Pro293 in PHBH). Final (2Fo−Fc) SIGMAA weighted electron density map corresponding to FAD and the unknown density was contoured at 1.5σ (blue) and 3σ (cyan). Figure was made using CCP4mg20.
The interface between antiparallel β-sheets (strands β5–6, β16–20) of the second domain and the isoalloxazine ring of FAD comprises the binding site for the substrate. We observed additional strong electron density in the (2Fo−Fc) SIGMAA weighted composite maps near the isoallozazine ring of FAD that corresponds to the predicted substrate binding site though the identity of the molecule remains unknown (Fig 2D). The unknown electron density was found for both the P21 and P1 (data not shown) crystal forms of TetX2. Although, we were unable to determine the identity of the unknown molecule that gives rise to the strong density seen in the composite electron density map, we speculate based on its position near the isoalloxazine ring of FAD and similar location to the substrate of PHBH that this molecule is a natural E. coli metabolite that co-purified with TetX2. The TetX2 structure bound to an unknown substrate is in a ‘closed’ state and the strong yellow color of the crystals suggests that the flavin is oxidized and therefore unable to carry out further catalysis. This might also explain the extended conformation of FAD which resembles more the FAD conformation of PHBH with substrate bound rather than the FAD in the unbound PhzS structure.
Comparison of the active sites between TetX2 and PHBH
Superposition of TetX2 to the well characterized PHBH (Fig. 2C) (PDB ID: 1cj3) and Pseudomonas aeruginosa PhzS (PDB ID: 3c96) results in an r.m.s.d. of 4.1 Å and 2.9 Å, respectively. As shown in Fig. 2C, there are substantive differences in the structure compared to PHBH in the N-terminal domain consistent with the modest sequence identity although the general topology is maintained. The overall comparison of the active sites between crystal structures of TetX2 and substrate bound PHBH shows that the FAD cofactor is oriented in a similar position in both proteins (Fig. 2D) but that the specific residues surrounding the predicted substrate binding site of TetX2 compared to PHBH are poorly conserved, which is not surprising considering that the enzymes bind significantly different substrates. TetX2 maintained a highly conserved proline (Pro318) that is analogous to Pro293 in PHBH that interacts with the phenolic oxygen of the substrate of PHBH. This residue has been suggested to be important for maintaining the proper FAD conformation18. The location of the unknown density in the TetX2 structure closely resembles the position of the substrate in PHBH. Arg213 and Met215 in the TetX2 structure are in positions to make hydrogen bonds to the unknown substrate in a manner analogous to residues Ser212 and Tyr201 in PHBH. In addition, the sidechain of His234 and the backbone amide of Gly58 can hydrogen bond with the substrate similarly to Arg214 in PHBH. We speculate that Phe224 is similar to Tyr222 in PHBH, which is also important for substrate binding. A significant shift of a loop (residues 54–59) between the substrate binding site and the isoalloxazine ring of FAD is observed in TetX2 when compared to PHBH loop (residues 42–47), which results in an expanded binding pocket that might be required to accommodate a large substrate, such as tetracycline. While the natural substrate of TetX2 remains unknown, the ability of TetX2 to inactivate tetracyclines in a flavin dependent manner together with the structure presented here confirm the role of TetX2 as a Class A oxidoreductase. The TetX2 structure provides the basis for future structure based drug design and physicochemical characterization towards this mechanism of broad tetracycline resistance.
Acknowledgments
The authors would like to thank Jay Nix for collecting data for SAD experiment at ALS. This work was supported by the NIH (R01AI080714) and The Welch Foundation (C1584) to Y.S. The Rice University Crystallographic Core Facility is supported by a Kresge Science Initiative endowment grant.
References
- 1.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speer BS, Shoemaker NB, Salyers AA. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clinical microbiology reviews. 1992;5(4):387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speer BS, Bedzyk L, Salyers AA. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. Journal of bacteriology. 1991;173(1):176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wangrong Yang IFM, Koteva Kalinka P, Bareich David C, Hughes Donald W, Wright Gerard D. TetX Is a Flavin-dependent Monooxygenase Conferring Resistance to Tetracycline Antibiotics. The Journal of Biological Chemistry. 2004;279(50):52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- 5.Moore IF, Hughes DW, Wright GD. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry. 2005;44(35):11829–11835. doi: 10.1021/bi0506066. [DOI] [PubMed] [Google Scholar]
- 6.Whittle G, Hund BD, Shoemaker NB, Salyers AA. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Applied and environmental microbiology. 2001;67(8):3488–3495. doi: 10.1128/AEM.67.8.3488-3495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Berkel WJ, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. Journal of biotechnology. 2006;124(4):670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 8.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 9.Otwinowski ZaMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 10.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 13.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(21):8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. Journal of molecular biology. 1986;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 15.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. Journal of molecular biology. 1990;212(1):135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 16.Eppink MH, Schreuder HA, Van Berkel WJ. Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding. Protein Sci. 1997;6(11):2454–2458. doi: 10.1002/pro.5560061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenhagen BT, Shi K, Robinson H, Gamage S, Bera AK, Ladner JE, Parsons JF. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry. 2008;47(19):5281–5289. doi: 10.1021/bi702480t. [DOI] [PubMed] [Google Scholar]
- 18.Palfey BA, Basu R, Frederick KK, Entsch B, Ballou DP. Role of protein flexibility in the catalytic cycle of p-hydroxybenzoate hydroxylase elucidated by the Pro293Ser mutant. Biochemistry. 2002;41(26):8438–8446. doi: 10.1021/bi012073g. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]