Abstract
Modification of histones is critical for the regulation of all chromatin-templated processes. Yeast Rtt109 is a histone acetyltransferase (HAT) that acetylates H3 lysines 9, 27 and 56. Rtt109 associates with and is stabilized by Nap1 family histone chaperone Vps75. Our data suggest Vps75 and Nap1 have some overlapping functions despite their different cellular localization and histone binding specificity. We determined that Vps75 contains a classical nuclear localization signal and is imported by Kap60–Kap95. Rtt109 nuclear localization depends on Vps75, and nuclear localization of the Vps75-Rtt109 complex is not critical for Rtt109-dependent functions, suggesting Rtt109 may be able to acetylate nascent histones before nuclear import. To date, the effects of VPS75 deletion on Rtt109 function had not been separated from the resulting Rtt109 degradation; thus, we used an Rtt109 mutant lacking the Vps75-interaction domain that is stable without Vps75. Our data show that in addition to promoting Rtt109 stability, Vps75 binding is necessary for Rtt109 acetylation of the H3 tail. Direct interaction of Vps75 with H3 likely allows Rtt109 access to the histone tail. Furthermore, our genetic interaction data support the idea of Rtt109-independent functions of Vps75. In summary, our data suggest that Vps75 influences chromatin structure by regulating histone modification and through its histone chaperone functions.
Keywords: Vps75, Rtt109, Nap1, nuclear import, Kap60, Gcn5, histone acetylation, histone chaperone, HAT
INTRODUCTION
Eukaryotic DNA is wrapped around histone proteins forming a compact structure called chromatin. Tight compaction allows the packaging of the entire genome into a single nucleus, but it also inhibits DNA access. Cells have developed complex mechanisms for regulating chromatin structure to allow DNA access as needed. Chromatin structure is dynamic; nucleosomes can be assembled and disassembled and individual histone proteins can be removed and exchanged. In addition, covalent modifications such as acetylation, phosphorylation and methylation may occur on each histone (1, 2). Combinatorially or individually, these modifications affect the compaction of the chromatin by altering the affinity of histones for DNA and also regulate which proteins are recruited to specific areas of the chromatin (3, 4). These processes are carried out by the combined activities of ATP-dependent chromatin remodelers and non-enzymatic histone chaperones (5–7). Histone chaperones are required to prevent the highly basic histones from making inappropriate contacts with DNA or other proteins (8). They facilitate histone transfer to remodeling factors and can act as buffers holding excess histones until needed. Histone chaperones are important regulators of chromatin assembly, and some are involved in transport of nascent histones (9, 10).
Members of the evolutionarily conserved Nap1/SET superfamily of histone chaperones are multifunctional proteins (11, 12). The founding member, yeast Nap1, binds primarily to histones H2A and H2B and promotes their import into the nucleus by increasing the affinity of the histones for their import factor, the karyopherin Kap114 (13, 14). Nap1 can assemble and disassemble chromatin in vitro, is recruited to active genes and has been shown to regulate transcription of 10% of the genome in vivo (11, 15, 16). In human cells, the family consists of SET, TSPY, TSPYL proteins and several Nap1-like proteins (12). SET was discovered due to its involvement in a translocation that has been implicated in the development of leukemia (12, 17). Many functions have been attributed to SET, suggesting it is involved in transcription, cell cycle control and apoptosis (12, 18). Vps75 is a second Nap1 family chaperone in yeast. Structural studies have shown that Vps75 is more closely related to human SET than to yeast Nap1, implying that SET and Vps75 may have conserved functions (19). Initial studies have shown that Vps75 is a histone H3/H4 chaperone that associates with a histone acetyltransferase (HAT), Rtt109, in vivo (20–22). Rtt109 is the structural homolog of the human HAT p300 (23–25). Strikingly, the H3/H4 chaperone SET is part of the INHAT complex, which inhibits the activity of the human HAT p300 (26). Thus, determining the function of yeast Vps75 may help elucidate the function of the human oncoprotein SET.
Saccharomyces cerevisiae Vps75 was originally identified in a screen for mutants that exhibited defects in vacuolar protein sorting (27). Several groups determined that Rtt109 was the HAT responsible for acetylation of histone 3 lysine 56 (H3 K56ac), a modification to nascent H3 that is of particular interest due to its location on the globular domain of the histone at the entry/exit point of the DNA on the nucleosome (20, 28–31). Rtt109 was found to associate with two H3/H4 histone chaperones in vivo: Asf1 or Vps75, and in vitro, acetylation of H3 by Rtt109 was increased by the addition of either Asf1 or Vps75 (20, 22, 28, 32). In vivo, Rtt109 and Asf1 are essential for H3 K56 acetylation, but surprisingly, Vps75 is not. Furthermore, significant sensitivity to genotoxic drugs is observed in rtt109Δ, asf1Δ and H3 K56R mutant strains but not in vps75Δ (20, 28, 29). The most noticeable phenotype of vps75Δ yeast is a decrease in cellular levels of Rtt109 protein due to degradation, suggesting that Vps75 acts as an Rtt109 chaperone to protect and stabilize Rtt109 (33). Lysines 9 and 27 of H3 (H3 K9 and K27) are other sites of acetylation found on most nascent H3 (34). H3 K9 and 27 are primarily acetylated by another HAT, Gcn5; however, studies using gcn5Δ yeast have demonstrated that Rtt109 also acetylates H3 K9 and K27 (33, 35). Whereas H3 K56ac is not dependent on Vps75, acetylation of H3 K9 and K27 by Rtt109 is Vps75 dependent. In the absence of Gcn5, the additional deletion of RTT109 or VPS75 causes complete loss of H3 K9ac and K27ac and a growth defect (33, 35). Why Vps75 is critical for Rtt109 HAT activity on H3 K9 and K27 but not on H3 K56 is not known. It is possible that in the absence of Vps75, Rtt109 is degraded and the ensuing low level of Rtt109 is sufficient to acetylate only its primary target lysine, H3 K56. An alternative hypothesis is that beyond to binding to and stabilizing Rtt109, Vps75 has an additional function in promoting or regulating the acetylation of H3 K9 and K27 by Rtt109 through different mechanisms, which we address here.
We wanted to determine whether Vps75 might function like Nap1 in promoting nuclear import of histones or other proteins. Proteins are imported into the nucleus in complex with transport proteins called karyopherins or importins (36, 37). These proteins bind their cargoes via cognate nuclear localization signals (NLS) in the cytoplasm and transport them through the nuclear pore complex. Inside the nucleus, the interaction of the karyopherin with RanGTP leads to release of the cargo, leaving the karyopherin free to be recycled (38, 39). Karyopherins can bind their cargoes directly, but the best characterized karyopherin, importin β (Kap95 in budding yeast) can use an adaptor protein called karyopherin α (Kap60) to bind the NLS. The Kap60–Kap95 heterodimer recognizes a classical NLS, which usually comprises a short cluster of basic amino acids (36, 38, 40). Here we present evidence that Vps75 is imported into the nucleus via a classical NLS and the karyopherin Kap60. Deletion of VPS75 or expression of mislocalized Vps75 NLS mutants caused partial mislocalization of the HAT Rtt109. Surprisingly, when Rtt109 was mislocalized in the presence of the Vps75 NLS mutants, it remained functional, emphasizing the importance of the Rtt109-Vps75 complex. In order to determine the ways in which Vps75 may promote Rtt109 activity beyond preventing degradation of Rtt109, we used a stable mutant form of Rtt109 to reveal a novel requirement for Vps75 in Rtt109 acetylation of H3 K9. We speculate that a direct interaction between Vps75 and histone H3 allows Rtt109 access to the H3 tail for acetylation of H3 K9.
RESULTS
Vps75 and Nap1 have overlapping and distinct cellular functions
Vps75 and Nap1 are structurally related proteins and members of the evolutionarily conserved Nap1 superfamily of histone chaperones (12, 41). Nap1 is a nucleo-cytoplasmic shuttling phosphoprotein whose steady state localization is predominantly cytoplasmic (14, 42). We compared the localization of Vps75 to that of Nap1 in living cells using fluorescent protein tags. Unlike Nap1-GFP2, the steady state localization of Vps75-mCherry was almost exclusively nuclear (FIG 1A). Nap1 is a histone binding protein that preferentially binds H2A and H2B, whereas Vps75 has been reported to bind both H3/H4 and H2A/H2B (13, 41, 43, 44). We tested the ability of recombinant Vps75 to bind purified histones and in our in vitro assays Vps75 preferentially bound to H3 and H4 (FIG 1B). We determined whether Vps75 and Nap1 had overlapping functions in vivo by testing for a genetic interaction between VPS75 and NAP1. The growth of wild type, vps75Δ, nap1Δ and vps75Δ nap1Δ strains was compared under various conditions (FIG 1C). Deletion of VPS75 has been reported to confer a growth advantage on media with acetic acid, and this effect was explained as an alteration in the transcriptional regulation of acid-responsive genes (45). Our data agree with this finding but also reveal a similar growth advantage conferred by deletion of NAP1; however, deletion of both genes has no additional effect (FIG 1C). No synthetic effects on growth were observed in the vps75Δ nap1Δ strain when grown at 30°C or 37°C, on CSM lacking inositol, with galactose or with 15µg/ml benomyl (FIG 1C and data not shown). In the presence of hydroxyurea (HU), nap1Δ and vps75Δ grew similarly to wild type yeast, whereas nap1Δ vps75Δ grew significantly slower (FIG 1C). The genetic interaction of VPS75 and NAP1 on HU implies a synthetic defect in replication or DNA damage repair. Therefore, the related histone chaperones Vps75 and Nap1 have distinct localization and histone binding preferences, yet they have some similar and overlapping functions in vivo.
FIG 1.
Vps75 and Nap1 have shared and distinct roles in vivo. (A) Nap1-GFP2 and Vps75-mCherry were observed by fluorescent imaging in the same cells. Coincident DIC (differential interference contrast) image is also shown. (B) Purified chicken erythrocyte histones were incubated with immobilized GST or GST-Vps75. The bound material and 10% inputs were separated by SDS-PAGE and visualized by Coomassie Blue staining. (C) Ten-fold serial dilutions of the indicated strains were grown on CSM (left three panels) or YPD (right two panels) lacking inositol or with hydroxyurea or acetic acid added as specified at 30°C (or 37°C where indicated) for 3 days. WT represents wild type.
Vps75 has a classical NLS and is imported by Kap60–Kap95
We wanted to determine whether Vps75, in an analogous manner to Nap1, promotes the nuclear import of histones or other nuclear proteins (14). Thus, analysis of the nuclear import pathways of Vps75 would be critical for understanding its function. Vps75 contains three sequences that fit the minimal consensus for a classical nuclear localization signal, K(K/R)x(K/R), and one of these sequences, 257PSSKKRKV264, comprises the last 8 carboxy terminal residues of the protein (FIG 2A) (40, 46). Vps75-GFP2 fusion plasmids were created and while Vps75 was localized to the nucleus, both Vps75(1–222)-GFP2 and Vps75(1–256)-GFP2, in which only the last 8 residues were deleted, were completely mislocalized to the cytoplasm (FIG 2B). To verify that the 260KKRK263 sequence indeed represents the NLS, the first two lysines were substituted with glycines in the context of the whole protein, and Vps75(K260G, K261G)-GFP2 was similarly mislocalized (FIG 2B). Lastly, to demonstrate that this sequence was sufficient to serve as an NLS, the last 22 amino acids of Vps75 were fused to GFP2, and whereas GFP2 was localized throughout the cell, Vps75(243–264)-GFP2 accumulated in the nucleus (FIG 2C).
FIG 2.
Vps75 import depends on a classical nuclear localization signal and is mediated by Kap60. (A) Schematic of Vps75 with amino acid numbers, domains and NLS labeled. (B) The indicated GFP fusion proteins were expressed in vps75Δ, induced for 1 hour and observed by fluorescent imaging. KG,KG represents Vps75(K260G, K261G). DIC image is also shown (right panel). (C) Vps75(243–264)-GFP2 and GFP2 were observed in vps75Δ by fluorescent imaging after 1 hour induction. DIC image is also shown (right panel). (D) Recombinant MBP-Vps75 wild type, MBP-Vps75 truncated forms (indicated by V followed by amino acid numbers), MBP-Vps75(K260G, K261G) (abbreviated V(KG,KG)) and MBP-LacZ (negative control) were incubated with immobilized GST, GST-Kap60 and GST-Rtt109. The bound material from binding assays or 25% input samples were separated by SDS-PAGE and western blotted with anti-MBP antiserum (middle and right panels). 25% input samples of GST proteins were stained with Coomassie Brilliant Blue (CBB) (Left panel). (E) Vps75-GFP2, Vps75(243–264)-GFP2 or Yra1-GFP2 was expressed in the indicated strains, which were grown at room temperature (RT) or 30°C or shifted to 37°C for 4 hours including 2h induction in -met and observed by fluorescent imaging. (F) Vps75-GFP2 expression was induced for 2h in the strains indicated.
Import of proteins with classical NLSs is mediated by the karyopherin heterodimer Kap60–Kap95 and, by definition, the classical NLS binds directly to Kap60 (37, 40). We wanted to assess whether Vps75 and Kap60 interacted directly and thus tested the binding of these proteins in vitro. Vps75 bound to GST-Kap60 but not to GST alone, demonstrating a direct physical interaction between Kap60 and Vps75 (FIG 2D). To determine if this binding was dependent on the NLS-containing region, recombinant Vps75 mutants Vps75(1–222) and Vps75(K260G, K261G), both of which lack the putative NLS, were incubated with immobilized GST-Kap60 or GST-Rtt109 as a control. Whereas wild type Vps75, Vps75(1–222) and Vps75(K260G, K261G) bound to Rtt109, we only observed significant binding of wild type Vps75 to Kap60 (FIG 2D, left panel). We also expressed recombinant Vps75(243–264), which comprises the last 22 amino acids of Vps75 including the NLS. Binding of Vps75(243–264) to GST-Rtt109 and GST-Kap60 was tested, and although the interaction was weaker overall and required longer exposure times, the western blot revealed that Vps75(243–264) specifically interacted with Kap60 (FIG 2D, right panel). Together, these data indicate that Vps75 nuclear import depends on a classical NLS, 260KKRK263, and that the NLS-containing region allows direct physical interaction of Vps75 and the karyopherin Kap60.
The identification of a classical NLS led us to predict that Kap60–Kap95 would be the primary import factors for Vps75, and we analyzed the role of different karyopherins in the nuclear import of Vps75 in vivo. The localization of Vps75-GFP2 was assessed in strains each with a single deletion or temperature sensitive mutant allele of one of the 12 yeast importins. The most significant mislocalization of Vps75-GFP2 was observed in the kap60-ts strain at 37°C (FIG 2E). As a control we compared Vps75 localization with that of the nuclear mRNA annealing factor, Yra1, which is imported by Kap121 and Kap123 (47). Whereas Yra1-GFP2 appeared completely nuclear in kap60-ts yeast at the permissive and nonpermissive temperatures, both the full length Vps75-GFP2 and the truncation containing the classical NLS, Vps75(243–264)-GFP2, were somewhat mislocalized at room temperature and significantly mislocalized at 37°C (FIG 2E). Vps75-GFP2 was slightly more concentrated in the nucleus than in the cytoplasm of some kap60-ts cells even at 37°C, suggesting not all Kap60 function was lost and/or that full length Vps75 may have other, secondary routes into the nucleus. Vps75-GFP2 and Vps75(243–264)-GFP2 were also more cytoplasmic overall in kap95-ts at 37°C than in wild type cells, but localization in this strain was much more heterogeneous, ranging from cells with only nuclear GFP to cells in which the nuclear and cytoplasmic GFP appeared equal (data not shown). In the other importin mutant strains (kap114Δ, kap123Δ, kap121–34, kap108Δ, kap119Δ, kap122Δ, kap104Δ, kap111Δ, kap142Δ and kap120Δ) Vps75-GFP2 localization remained nuclear as in wild type yeast (examples shown in FIG 2F, other data not shown). In summary, our findings suggest that the importins Kap60 and Kap95 mediate the major import pathway of Vps75 via a C-terminal classical NLS.
Vps75 promotes the nuclear localization of Rtt109
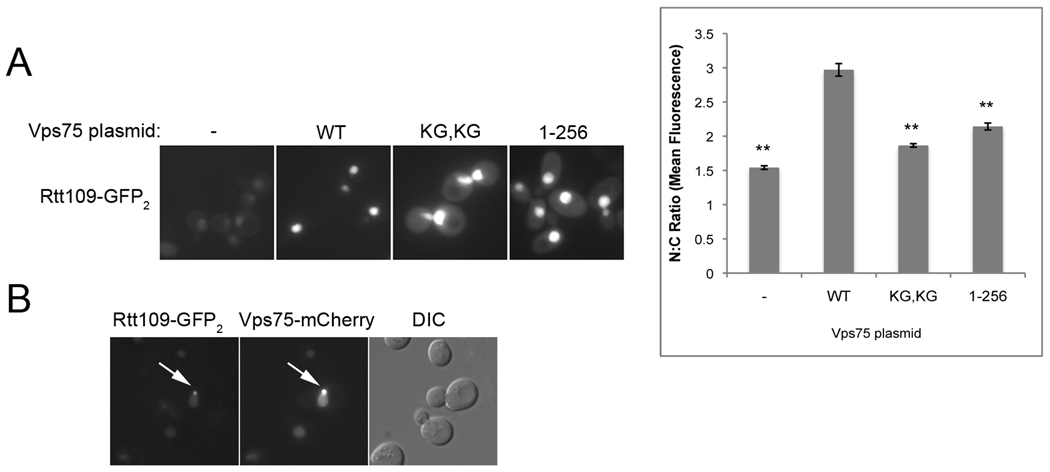
Nap1 assists in the import of histones, as evidenced by the mislocalization of histone NLS-GFP fusions in nap1Δ yeast (14). Alternatively, when we compared the localization of the H3 NLS-GFP2 reporter in wild type and vps75Δ strains, we did not detect any difference, implying that Vps75 does not play a critical role in H3/H4 import (data not shown). It remained possible that Vps75 functions by promoting the import of a non-histone nuclear protein such as the HAT Rtt109, which it binds with high-affinity (22, 48). To test whether Rtt109 import was dependent on Vps75, Rtt109-GFP2 was expressed in rtt109Δvps75Δ yeast with or without plasmid-borne Vps75. In the presence of Vps75, Rtt109-GFP2 was observed exclusively in the nucleus, but in the absence of Vps75 some mislocalization to the cytoplasm was clearly detected (FIG 3A). Vps75 prevents the degradation of Rtt109, and the amount of Rtt109-GFP2 observed was much lower in the cells lacking Vps75. In order to compare Rtt109-GFP2 localization in these strains, the ratio of mean fluorescence intensity in the nucleus to the cytoplasm (N:C) was quantified. Rtt109 had a significantly lower N:C ratio in rtt109Δvps75Δ than in the same strain with a wild type Vps75 plasmid (FIG 3A graph). These data suggested that although Vps75 was not essential for Rtt109 nuclear localization, Vps75 improved the efficiency of Rtt109 import or helped maintain Rtt109 once inside the nucleus.
FIG 3.
Vps75 promotes nuclear localization of Rtt109. (A) Rtt109-GFP2 was expressed in rtt109Δvps75Δ containing either pRS316 (−) or plasmids expressing the indicated mutants of Vps75 and observed by fluorescent imaging. KG, KG represents Vps75(K260G, K261G). The mean fluorescence intensity of a defined pixel area was measured in the nucleus (N) and the cytoplasm (C) and used to calculate the N:C ratio. The mean ratio for more than 190 cells was calculated for each strain and graphed along with standard error. ** p≪ 0.0001 compared to wild type (WT). (B) Rtt109-GFP2 was expressed in VPS75-mCherry yeast and both were observed by fluorescent imaging. DIC image is also shown. Arrows indicate colocalizing fluorescent proteins in one of these foci.
To further explore the possible role of Vps75 in Rtt109 nuclear import, we analyzed the localization of Rtt109-GFP2 in the presence of Vps75 NLS mutants Vps75(1–256) and Vps75(K260G, K261G), which are mislocalized to the cytoplasm (FIG 2B). Expression levels of Rtt109-GFP2 in the presence of Vps75(1–256) or Vps75(K260G, K261G) were similar to that observed with wild type Vps75 and were much greater than the level observed in the absence of Vps75 (FIG 3A). This suggested Vps75 can stabilize Rtt109 regardless of whether it is localized to the nucleus or cytoplasm. Rtt109-GFP2 was also apparent in the cytoplasm of cells expressing either of the Vps75 NLS mutants (FIG 3A). This localization was reflected in a statistically significant decrease in the N:C ratio of Rtt109-GFP2 in these strains compared to that in yeast with wild type Vps75 (FIG 3A graph). We had tested the mislocalization of Rtt109 in the karyopherin mutant strains but did not identify any Kap deletion that gave a strong phenotype (data not shown). Consequently the mislocalization of Rtt109 in Vps75 NLS mutants was greater than that seen in any of the Kap mutant strains (data not shown). The effect of Vps75(1–256) and Vps75(K260G, K261G) on Rtt109 stability and localization suggested that Vps75 plays a role in Rtt109 import or nuclear retention and that the Vps75 NLS mutants and Rtt109 interact in the cytoplasm.
As we examined the localization of Rtt109 and Vps75, we noticed not only strong nuclear signal but that in some cells they accumulated in bright subnuclear foci. To establish whether Vps75 and Rtt109 localized in the same foci, we observed both mCherry tagged Vps75 and Rtt109-GFP2 in cells and determined that the proteins colocalized in the same subnuclear foci (FIG 3B). Interestingly, although both proteins are localized in the same foci, Rtt109 targeting to these foci did not require Vps75, nor did Vps75 targeting require Rtt109 (data not shown).
The nuclear localization of Vps75 is not critical for its Rtt109-dependent functions
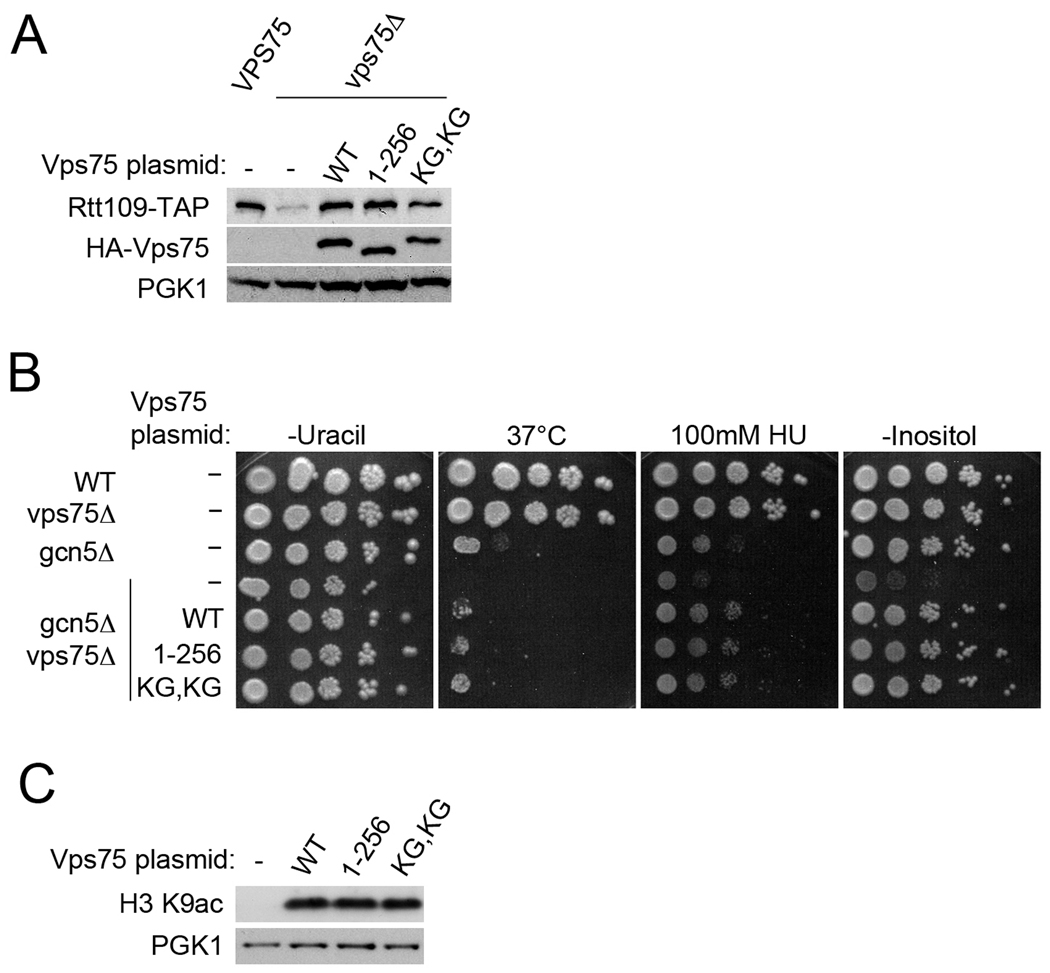
Our microscopy data indicated that both the wild type and the cytoplasmic versions of Vps75 stabilized Rtt109. To confirm this finding using a biochemical approach, we made whole cell lysates from an RTT109-TAP vps75Δ strain expressing either wild type Vps75 or a cytoplasmic mutant, Vps75(1–256) or Vps75(K260G, K261G). In agreement with published data, deletion of VPS75 significantly decreases the amount of Rtt109-TAP detected in lysates (33). Expression of Vps75, Vps75(1–256) and Vps75(K260G, K261G) restored Rtt109-TAP levels to those observed with endogenous VPS75 (FIG 4A). These results confirmed the ability of Vps75 to prevent degradation of Rtt109 regardless of its localization.
FIG. 4.
Cytoplasmic Vps75 NLS mutants are functional in vivo. (A) Whole cell lysates from RTT109-TAP and RTT109-TAP vps75Δ strains transformed with the indicated Vps75 plasmids were western blotted using antibodies against IgG (for TAP), HA, and PGK1 (loading control). KG, KG represents Vps75(K260G, K261G). (B) Ten-fold serial dilutions of the indicated strains transformed with Vps75 plasmids as shown were grown on CSM lacking uracil at 30°C (or 37°C where indicated) with 100mM HU or media lacking inositol as indicated. (C) Whole cell lysates from gcn5Δvps75Δ transformed with Vps75 plasmids as indicated were western blotted using antibodies against H3 K9ac and PGK1 (loading control).
To determine whether the localization of Vps75 was important for functions other than stabilizing Rtt109, we analyzed Rtt109-mediated acetylation of H3 K9. In gcn5Δ, acetylation of lysines 9 and 27 on the H3 tail is known to decrease significantly, however, Rtt109-dependent acetylation of these sites can be observed (33, 35). Rtt109 requires Vps75 for the acetylation of H3 K9 and K27, whereas Vps75 is not essential for Rtt109-mediated acetylation of H3 K56. Thus, when GCN5 is deleted, the additional deletion of VPS75 causes a synthetic growth defect and loss of H3 K9ac and K27ac (33, 35). To assess Vps75 function, the gcn5Δvps75Δ strain was transformed with plasmids encoding wild type VPS75, vps75(1–256), and vps75(K260G,K261G). Comparison of the growth of these strains indicated that expression of Vps75, Vps75(1–256) or Vps75(K260G,K261G) reversed the growth defect of gcn5Δvps75Δ at 37°C, on hydroxyurea (HU) containing plates and in the absence of inositol (FIG 4B). Whole cell lysates were made from the same transformed yeast strains and were used to analyze H3 K9ac by western blot. As expected, H3 K9ac was not detected in the gcn5Δvps75Δ background, however, similar levels of H3 K9ac were observed in strains expressing Vps75 and both NLS mutants, Vps75(1–256) and Vps75(K260G,K261G) (FIG 4C). These results suggested that the nuclear localization of Vps75 is not critical for its function in Rtt109-mediated acetylation of lysine 9 on the H3 tail. As mislocalized Vps75 sequestered some of the Rtt109 in the cytoplasm and there was no resulting decrease in Rtt109 function, it is possible that the two proteins do not need to be in the nucleus but rather need to be in the same compartment to function, highlighting the importance of the interaction between Vps75 and Rtt109.
Rtt109ΔL is a stable, functional HAT that does not bind Vps75 in vivo
To understand the role of the Vps75-Rtt109 interaction in Rtt109 function, we looked for mutants that would abrogate binding in vivo. We tested two previously published mutants of Vps75 that either did not bind Rtt109, Vps75(E218K, D222K), or had reduced binding to Rtt109, Vps75(R173E, K177E) in vitro (19). When expressed in yeast, however, immunoprecipitation experiments indicated robust binding of both mutants to Rtt109 under our conditions (data not shown). We also analyzed a published mutant of RTT109, Rtt109ΔL, in which a proteolytically sensitive region (Rtt109 residues 128–170) was replaced with a short linker (GSGG) to create a more stable protein for crystallization studies (FIG 5A) (49). The deleted region appeared to be responsible for Vps75 binding in vitro, but Rtt109ΔL was shown to be catalytically active (49). These results are consistent with findings for a similar mutant Rtt109(Δ130–179) (50). We tested whether Rtt109ΔL was deficient in Vps75 binding in vivo. We expressed Rtt109ΔL in yeast, immunoprecipitated the protein from yeast lysates and western blotted coprecipitating proteins for Vps75. Abundant copurification of Vps75 was detected with Rtt109 whereas no detectable Vps75 copurified with Rtt109ΔL (FIG 5B, left). We confirmed these results by performing the immunoprecipitation in reverse (FIG 5B right). We also observed that Rtt109ΔL-GFP2 was partially mislocalized to the cytoplasm, consistent with its lack of interaction with Vps75 in vivo (data not shown).
FIG 5.
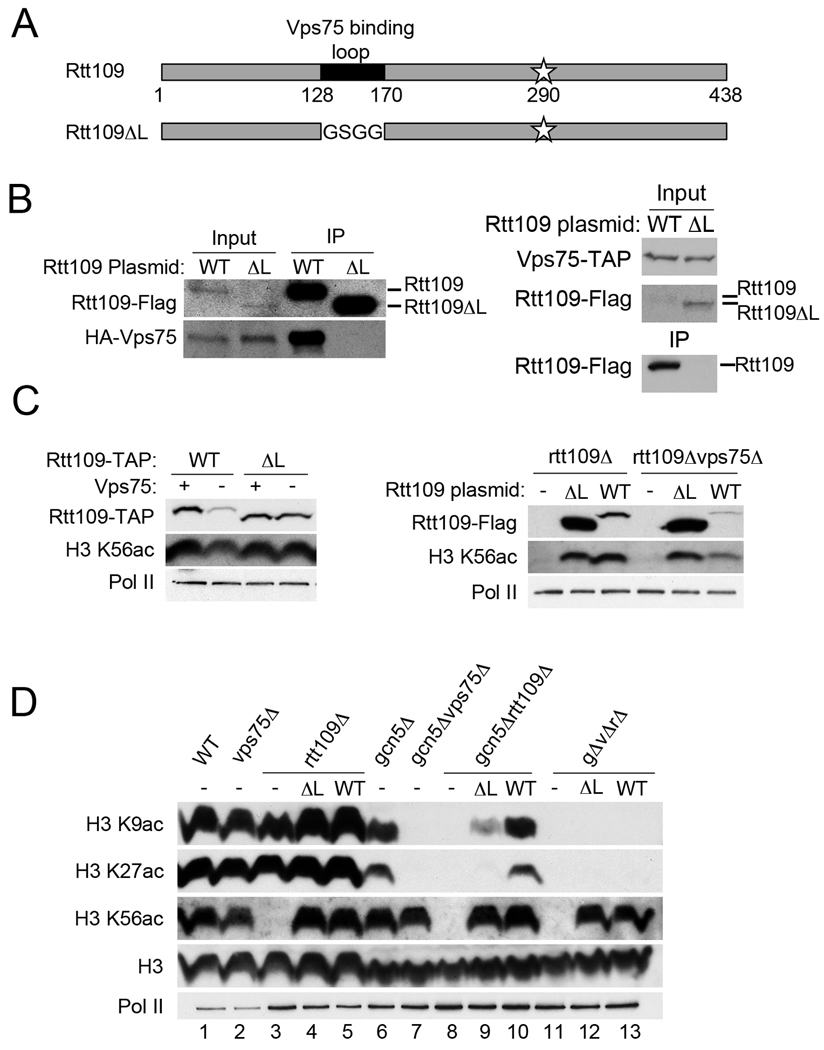
A stable mutant of Rtt109 requires the presence of Vps75 for H3 K9ac despite lack of Vps75-Rtt109 complex formation. (A) Schematic of Rtt109 and Rtt109ΔL showing Vps75 binding region (black) or linker and site of autoacetylation (star). Amino acid numbers are indicated. (B) Lysates from rtt109Δvps75Δ strains expressing HA-Vps75 and Rtt109-Flag (WT) or Rtt109ΔL-Flag (ΔL) plasmids were immunoprecipitated using anti-Flag sepharose and associated proteins were western blotted using antibodies against Flag and HA. Inputs of 0.5% of material are shown (left). Lysates from Vps75-TAP rtt109Δ yeast expressing Rtt109-Flag or Rtt109ΔL-Flag were immunoprecipitated using IgG sepharose and associated proteins were western blotted with antibodies against IgG and Flag. Inputs of 2% are shown (right). (C) RTT109-TAP vps75Δ (WT) or rtt109ΔL-TAP vps75Δ (ΔL) expressing (+) or deleted for (−) VPS75 were western blotted using antibodies against IgG, H3 K56ac and Pol II (loading control) (left). Whole cell lysates from rtt109Δ or rtt109Δvps75Δ strains expressing Rtt109ΔL, Rtt109 WT or p415 ADH1 (−) were western blotted using antibodies against Flag, H3 K56ac and Pol II (loading control) (Right). (D) Lysates from indicated strains expressing Rtt109ΔL (ΔL) or Rtt109 (WT) plasmids were western blotted with antibodies against H3 K9ac, H3 K27ac, H3 K56ac, H3 and Pol II. (gΔvΔrΔ represents gcn5Δvps75Δrtt109Δ).
It has been assumed that the regulation of Rtt109 stability by Vps75 occurs through physical interaction with the proteolytically sensitive region of Rtt109 but this has not been experimentally demonstrated in vivo. Loss of VPS75 would not be predicted to decrease the amount of Rtt109ΔL as it does not bind Vps75. To test this hypothesis, whole cell lysates were made from RTT109-TAP, RTT109-TAP vps75Δ, rtt109ΔL-TAP and rtt109ΔL-TAP vps75Δ strains. Unlike the Rtt109-TAP protein, which decreased when VPS75 was deleted, the Rtt109ΔL-TAP protein was equivalent in cells either expressing or deleted for VPS75 (FIG 5C, left). Thus the Rtt109ΔL protein did not bind to Vps75 in vivo and was stable even in the absence of Vps75. This suggested that the interaction between Vps75 and Rtt109 amino acids 128–170 protects Rtt109 from degradation in vivo, as in vitro.
As Rtt109ΔL is stable and cannot interact with Vps75, we wanted to use this mutant to determine which functions of Rtt109 require Vps75 binding. We tested whether Rtt109ΔL is capable of acetylating the major known target of Rtt109, H3 K56. Western blotting lysates with an antibody specific for H3 K56ac revealed that Rtt109ΔL was catalytically active, and similar amounts of H3 K56ac were detected in cells with either Rtt109 or Rtt109ΔL (FIG 5C, left). Furthermore, loss of VPS75 did not affect the ability of Rtt109ΔL to acetylate H3 K56 as there was no detectable difference in the level of H3 K56ac in rtt109ΔL-TAP and rtt109ΔL-TAP vps75Δ strains. In contrast there was a slight decrease in H3 K56ac in RTT109-TAP vps75Δ compared to that in RTT109-TAP (FIG 5C, left). Thus, Rtt109ΔL is both stable and catalytically active in vivo in the presence or absence of Vps75. The increased stability of Rtt109ΔL compared to Rtt109 in the absence of Vps75 was confirmed using Rtt109-Flag or Rtt109ΔL-Flag proteins expressed from plasmids (FIG 5C, right). We observed that Rtt109ΔL-Flag was significantly more abundant than Rtt109-Flag, although they were expressed from the same promoter and plasmid. Even so, the overall amount of H3 K56ac was equivalent in yeast expressing either Rtt109ΔL or Rtt109, suggesting both mutant and wild type proteins achieved similar maximal levels of bulk acetylation in vivo (FIG 5C, right).
Stable Rtt109 requires Vps75 to acetylate H3 K9
Studies using VPS75 deletion strains have indicated that acetylation of the H3 tail at lysines 9 and 27 by Rtt109 is dependent on Vps75 (33, 35). The properties of Rtt109ΔL allowed us to test whether the critical role of Vps75 in promoting H3 tail acetylation is to maintain adequate cellular levels of Rtt109 by preventing its degradation, or whether Vps75 plays some other direct role. If Vps75 contributes to Rtt109 acetylation solely through promoting its stability, then the stable mutant, Rtt109ΔL, should be able to acetylate these H3 tail lysines in the absence of Vps75. If Vps75-Rtt109 binding is required, Rtt109ΔL may not be able to acetylate H3 K9 and K27 at all. Alternatively, if Vps75 plays another role in Rtt109 acetylation of H3 K9 and K27 then Rtt109ΔL may require the presence of Vps75 to acetylate the H3 tail. In order to study acetylation of H3 K9 and K27 by Rtt109, it was necessary to delete the major H3 tail HAT, GCN5, and thus the deletion strains gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ were generated. Yeast strains were transformed with plasmids expressing Rtt109-Flag or Rtt109ΔL-Flag, and the corresponding whole cell lysates were probed with antibodies against acetylated H3 K9 or K27. In agreement with previous publications, we observed a decrease in both acetylation marks in gcn5Δ compared to wild type, rtt109Δ and vps75Δ and a complete loss of both acetylation marks in gcn5Δ vps75Δ and gcn5Δ rtt109Δ (FIG 5D, lanes 1–3 and 6–8) (33, 35). Furthermore, there was no H3 K9ac or K27ac in the triple deletion strain gcn5Δvps75Δrtt109Δ, and because wild type Rtt109 requires Vps75 for acetylation of the H3 tail, expression of Rtt109 in this strain did not restore acetylation of H3 K9 or K27 (FIG 5D, lanes 11 and 13). In the gcn5Δrtt109Δ strain, where Vps75 is present, expression of wild type Rtt109 resulted in acetylation of H3 K9 and K27 whereas expression of the Vps75 binding deficient mutant Rtt109ΔL yielded a dramatic decrease in H3 K9 acetylation and no detectable H3 K27 acetylation (FIG 5D, lanes 9–10). This suggests that physical interaction of Rtt109 with Vps75 is required for efficient acetylation of the H3 tail, and that the role of Vps75 extends beyond maintaining Rtt109 stability. As we could detect a small amount of H3 K9ac in gcn5Δrtt109Δ yeast expressing Rtt109ΔL, we assessed whether this acetylation was dependent on Vps75 by comparing it to Rtt109ΔL acetylation of H3 K9 in gcn5Δvps75Δrtt109Δ. As seen in lane 12, Rtt109ΔL did not detectably acetylate the H3 tail when VPS75 was deleted, further supporting an important role for Vps75 in Rtt109 HAT activity (FIG 5D).
Loss of H3 acetylation correlates with growth defects
To further understand the function of Vps75 in promoting H3 tail acetylation and determine the significance of losing this acetylation in vivo, we compared growth of the mutant strains expressing Rtt109 and Rtt109ΔL under a variety of conditions. Overall, gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ grew the least well, which was expected because they lack two partially redundant, important HATs, and their growth was improved by expression of either Rtt109 or Rtt109ΔL (FIG 6A). When the growth assay data were compared to the acetylation status of H3 K9, K27 and K56 in the same strains (data from FIG 5D is summarized in chart form beside the images in FIG 6A), correlations between different growth conditions and particular acetylation marks emerged. Sensitivity to HU indicates impairment in DNA damage repair pathways, and previous reports have demonstrated HU sensitivity in the rtt109Δ and H3 K56R strains (20, 44). Not surprisingly, we observed that the three strains lacking Rtt109 (rtt109Δ gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ) and therefore had no detectable H3 K56ac were extremely sensitive to HU (FIG 6A). Expression of either Rtt109 or Rtt109ΔL effectively reversed these slow growth phenotypes in gcn5Δvps75Δrtt109Δ and gcn5Δrtt109Δ consistent with the ability of Rtt109ΔL to acetylate K56 efficiently (FIG 6A).
FIG 6.
Vps75 has Rtt109-independent functions. (A) Ten-fold serial dilutions of the indicated strains (gΔvΔrΔ represents gcn5Δvps75Δrtt109Δ) expressing Rtt109ΔL (ΔL), Rtt109 (WT) or p415 ADH1 (−) plasmids were grown on plates lacking leucine at 30°C (or 37°C where indicated), or with 50mM HU, or media lacking inositol for 3 days. Chart summarizes acetylation status of strains taken from FIG 5D data where + represents detectable acetylation and – represents no acetylation, +/− represents low level of acetylation. (B, C) Ten-fold serial dilutions of the indicated strains were grown on CSM or indicated media at 30°C or 37°C for 3 days (B) or 2 days (C). (D) Models of potential Vps75 functions. Top and middle rows: curved arrows indicate HAT activity; star represents acetylation; X represents any histone-modifying enzyme. Bottom row depicts general histone exchange and chromatin assembly functions of Vps75, which may be Rtt109-dependent or independent. See Discussion for details.
High temperature is a general stressor for cells and can exacerbate growth defects. The strains that grew the slowest at 37°C were those that had no detectable H3 K9ac, whereas the ones that grew the fastest had wild type levels of H3 K9ac. The two strains that had only moderately decreased levels of H3 K9ac, gcn5Δrtt109Δ expressing Rtt109 and gcn5Δ exhibited moderate growth at 37°C (FIG 6A). Strikingly, gcn5Δrtt109Δ strains grew equivalently on all media tested whether expressing Rtt109ΔL or Rtt109, despite the differences in acetylation between these two strains (FIG 6A). The former has a much lower level of H3 K9ac and no H3 K27ac. In contrast, when comparing growth of gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ strains expressing Rtt109ΔL, we determined that loss of Vps75 led to a significant growth defect and this was exacerbated at 37°C. This correlated with our finding that although stable, Rtt109ΔL cannot acetylate H3 K9 in the absence of Vps75. These data imply that achieving a certain level of H3 K9ac in the cell may an important determinant in the different growth rates of these strains at 37°C. Sensitivity to media lacking inositol usually correlates with a transcriptional defect and the pattern of growth on media lacking inositol was similar to that seen at 37°C although not as severe, further suggesting that H3 K9ac levels are important for transcription (FIG 6A) (51).
Genetic interactions support Rtt109-independent functions of Vps75
The gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ strains were the only strains that lack both HAT activities and thus all three acetylation marks, H3 K9, K27 and K56, yet the triple deletion strain grew slower than gcn5Δrtt109Δ in the absence of inositol, suggestive of an important function for Vps75 beyond promoting Rtt109 activity (FIG 6A). It has been reported previously that Vps75 can be recruited to active genes in the absence of Rtt109 consistent with an Rtt109-independent function (45). We wanted to explore this phenomenon further by comparing growth of the untransformed strains on complete synthetic medium (CSM), which gave better growth rates overall than the selective media used in the previous assays (FIG 6B). Any Rtt109-independent functions of Vps75 are most likely redundant with other factors because of the lack of significant growth defects of the vps75Δ strain. The gcn5Δvps75Δ strain grew more slowly than the gcn5Δ strain at 37°C, most likely due to further loss of H3 K9 and K27 acetylation through loss of Rtt109 activity (FIG 6B). Because of the lack of H3 K9ac, K27ac and K56ac, we would have expected growth of gcn5Δrtt109Δ and gcn5Δvps75Δrtt109Δ strains to be slow but equivalent. However, on all plates except the control, gcn5Δvps75Δrtt109Δ grew significantly slower than gcn5Δrtt109Δ, suggesting that Vps75 is important for cellular functions beyond those of the two histone-modifying enzymes, Gcn5 and Rtt109 (FIG 6B). Furthermore, the defects observed on medium lacking inositol and containing galactose were consistent with Vps75 having an important role in transcription.
VPS75 and ASF1 encode the two Rtt109-specific histone chaperones (20, 28, 29). It has been demonstrated that deletion of ASF1 causes loss of Rtt109-mediated acetylation of H3 K9 and K56 (20, 33, 52). Deletion of RTT109 and VPS75 in yeast yields Rtt109-dependent defects but no synthetic growth defect. Therefore we wondered if there might be a genetic interaction between ASF1 and VPS75. As seen in Figure 6C, deletion of ASF1 and VPS75 did indeed result in synthetic growth defects, especially at 37°C. Thus, the genetic interaction of ASF1 and VPS75 provides further evidence for functions of Vps75 beyond the Rtt109-dependent acetylation of H3 K9 and K56.
DISCUSSION
In this report, we have identified the classical NLS of Vps75 and present evidence that Vps75 is imported primarily by Kap60–Kap95. Our studies also revealed a role for Vps75 in proper nuclear localization of Rtt109. Using a stabilized Rtt109 mutant that is defective in binding to Vps75, we determined that the Vps75-Rtt109 complex is needed for acetylation of the H3 tail by Rtt109. Furthermore, our demonstration that the stable Rtt109ΔL cannot acetylate H3 K9 in the absence of Vps75 proves that Vps75 does more than simply prevent degradation of Rtt109. We propose a model in which the interaction of Vps75 with histone H3 promotes acetylation of H3 K9 by Rtt109.
Vps75 and Nap1 have different localization but some overlapping function
At steady state, Nap1 is cytoplasmic and Vps75 is nuclear. Although their overall structures are similar, the determinants of localization are not shared (19, 41). The region of Nap1 containing the nuclear export signal (NES) is completely lacking in Vps75, and the region of Vps75 we have identified as the classical NLS is not present in Nap1 (14). Instead, Kap114 binds a central region of Nap1 and mediates its nuclear import (53). We determined that Vps75 is primarily imported by Kap60–Kap95, but other karyopherins likely play a minor role. It is not known whether Vps75 shuttles in and out of the nucleus; if so, it would likely contain an, as yet undiscovered, NES. It is also not known whether Vps75 promotes histone import in the same way as Nap1. If Vps75 played a major role in H3 and H4 import, we might have expected Vps75 to primarily use the H3–H4 import factor, Kap123 (54, 55). The best understood functions of Vps75 and Nap1 are distinct and involve different histones: Vps75 primarily interacts with H3/H4 whereas Nap1 primarily interacts with H2A and H2B (13, 43, 44). In addition, Vps75 binds a HAT and plays a direct role in promoting histone post-translational modification whereas no such function has been demonstrated for yeast Nap1. Despite these differences in function, our genetic interaction data demonstrate that Vps75 and Nap1 both act in repressing acid-responsive genes and have redundant functions in the response to replication stress and/or DNA damage, highlighting their roles in transcription and replication. These findings suggest that Nap1 family chaperones have evolved to have both overlapping and distinct functions.
Effect of Vps75 on Rtt109 localization
We show for the first time that the nuclear localization of Vps75 is critical for the correct localization of Rtt109. In the presence of completely mislocalized Vps75 NLS mutants, Rtt109 can be seen in the cytoplasm. It is possible that cytoplasmic Vps75 sequesters Rtt109, however, the fact that the Rtt109 N:C ratio is as low, or lower, in vps75Δ cells suggests that Vps75 is actually required for normal nuclear localization of Rtt109. Vps75 may promote the import of Rtt109, or alternatively Vps75 could function in the nuclear retention of Rtt109. The idea that Vps75 and Rtt109 interact outside of the nucleus is an appealing model, because it would explain how nascent Rtt109 is protected from degradation. In this model, Vps75 would bind to newly synthesized Rtt109 in the cytoplasm to simultaneously promote efficient import of Rtt109 and to protect it from degradation throughout the transport process. We also detected Rtt109 and Vps75 in overlapping foci inside the nucleus. We speculated that these foci may be related to the roles Rtt109 and Vps75 play in DNA repair and genomic integrity (21, 28, 30, 56–59). Cells lacking Rtt109 are hypersensitive to DNA damaging agents and have increased numbers of Rad52 foci (59). In human cells it has been shown that H3 K56ac is enriched at DNA damage foci and there is evidence that H3 K56ac persists at the sites of damage in yeast also (60, 61). If the subnuclear foci we observed are actually sites of DNA damage, the fact that Vps75 and Rtt109 are independently targeted to them could indicate a common function in DNA damage repair. Their role could potentially be histone modification, such as H3 K9ac or K56ac, or regulating histone exchange at those sites of damage. Future experiments will determine whether they are sites of DNA damage and colocalize with Rad52 or other DNA damage repair proteins.
Cytoplasmic Vps75 mutants restore Rtt109 function
Our experiments have demonstrated that the Vps75 NLS mutants functioned equivalently to WT Vps75 in the stabilization of Rtt109, in Rtt109-mediated acetylation of H3 K9 and in reversing gcn5Δvps75Δ growth defects. In these strains, however, much of the Vps75 and a proportion of Rtt109 were observed in the cytoplasm. While we cannot rule out the possibility that there were enough Vps75-Rtt109 complexes remaining in the nucleus to carry out their functions, it is possible that cytoplasmic Vps75-Rtt109 complexes were active and functional. The idea that Rtt109 in complex with Vps75 might be able to acetylate H3 in either the nucleus or the cytoplasm is consistent with the Rtt109 targets H3 K9, K27 and K56 being early marks on nascent histones before incorporation into chromatin and the demonstration that Rtt109 acetylates only non-nucleosomal histones (34, 61, 62). This may be analogous to the acetylation of H4 K5 and K12 by Hat1, which occurs in the cytoplasm (63, 64).
Vps75-Rtt109 binding promotes Rtt109 activity
Vps75 was initially thought to be one of two H3/H4 chaperones needed for Rtt109 catalytic activity, because in vitro either Vps75 or Asf1 could promote Rtt109 acetylation of H3 (20). However, as both Asf1 and Vps75 are required for Rtt109-mediated acetylation of the H3 tail in vivo, it is unlikely that they are performing the same specific function. This suggests that Vps75 has functions that are not redundant with Asf1.
One of the primary functions unique to Vps75 is protection of Rtt109 from degradation. Surprisingly, this function is not necessary for Rtt109-mediated acetylation of H3 K56, as in the absence of Vps75 there is sufficient Rtt109 for robust H3 K56 acetylation. In contrast, Vps75 is essential for Rtt109-mediated acetylation of H3 K9 and K27, and this could potentially be explained by its role in Rtt109 stability. However, even when Rtt109 is stable due to deletion of the Vps75 binding loop, our data reveal that Vps75 binding to Rtt109 serves an important purpose beyond protecting it from degradation. Rtt109 that cannot interact with Vps75 loses the ability to efficiently acetylate H3 K9 and H3 K27. This suggests the formation of the Vps75-Rtt109 complex is necessary for efficient HAT activity, which could be due to holding Rtt109 in an active conformation (65). While this manuscript was under revision, a new X-ray crystal structure of the Rtt109-Acetyl CoA-Vps75 complex was published, which showed a symmetrical ring made of two molecules each of Rtt109 and Vps75 with a hole in the middle for substrate binding (66). This structure and other data presented in the manuscript provide further support that that, in vivo, Vps75 binding is necessary for H3 tail acetylation by Rtt109 and that Rtt109(128–170) contains the key binding domain for Vps75 (66).
We did observe a small amount of H3 K9 acetylation with Rtt109ΔL; however, this only occurred in the presence of Vps75. In the original in vitro studies with recombinant proteins, no binding was detected between Rtt109ΔL and Vps75, and we verified this lack of interaction in yeast lysates (49). The small amount of H3 K9 acetylation observed could be due to Rtt109ΔL interaction with Vps75 at levels that are below the detection limit of our coimmunoprecipitation assays. This would explain why the activity of Rtt109ΔL was dependent on the presence of Vps75. Consistent with this idea we did detect very low levels of binding with recombinant Rtt109ΔL in our in vitro binding assays, and a similar mutant, Rtt109Δ130–179, was shown to bind Vps75 at low levels in vitro (data not shown) (50). Although less likely, we cannot exclude the alternative explanation that Rtt109ΔL does not interact with Vps75 in vivo, but can acetylate H3 K9 in a Vps75 binding-independent manner, albeit much less efficiently than wild type Rtt109 in complex with Vps75. This activity is still dependent on the presence of Vps75 in the cell. In either situation it is likely that Vps75 interacts with the histones and helps present them as substrates to Rtt109. Thus, we propose a model (FIG 6D top) in which the H3 tail typically exists in a conformation such that Gcn5 but not Rtt109 can acetylate K9 and 27. Direct physical interaction of Vps75 with H3 could then cause a conformational change in the histone tail, allowing Rtt109 access to H3 lysines 9 and 27 for acetylation. Therefore, significant acetylation of H3 K9 and K27 in gcn5Δ would require both Vps75 and Rtt109. In addition, our data demonstrate that Rtt109-Vps75 binding is essential for H3 K27 acetylation. The ability of Rtt109ΔL to acetylate a modest amount of H3 K9 but no K27 is most likely explained by studies that have demonstrated that H3 K27 is a less efficient in vitro substrate for Rtt109 than H3 K9 (65).
Vps75 is important for cell growth in the absence of RTT109 and GCN5
Our genetic data suggest that Vps75 has functions that are independent of Rtt109, consistent with histone chaperones being multifunctional proteins and with published data (9, 10, 45). Previous evidence for Rtt109-independent functions of Vps75 includes comparison of the epistatic miniarray profiles of Rtt109 and Vps75, the fact that Vps75 regulates transcription of different genes from those regulated by Rtt109 and that recruitment of Vps75 to chromatin is not dependent on Rtt109 (45). We show for the first time genetic interactions between VPS75 and ASF1 and between GCN5, VPS75 and RTT109, which provide further support for Rtt109-independent functions of Vps75. In the absence of Gcn5 and Rtt109, chromatin lacks key histone acetylation marks, primarily on the H3 tail, which causes increased sensitivity to loss of Vps75. While this effect could be direct or indirect, it is tempting to speculate that this sensitivity may be related to other histone modifications. It is possible that the interaction of Vps75 with H3 is of more general importance, and Vps75 also assists other HATs or histone modifying enzymes in gaining access to the H3 tail (see model, FIG 6D, middle). These activities might be more critical when GCN5 and RTT109 are deleted, because there is some redundancy between histone modifications. Without the acetylation marks provided by Gcn5 and Rtt109, the other modifications that may be promoted by Vps75 could become more important for regulation of processes such as DNA replication, repair and transcription.
Alternatively, the dependence on Vps75 in the gcn5Δrtt109Δ background might relate to the histone exchange and chromatin assembly functions of Vps75 rather than promoting other histone modifications (FIG 6D bottom). Changes to chromatin can occur through exchange of histones with different modifications or through exchange of canonical histones with histone variants (5). In these situations, histone chaperones and chromatin remodeling enzymes must work together to ensure that properly modified histones or histone variants are being exchanged at the correct regions of the chromatin. Vps75, like other histone chaperones, may help promote the exchange of histones with specific modifications, but these functions of Vps75 are only beginning to be elucidated (FIG 6D bottom). There is evidence that Vps75 regulates replication-independent histone turnover (45, 67). Although these experiments were not performed in the absence of Rtt109 or any other HAT, these histone exchange functions of Vps75 could certainly be impacted by loss of two major HATs and the modifications they create. Thus, it might be in this way that deletion of VPS75, GCN5 and RTT109 results in synthetic sickness. In addition, the single deletion of the H3/H4 chaperone Asf1 results in the loss of Rtt109-mediated acetylation of both H3 K9 and K56. Therefore, the growth defect observed when both ASF1 and VPS75 are deleted further suggests that Vps75 has roles in the cell beyond the regulation of Rtt109-mediated acetylation of these sites (33, 52).
In summary, we have shown that the histone chaperone Vps75 is a nuclear protein that plays several important roles in the regulation of Rtt109 acetylation activity. Through direct interaction, Vps75 ensures that Rtt109 is efficiently localized in the nucleus, stabilizes Rtt109 and presents histones to Rtt109 for acetylation. Many questions remain regarding these proteins in yeast: if Vps75, not Asf1, is presenting the H3 tail to Rtt109, what essential role is Asf1 playing? Are Gcn5 and Rtt109 redundant in acetylating H3 K9 and K27 or is Rtt109 acting on a subset of histones during S phase? As Rtt109 can acetylate H3 K56 in vps75Δ yeast, is it then dimeric, monomeric or in complex with Asf1? We also show that Vps75 has Rtt109-independent functions, most likely related to its ability to chaperone histones during the remodeling of chromatin, suggesting Vps75 is a multifunctional protein that influences multiple chromatin-templated processes.
Materials and methods
Yeast strains and plasmids
All yeast strains are haploid S. cerevisiae and derived from S288C/BY4741 except Kap mutant strains, which are derived from the following strains in W303, DF5 and BY4741 backgrounds: kap60-ts (srp1–31) (68); kap95-ts (a kind gift of Dr. John Aitchison), kap114Δ (69), kap123Δ (70), kap121–34 (71), kap108Δ (72), kap119Δ (73), kap122Δ (Open Biosystems), kap104Δ (74), kap111Δ (75), kap142Δ (76) and kap120Δ (a generous gift of Drs. Susana Chaves and Günter Blobel). Standard procedures for yeast manipulation were used. The following strains were obtained from the Open Biosystems deletion collection in which ORFs were replaced with KanMX: vps75Δ, gcn5Δ, rtt109Δ, asf1Δ and nap1Δ. vps75::NatMX and nap1::NatMX strains were generated from Y2454 (MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 MFA1pr-HIS3 can1Δ0) and mated with the strains above to produce the following haploid strains: nap1::NatMX vps75::KanMX; gcn5::KanMX vps75::NatMX; gcn5::KanMX rtt109::HIS3; gcn5::KanMX vps75::NatMX rtt109::HIS3; and rtt109::KanMX vps75::NatMX. The strains asf1Δvps75Δ, VPS75-TAP rtt109::KanMX and RTT109-TAP vps75::KanMX were generous gifts of Drs. Fillingham and Greenblatt (33). VPS75-TAP and RTT109-TAP strains were obtained from OpenBiosystems. rtt109ΔL-TAP was made by genomic replacement of the wild type allele with rtt109ΔL, which was amplified from a plasmid generously provided by Drs. Stavropoulos and Blobel (49). Vps75-mCherry strain was created using plasmid pFA6a-mCherry-Sp HIS5 (a kind gift of Drs. Westfall and Thorner, similar to pPW58 described in (77)). Plasmids pGEX-Rtt109 and pGEX-Kap60 were made by insertion of the coding sequences into the plasmid pGEX-4T1. For MBP tagged proteins, pMal-Vps75 (wild type, 1–222, 1–256, 243–264 and K260G,K261G) plasmids were made by inserting VPS75 coding sequences into pMal-c2X (NEB). Plasmid sequencing revealed mutation of a single base, A267T, which caused the semiconservative amino acid substitution E89D in all Vps75 plasmids. The plasmids pGFP2-C-FUS, pNap1-GFP2 and pYra1-GFP2 were previously described and include the inducible MET25 promoter (13, 14, 16). Rtt109-GFP2 and Vps75-GFP2 plasmids (wild type, 1–222, 1–256, 243–264 and K260G,K261G) were made by inserting RTT109 or VPS75 coding sequences into pGFP2-C-FUS. The HA-Vps75 plasmid pRS316-VPS75 promoter-HA-VPS75-CYC1 terminator was created by inserting the promoter region 480bp upstream of VPS75 start, HA-VPS75 (VPS75 coding sequence or mutant versions preceded by a single HA epitope) and the CYC1 terminator into pRS316. Rtt109-Flag or Rtt109ΔL-Flag plasmids were created by insertion of the appropriate RTT109 sequence with a C-terminal Flag epitope into p415-ADH1 promoter. All plasmids were sequenced.
Growth assays
Liquid cultures were grown to mid/late log phase, equalized by OD600, then 10 fold serial dilutions were plated. All plates have dextrose (except where galactose is indicated) and were grown at 30°C unless otherwise noted and with added drugs or lacking inositol as noted. Pictures were taken of all plates in each set at the same time.
Microscopy
Microscopy of strains expressing GFP plasmids or of the VPS75-mCherry strain was performed using a Nikon Microphot-SA microscope (Melville, NY) and imaging was done using OpenLab software (Improvision, Lexington, MA). All images for each fluorophore in each set of panels were photographed with identical exposure settings and manipulated identically in Adobe Photoshop. Induction of the GFP reporter in media lacking methionine for the times noted in figure legends was performed for some strains. Images used for quantitation of N:C ratios were taken at a consistent exposure time that minimized pixel saturation and captured in 12-bit format. Boxes with an area of 36 pixels were drawn in the nucleus and cytoplasm (excluding vacuoles) of each cell (number of cells used for each >190) and the mean pixel value in each was measured and used to calculate N:C ratio of fluorescence intensity. The mean N:C ratio for each strain is reported plus the standard error which was calculated using a 2-tailed student’s t-Test.
Purification of recombinant proteins and in vitro binding assays
Bacterially expressed GST-Rtt109 and GST-Kap60 were purified on glutathione sepharose and MBP-LacZ, MBP-Vps75 were purified on amylose agarose essentially as described (78). In vitro binding assays were performed essentially as described with recombinant proteins at concentrations indicated below (78). In the histone binding assay, 4µg chicken erythrocyte histones were used per binding assay and GST and GST-Vps75 were used at 1µM and 0.5µM, respectively. In the Kap60 and Rtt109 binding assays, GST and GST-Rtt109 were used at 0.5µM and GST-Kap60, MBP-LacZ, MBP-Vps75, MBP-Vps75(K260G, K261G), MBP-Vps75(1–222) and MBP-Vps75(243–264) were used at 0.25µM, and the blot was probed with anti-MBP antiserum (NEB).
Immunoprecipitations
For Rtt109-Flag and Rtt109ΔL-Flag: Cultures of rtt109Δvps75Δ yeast expressing HA-Vps75 and either Rtt109-Flag or Rtt109ΔL-Flag were grown in complete synthetic medium lacking leucine and uracil to an optical density at 600nm (OD600) of ~1.6. Sixty five OD600 units were collected of each, washed and spheroplasted using 10% glusulase. Lysates were prepared by sonication of cells in 8% polyvinylpyrrolidone, 0.025% Triton X-100, 5mM dithiothreitol with protease inhibitors and incubated with Flag agarose after removal of an input sample. After washing Flag agarose, bound proteins were eluted using SDS-PAGE loading buffer at 37°C. SDS-PAGE resolution of eluted proteins was followed by transfer to PVDF membrane and Western blotting using antibodies against Flag and HA. Immunoprecipitation of Vps75-TAP was performed similarly except as follows: VPS75-TAP rtt109Δ yeast expressing Rtt109-Flag and Rtt109ΔL-Flag plasmids were grown in CSM minus leucine, the resultant lysates were incubated with IgG sepharose and Western blots were performed using antibodies against IgG and Flag. Antibodies were obtained from the following sources: Flag M2 from Sigma, HA 12CA5 from UVA hybridoma center, H3 and H3 K9ac from Abcam, H3 K27ac from Active Motif, H3 K56ac from Millipore/Upstate, Pol II (recognizes Rpo21 subunit of RNAP II) from Covance, IgG from MP Biomedicals/Cappel, and PGK1 from Invitrogen.
ACKNOWLEDGEMENTS
We would like to thank Pete Stavropoulos and Günter Blobel for the Rtt109ΔL plasmid, Jeffrey Fillingham and Jack Greenblatt for yeast strains, Patrick Westfall and Jeremy Thorner for the mCherry plasmid and Brian Del Rosario and Ann Beyer for helpful comments on the manuscript. This work was supported by research grant R01 GM65385 from the National Institutes of Health.
REFERENCES
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142(5):682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelato KA, Fischle W. Role of histone modifications in defining chromatin structure and function. Biol Chem. 2008;389(4):353–363. doi: 10.1515/BC.2008.048. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochim Biophys Acta. 2009;1789(1):26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18(3):282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocha W, Verreault A. Clothing up DNA for all seasons: Histone chaperones and nucleosome assembly pathways. FEBS Lett. 2008;582(14):1938–1949. doi: 10.1016/j.febslet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16(2):104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37(6):834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14(11):997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 10.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65(3):414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zlatanova J, Seebart C, Tomschik M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 2007;21(7):1294–1310. doi: 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 12.Park YJ, Luger K. Structure and function of nucleosome assembly proteins. Biochem Cell Biol. 2006;84(4):549–558. doi: 10.1139/o06-088. [DOI] [PubMed] [Google Scholar]
- 13.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J Cell Biol. 2001;153(2):251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21(23):6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohkuni K, Shirahige K, Kikuchi A. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2003;306(1):5–9. doi: 10.1016/s0006-291x(03)00907-0. [DOI] [PubMed] [Google Scholar]
- 16.Del Rosario BC, Pemberton LF. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol Cell Biol. 2008;28(7):2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M. Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci U S A. 2007;104(11):4285–4290. doi: 10.1073/pnas.0603762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y, Meeth K, Jiang E, Luo C, Marmorstein R. Structure of Vps75 and implications for histone chaperone function. Proc Natl Acad Sci U S A. 2008;105(34):12206–12211. doi: 10.1073/pnas.0802393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25(5):703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282(39):28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 22.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 23.Bazan JF. An old HAT in human p300/CBP and yeast Rtt109. Cell Cycle. 2008;7(12):1884–1886. doi: 10.4161/cc.7.12.6074. [DOI] [PubMed] [Google Scholar]
- 24.Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16(10):1503–1510. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Tang Y, Cole PA, Marmorstein R. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008;18(6):741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 27.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13(7):2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315(5812):649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315(5812):653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 30.Schneider J, Bajwa P, Johnson FC, Bhaumik SR, Shilatifard A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J Biol Chem. 2006;281(49):37270–37274. doi: 10.1074/jbc.C600265200. [DOI] [PubMed] [Google Scholar]
- 31.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446(7137):806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 32.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15(9):948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28(13):4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383(6597):269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 35.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 2010;37(4):469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6(3):187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 37.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 38.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112(4):441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 39.Lusk CP, Makhnevych T, Wozniak RW. New ways to skin a kap: mechanisms for controlling nuclear transport. Biochem Cell Biol. 2004;82(6):618–625. doi: 10.1139/o04-111. [DOI] [PubMed] [Google Scholar]
- 40.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008;15(9):957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calvert ME, Keck KM, Ptak C, Shabanowitz J, Hunt DF, Pemberton LF. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Mol Cell Biol. 2008;28(4):1313–1325. doi: 10.1128/MCB.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16(6):3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem. 2007;282(17):12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 45.Selth LA, Lorch Y, Ocampo-Hafalla MT, Mitter R, Shales M, Krogan NJ, Kornberg RD, Svejstrup JQ. An rtt109-independent role for vps75 in transcription-associated nucleosome dynamics. Mol Cell Biol. 2009;29(15):4220–4234. doi: 10.1128/MCB.01882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J Biol Chem. 2001;276(2):1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 47.Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21(13):4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albaugh BN, Kolonko EM, Denu JM. Kinetic mechanism of the Rtt109-Vps75 histone acetyltransferase-chaperone complex. Biochemistry. 2010;49(30):6375–6385. doi: 10.1021/bi100381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci U S A. 2008;105(34):12236–12241. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, Marmorstein R. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15(7):738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13(12):1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 52.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103(18):6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosammaparast N, Del Rosario BC, Pemberton LF. Modulation of histone deposition by the karyopherin kap114. Mol Cell Biol. 2005;25(5):1764–1778. doi: 10.1128/MCB.25.5.1764-1778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blackwell JS, Jr, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J Biol Chem. 2007;282(28):20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 55.Mosammaparast N, Guo Y, Shabanowitz J, Hunt DF, Pemberton LF. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J Biol Chem. 2002;277(1):862–868. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- 56.Jessulat M, Alamgir M, Salsali H, Greenblatt J, Xu J, Golshani A. Interacting proteins Rtt109 and Vps75 affect the efficiency of non-homologous end-joining in Saccharomyces cerevisiae. Arch Biochem Biophys. 2008;469(2):157–164. doi: 10.1016/j.abb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Peterson CL. Genome integrity: a HAT needs a chaperone. Curr Biol. 2007;17(9):R324–R326. doi: 10.1016/j.cub.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Downs JA. Histone H3 K56 acetylation, chromatin assembly, and the DNA damage checkpoint. DNA Repair (Amst) 2008;7(12):2020–2024. doi: 10.1016/j.dnarep.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 59.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3(12):e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436(7048):294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 62.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282(19):14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 63.Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273(20):12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 64.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87(1):85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 65.Kolonko EM, Albaugh BN, Lindner SE, Chen Y, Satyshur KA, Arnold KM, Kaufman PD, Keck JL, Denu JM. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc Natl Acad Sci U S A. 2010;107(47):20275–20280. doi: 10.1073/pnas.1009860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Y, Holbert MA, Delgoshaie N, Wurtele H, Guillemette B, Meeth K, Yuan H, Drogaris P, Lee EH, Durette C, Thibault P, Verreault A, Cole PA, Marmorstein R. Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure. 2011;19(2):221–231. doi: 10.1016/j.str.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4(11) doi: 10.1371/journal.pgen.1000270. e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci U S A. 1995;92(17):7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pemberton LF, Rosenblum JS, Blobel G. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J Cell Biol. 1999;145(7):1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89(5):715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 71.Leslie DM, Grill B, Rout MP, Wozniak RW, Aitchison JD. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol Cell Biol. 2002;22(8):2544–2555. doi: 10.1128/MCB.22.8.2544-2555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139(7):1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albertini M, Pemberton LF, Rosenblum JS, Blobel G. A novel nuclear import pathway for the transcription factor TFIIS. J Cell Biol. 1998;143(6):1447–1455. doi: 10.1083/jcb.143.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274(5287):624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 75.Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139(7):1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida K, Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152(4):729–740. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Westfall PJ, Thorner J. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot Cell. 2006;5(8):1215–1228. doi: 10.1128/EC.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Straube K, Blackwell JS, Jr, Pemberton LF. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic. 2010;11(2):185–197. doi: 10.1111/j.1600-0854.2009.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]