Abstract
We describe here a method, Quarc, for accurately quantifying structural changes in organs, based on serial MRI scans. The procedure can be used to measure deformations globally or in regions of interest (ROIs), including large-scale changes in the whole organ, and subtle changes in small-scale structures. We validate the method with model studies, and provide an illustrative analysis using the brain. We apply the method to the large, publicly available ADNI database of serial brain scans, and calculate Cohen’s d effect sizes for several ROIs. Using publicly available derived-data, we directly compare effect sizes from Quarc with those from four existing methods that quantify cerebral structural change. Quarc produced a slightly improved, though not significantly different, whole brain effect size compared with the standard KN-BSI method, but in all other cases it produced significantly larger effect sizes.
Keywords: Nonlinear image registration, Regional change quantification, MRI biomarkers
Introduction
Structural change in organs takes place on many time scales, and can often be seen on Magnetic Resonance Imaging (MRI) scans. In the brain, for example, normal aging, neonatal development, and many neurological disorders are associated with structural change. Individual disorders will have their own patterns of tissue deformation and evolution of tissue atrophy or hypertrophy, though with variability across subjects. To discriminate pathologies, especially for early diagnosis, patterns in the onset and development of change need to be understood quantitatively. This requires precise in vivo detection and measurement of structural changes in individuals. Alzheimer’s disease (AD), for example, is believed to be irreversible, so early diagnosis is key for therapeutic e orts aimed at slowing or halting its development to be successful (DeKosky and Marek, 2003). Progressive atrophy in AD arises from dendrite and neuron loss (Bobinski et al., 2000; Braak and Braak, 1991), and can be seen on structural MRI scans (Atiya et al., 2003; Ramani et al., 2006). Currently, however, longitudinal structural scans are typically not used for diagnosis, a situation that does not reflect inherent or technological limitations in MRI. To the contrary, tremendous anatomical detail can be obtained from MRI, but is by no means fully exploited clinically—even something as relatively simple as rigid body alignment of a subject’s serial scans is not routinely performed, yet its utility is obvious (Bradley and Bydder, 1997).
We report here on a numerical method based on serial MRI scans that allows for early detection of anatomical changes (Murphy et al., 2010), including those of small ROIs, and their quantification over time. This computational MR image processing application, called Quarc (quantitative anatomical regional change), involves image smoothing and minimization of a regularized merit function for the displacement field that locally registers the serial images (Ashburner et al., 1999). It has been applied extensively to serial pairs of scans in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (McEvoy et al., 2011, 2010; Fjell et al., 2010c,b,a; Murphy et al., 2010; McDonald et al., 2010; Holland et al., 2009; Fjell et al., 2009; McDonald et al., 2009; McEvoy et al., 2009), and is one of several methods already used in research that tackle the problem of volumetric change and local longitudinal registration, e.g., fluid registration (Christensen et al., 1996; Miller et al., 1993; Freeborough and Fox, 1998), Tensor Based Morphometry (TBM) (Leow et al., 2007; Yanovsky et al., 2009; Hua et al., 2010), Voxel based Morphometry (VBM) (Ashburner and Friston, 2000; Tzourio-Mazoyer et al., 2002), and the Boundary Shift Integral (BSI) (Freeborough and Fox, 1997; Leung et al., 2010). Here we describe the method in detail, validate it with model studies, and directly compare with several of the existing methods—using results of power calculations on Cohen’s d effect size estimates for detecting ROI change, relative to normal aging, in AD.
Methods
ADNI
All data used in the preparation of this article were obtained from the ADNI database (www.loni.ucla.edu/ADNI). ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. ADNI’s goal is to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. ADNI is the result of e orts of many co-investigators from a broad range of academic institutions and private corporations. ADNI has recruited 229 cognitively normal individuals to be followed for 3 years, 398 people with MCI to be followed for 3 years, and 192 with mild AD to be followed for 2 years (see www.adniinfo.org). The research protocol was approved by each local institutional review board and written informed consent is obtained from each participant.
Participants
The ADNI general eligibility criteria have been described elsewhere (Petersen et al., 2010). Briefly, subjects are not depressed, have a modified Hachinski score of 4 or less, and have a study partner able to provide an independent evaluation of functioning. Healthy control (HC) subjects have a Clinical Dementia Rating (CDR) of 0. Subjects with MCI have a subjective memory complaint, objective memory loss measured by education-adjusted scores on Wechsler Memory Scale Logical Memory II, a CDR of 0.5, preserved activities of daily living, and absence of dementia. Subjects with AD have a CDR of 0.5 or 1.0 and meet National Institute of Neurological Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association criteria for probable AD.
Subject Data Acquisition and Preparation
Subjects are scanned twice with a 3D MPRAGE protocol at 1.5T every six-months. Double scanning allows up to a increase in signal-to-noise. At each time point, to provide a faithful geometric representation of the subject, reduce noise, and be in a position to consistently determine over time what parts of the brain are growing, shrinking, or merely being displaced, and by how much, several processing steps are required. In general for any subject, no two images will be in register, they will be spatially distorted (see below), have different intensity ranges since there is no standard for scanners, and suffer from local intensity and contrast variability due to magnetic susceptibility of the subject and inhomogeneities in the radio-frequency magnetic field pulse (B1 RF-field) excitation and signal detection.
Before images can be registered in any way, they first must be corrected for deformation artifacts resulting from nonlinearities in the space-encoding gradient magnetic field in the scanner (Wald et al., 2001). The effect can be significant, with distortion of the field of view up to a few centimeters, depending on the scanner. The correction method is based on a spherical harmonic description of the gradient magnetic field that requires vendor-supplied coeffcients (Jovicich et al., 2006). Any remaining distortions in structural images are affne. Corrected images are written out in a 2563 voxel cube, where each voxel is a 1mm3 cube.
One of the images from a subject’s baseline pair is automatically segmented (Fischl et al., 2002), and a simply connected uniform brain mask, covering all sulci, is built from the segmentation by repeatedly smoothing with a Gaussian kernel. The mask is used to assist in registering, and the segmentation is used later to quantify ROI change.
At each subject-visit, gradient-corrected image pairs are rigid registered by minimizing a cost function:
| (1) |
N is the number of voxels in the image; is the location of the center of voxel i, whose intensity in the source image is denoted by , and in the target image is denoted ; α1, …, α6 are the translation and rotation parameters, with the current estimate of the rigid body transformation operator (Ashburner et al., 1997; Frackowiak et al., 2004a) which acts on the spatial coordinates of voxels in S; mi is the target mask value for voxel i (equal to 1 inside the skull, tapering to 0 over the length of a few voxels outside the skull and inferior to the brain. As this is a highly over-determined problem, the sum need only be carried out over every second or even fourth voxel along each dimension. Registered images are then averaged. For purposes of non-rigid affne registration, the brain mask is enlarged, again using Gaussian smoothing, to include the scalp (bright on T1 MPRAGE). Using the larger mask, each subject’s followup averaged image is fully (12-parameter) affne registered to the subject’s baseline. The larger mask is used because, when affne registering severely atrophied brains to baseline, care must be taken to include the skull (or more specifically, the sharply contrasting outer-skull/scalp interface) so as not to subsume an anatomical effect in a scanner artifact. The assumption underlying this is that the skull has not changed its overall shape and size during the followup period and that, in general, there will be non-rigid affne differences between scans acquired at serial scanning sessions. It is appropriate to use the full 12-parameters, without making any assumption that only 9-parameters are all that are required: if it so happens that only 9 are needed, only 9 will be fit if the affne registration algorithm is working correctly. Full affne registration can be carried out by minimizing a cost function similar to Eq. 1 with respect to a 12-parameter L: translations, rotations, uniaxial strains and shears. After this step, skulls (on T1 MPRAGE, black outer shell and thicker inner regions of medium intensity arising from cranial bone marrow) at followup times should match baseline skulls with high fidelity. Cubic interpolation, indistinguishable from sinc interpolation, is used throughout for resampling (Hajnal et al., 1995). The intensities are then globally rescaled by bringing the cumulative probability distribution of brain voxel intensities (within the brain mask) into close agreement with that of a standard, where cerebrospinal fluid ~25 and white matter ~150 (arbitrary scale).
Nonlinear Registration Cost Function
Nonlinear registration involves finding the individual three-dimensional displacements at each voxel that map the source image to the target (Frackowiak et al., 2004b). This is also carried out by minimizing a cost function, in general depending on 3N variables, where N is the total number of voxels in the image problem. A suitable cost function f is
| (2) |
It is assumed here that the images S and T have been affne aligned and spatially normalized (discussed in detail below). is the displacement at voxel i such that at the correct local registration, the resampled image S′ will look practically identical to T, where . For highest accuracy, the sum now involves all voxels. λ1 and λ2 are regularization parameters of the model that control the quality of the registration. They will generally have a wide range of valid values, but that range depends on the scale of intensities. With the standardized images in this study, we used λ1 = 0 and λ2 = 1500. Serial images that involve local change in T1, T2, or proton density, or that are topologically distinct, for example where a tumor is present in one but not the other, introduce additional complexity which requires modeling tissue contrast change and the use of mutual information (Studholme et al., 2006).
Minimization
Since the images being locally registered are already tightly affne registered, if each is heavily smoothed or blurred, e.g., by convolving with an isotropic Gaussian kernel of standard deviation ≃4–5 mm, they will look rather similar, even though the unsmoothed images may have relative internal deformations on the order of a centimeter. The minimization carried out at any level of image smoothing will be done most expeditiously if as much gradient information as possible is taken into account—conjugate directions in a second-order Newton’s method (Gershenfeld, 1999). The high smoothing and overall similarity of the images means a second-order Taylor expansion of f is a reasonable approximation—with enough smoothing one is in a concave basin of attraction. So one can perturb the quadratic form approximation for f around the current best estimate of the system displacement vector, and one has a large, sparse, symmetric linear algebra problem:
| (3) |
where is the Hessian of f at , is the gradient of f at , and the unknown quantity is the displacement around that nudges f toward the new global minimum at the new (lower) level of smoothing. The sparseness results from the coupling only of neighboring voxels through the derivative terms in H. There exist several effcient iterative methods for finding the solution of Eq. 3, e.g., conjugate gradients squared (CGS), generalized minimal residuals (GMRES), and biconjugate gradients stabilized method (Bi-CGSTAB) (van der Vorst, 1992, 2003). All results presented here were obtained using Bi-CGSTAB. The cumulative minimization thus carried out over a series of images of decreasing smoothness constitutes a single nonlinear registration step.
In summary, nonlinear registration is carried out as follows: (0) initialize the displacement field to zero, and the standard deviation σ of the smoothing kernel to 4–5 mm; (1) smooth the affne registered images; (2) set up the Hessian and gradient for the current net displacement field; (3) register the smooth images, i.e., calculate the refinement to the deformation field by solving Eq. 3; (4) update the deformation field: ; (5) decrease the width of the smoothing kernel by 0.25–0.5 mm. Repeat (1)–(5) until there is only a little smoothing, σ ≈ 1mm.
More accurate registration can be achieved by performing multiple nonlinear registration steps, as described above, but where the image being transformed is updated from the previous step, essentially greedy nonlinear registration. Note that successive displacement fields (and affne transformations) can be combined into a single deformation field so that only a single resampling from the starting image need take place at any stage, thus avoiding cumulative blurring. Fold-over in the deformation field, or negative Jacobian of the transformation, can be detected by reporting hexahedral volume elements with negative volume. It is not explicitly precluded by Eq. 2—a regularization term of the form λln(∣Ji∣), for example, where λ is a regularization parameter and Ji is the Jacobian of at , would facilitate this (Ashburner et al., 2000)—but can be avoided entirely in practice by sti ening the deformation field, i.e., by having a suffciently large λ2.
Local Intensity Normalization
Images are not always uniformly bright due both to variations in the local flip angle—nonuniform excitation caused mainly by RF-field (B1) inhomogeneities and the subject’s own magnetic susceptibility—and to nonuniform detection sensitivity of the receiver coil (Liang and Lauterbur, 2000), with the nonuniform brightness pattern potentially varying substantially from scan to scan. Use of head coils instead of body coils for brain imaging, though allowing better signal-to-noise ratio, can lead to images with pronounced intensity variation. If uncorrected, this will result in false expansion or contraction when performing nonlinear registration. Many methods have been developed over the years to ameliorate this situation, e.g., Sled et al. (1998), but subtle variation can remain that would corrupt sensitive measures of change. One can, however, with high accuracy make the images consistent (Lewis and Fox, 2004).
If the only assumption about the relative intensity variation between a pair of images is that it is spatially smooth, an accurate map of it can be made by minimizing a cost function of the form (in notation analogous to Eq. 2)
| (4) |
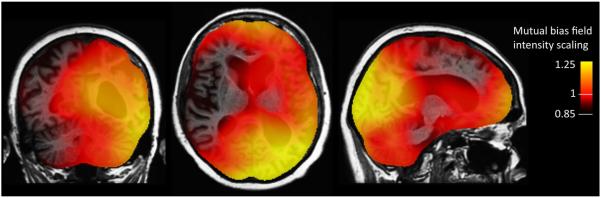
in the manner used for nonlinear registration, where bi is the intensity scaling factor at voxel i in image S that makes the intensity at that location closely agree with the intensity at voxel i in registered image T; γ1 and γ2 are regularization parameters. Since intensity nonuniformity in practice varies only very gently across images, this procedure promises to be very precise and robust. Intensity normalization and structural registration should be run iteratively in a self-consistent manner until convergence is reached—one helping perfect the other. Optimal γ1 and γ2 can be found simply by numerically exploring their impact on intensity normalization for geometrically almost identical images suffering from intensity distortion; typical values for the standard images used here are 0 and 105, respectively. Note that the smoothness of the intensity variation means that the sum in Eq. 4 need not be carried out over all voxels—every second voxel is reasonable. Results for a baseline and two-year followup scan-pair involving large structural change (e.g., left lateral ventricle expanded by 33%) are shown in Fig. 1.
Figure 1.
Mutual bias field correction: local intensity scaling of two-year followup image required to make anatomically identical points in it and the baseline image have similar intensities. The opacity of the intensity scaling field is reduced to show brain structure underneath.
For each ADNI subject used in this study, the 12-month time point was intensity normalized to the baseline image using this method, using up to five relaxation steps. The followup time point thus normalized could then directly be nonlinearly registered to the baseline image, using multiple (usually five) relaxation steps, with the incremental updates to the displacement field becoming exponentially smaller.
A single cost function, combining Eq. 2 and Eq. 4, that takes into account the coupled problems of structural change and mutual intensity inhomogeneity is
| (5) |
The b-u coupling makes the Hessian more complicated, and the problem space potentially grows to 4N dimensions, but this approach allows for the true displacement field to be calculated in a straightforward manner while simultaneously correcting for variations in intensity.
ROI Volume
The displacement field specifies where to locate voxel centers in the S image, in general turning cubic voxels into displaced irregular hexahedra. Given the corners of a hexahedron, which can be determined by trilinear interpolation of the displacements at the voxel centers, it is straightforward to calculate its volume—this can be done in several ways (Garg, 1998; Davies and Salmond, 1985), the most efficient being Grandy (1997), where it is given by one-sixth the sum of three determinants of 3 × 3 matrices built from the coordinates of the vertices. So the displacement field tells you how to locate the eight corner points of the hexahedral volume element in S that correspond to the corners of a particular cubic voxel in T. Though it is more usual in morphometric methods to calculate the fractional volume change by calculating the Jacobian of the transformation, it should be noted that here we directly calculate the hexahedral volumes, and knowing the original voxel volumes, the fractional volume change can immediately be found. Either way, a volume-change field is produced. Note also that hexahedral facets will in general not be planar, but volume can nevertheless be defined unambiguously leaving no interstitial gaps (Grandy, 1997).
Regions of interest in the brain can be specified by automated segmentation (Fischl et al., 2002). This method defines voxels as either fully in or not in a particular anatomical ROI—with current versions of automatic segmentation software, ROI boundaries do not traverse voxels.
The volume change for a whole anatomical ROI is obtained by averaging the volume changes for all the voxels identified as being in the ROI, ignoring a thin layer (about a voxel thick) on the boundary—to account for voxels being wholly misidentified, and voxels that inevitably will straddle tissue boundaries. Since the deformation field is designed to be uniform over the ROI, averaging over the interior of the ROI is reasonable.
Bias
To accurately quantify change, it is essential to control for potential bias that can arise in image analysis (Thompson and Holland, 2011). The problem of bias in image registration has been known since the early days of nonrigid morphometric methods (Christensen, 1999; Ashburner et al., 2000; Christensen and Johnson, 2001) and has received a great deal of attention recently, including the development of some general and implementation-specific solutions (Leow et al., 2007; Yanovsky et al., 2009; Reuter et al., 2010). Sources of bias include asymmetries in image smoothing and/or interpolation, and asymmetry in the image matching or regularization term in the cost function employed in image registration. Such bias can be accentuated to varying degrees depending on the interpolation and minimization scheme used (Yushkevich et al., 2010). Bias can also arise simply when averaging the volume-change field over an ROI, due to edge-effects between growing and shrinking regions.
Multiple relaxation steps, as described above, enable high-fidelity registration, as can be seen in Fig. 3 (F’-B), thereby maintaining high accuracy as regards inverse-consistency. However, to guard against potential measurement bias, the entire registration procedure is also run with the images switched, and the average of the volume-change fields in the baseline space is used for subsequent quantification of change.
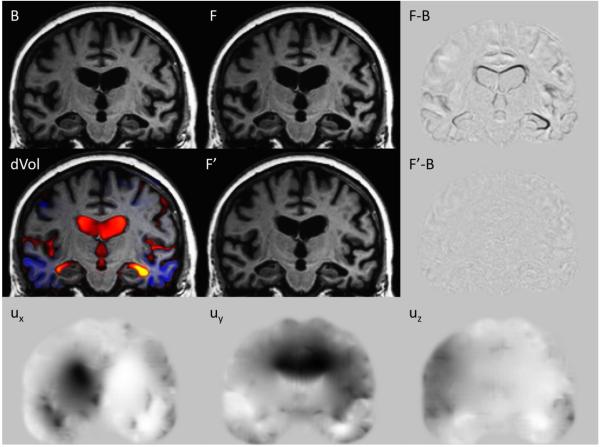
Figure 3.
Nonlinear registration for a subject initially diagnosed with MCI: Baseline B, two-year followup F, and the followup nonlinearly registered back to baseline, F’. F-B is the subtraction of the baseline from the followup, clearly showing the ventricular expansion, along with white matter and cortical shrinkage. The signal in F’-B is reduced close to the noise floor, showing that F’ is a high fidelity registration of F to B. Ux, Uy, and Uz, are the displacement fields effecting the registration; the scale is ±2 voxel lengths (gray background is zero, black is negative, white is positive). dVol is the volume-change field built from the displacement field, overlaid on the baseline image; yellow shows expansion of ~60%, and blue shows shrinkage of ~20% (dVol is reproduced in SI Fig. S3(A), which also provides the color-scale bar).
Results and discussion
Model Studies
To test the accuracy and robustness of the registration method, it was applied to models of two relatively extreme situations that are likely to arise in the context of brain registration: (A) large regions undergoing large volume change, e.g., ventricular expansion and contraction, and (B) small nestled regions involving several subregions of distinct intensities, undergoing small volume changes, e.g., a hippocampus on white matter in the temporal horn of a lateral ventricle. Nested spheres of different intensities were used to test both these cases, Fig. 2. The natural length unit in these studies is the length of a side of a cubic voxel, vl; in the neuroanatomies considered below, a voxel is cubic and of length 1 mm. The scale used for the models’ intensities was set by the standardized images of the brains: CSF ~25, hippocampus ~100, white matter ~150. Rician noise (Gudbjartsson and Patz, 1995; Pannalal, 1998) was added to the intensities, the two Gaussian components of which had a standard deviation of 5, approximately corresponding to noise in the brain images.
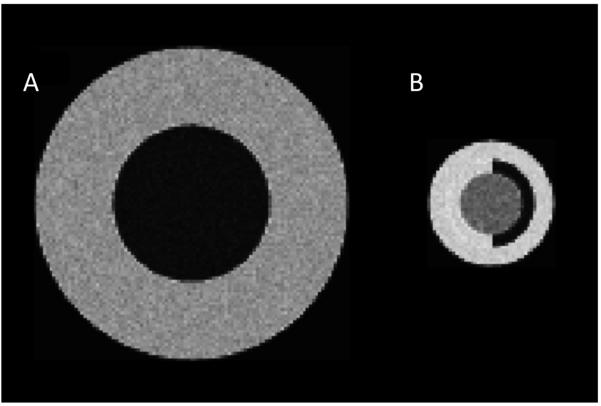
Figure 2.
Slices through spherical models, with Rician noise, undergoing volume change: (A) central sphere “ventricle” (radius R1 = 25 vl), outer shell “white matter” (radius R2 = 50 vl); (B) central sphere “hippocampus” (radius R1 = 10 vl), outer shell “white matter” (inner radius R2 = 14 vl, outer radius R3 = 20 vl), dark semi-shell “temporal horn of lateral ventricle”.
Each model consists of a pair of volume images to be mutually registered. For model-A, only one pair of images was studied. Fig. 2(A) shows a slice through one of these images: the inner sphere’s radius is 25 vl, and the outer radius is 50 vl. The inner sphere in the second image is 30% larger (a radius increase of 2.28 vl); the outer radius did not change. Noisy intensities were distributed around intensity magnitude 25 (inner sphere) and 120 (outer shell).
For model-B, a series of image pairs was studied. Fig. 2(B) shows a slice through one image for one of the pairs: the inner sphere’s radius is 10 vl, the inner radius of the outer shell is 14 vl, and the outermost radius is 20 vl. The inner sphere in the second image is 5% smaller (a radius decrease of 0.17 vl); the outer radii remained fixed. Note that the shell immediately surrounding the center sphere comprises two half-shells, one of which has the same intensity as the outer shell. Noisy intensities were distributed around intensity magnitude 100 (inner sphere), 150 (outer shell), and 25 (half-shell). The series of image pairs were similar except for the inner radius of the outer shell varying from 18 vl down to 11 vl, Table 1.
Table 1.
Mean (standard deviation) percent volume change for inner sphere in Fig. 2(b); actual volume change is −5%
|
R2 – R1 (vl) |
% vol change 1 relax step |
% vol change 2 relax steps |
% vol change 5 relax steps |
% vol change 1 relax step 3× zoom |
% vol change 2 relax steps 3× zoom |
% vol change 5 relax steps 3× zoom |
% vol change 10 relax steps 3× zoom |
|---|---|---|---|---|---|---|---|
| 8 | −3.0 (0.1) | −4.2 (0.2) | −5.3 (0.3) | −3.6 (0.1) | −4.7 (0.2) | −5.2 (0.2) | −5.2 (0.3) |
| 6 | −2.9 (0.2) | −4.2 (0.2) | −5.2 (0.3) | −3.5 (0.2) | −4.6 (0.2) | −5.1 (0.3) | −5.1 (0.3) |
| 4 | −2.8 (0.1) | −4.0 (0.2) | −5.2 (0.3) | −3.4 (0.1) | −4.6 (0.2) | −5.2 (0.2) | −5.2 (0.2) |
| 2 | −1.7 (0.1) | −2.7 (0.2) | −4.4 (0.3) | −3.0 (0.1) | −4.2 (0.2) | −5.1 (0.2) | −5.1 (0.2) |
| 1 | −0.9 (0.2) | −1.3 (0.2) | −1.4 (0.3) | −2.3 (0.1) | −3.4 (0.2) | −4.6 (0.3) | −5.1 (0.3) |
For each configuration, simulations were performed on twenty pairs of images—unique noisy instantiations—and the volume of the interior expanding or contracting sphere calculated. Since the volume change for a brain ROI is calculated by averaging the volume changes of all the voxels in the ROI (as identified by automatic segmentation), this method was also used for the models. The issue is how to handle the boundary voxels, which may be partly outside the ROI. Since the deformation field that effects registration is fairly uniform over the ROI, one only needs to average over voxels that are fully within the ROI—and the more of these that are included the more correctly the volume change thus calculated will reflect the actual volume change induced by the deformation field. If a mask is built from voxels identified as belonging in whole or in part to the ROI, this should be shrunk slightly, and the ROI volume change averaged over voxels within the shrunken mask. There will be slight variation in the estimated volume change, depending on the mask. More important, however, one can be consistent across ROIs by building the shrunken mask in a consistent way—e.g., by smoothing and thresholding a binary mask built from voxels identified as ROI tissue by automatic segmentation.
For model-A, where a 30% volume increase for the inner sphere is expected, a single nonlinear registration gives 27.8%, slightly underestimating the correct result: uniform expansion or contraction isn’t penalized but nonuniform deformation is, and this can happen near tissue boundaries. The gradient in the deformation field there effectively acts as a spring restricting deformation, thus leading to an underestimate of the true displacement. This underestimate could be reduced by using a smaller spring constant (λ2 in Eq. 2). However, a reduced spring constant means greater sensitivity to noise and other imperfections in the data, and so is not always desirable. A way around this limitation is to update the image undergoing transformation given the estimated deformation field, and then register the updated image to the original target—i.e., perform two relaxation steps. The calculated volume increase then is 29.7%. The standard deviation in both calculations is zero to one decimal place.
For situations as in model-B, the underestimation of the volume change is generally greater than in model-A, and up to 5 relaxation steps may be needed to get the correct result, as can be seen in the rows of Table 1 where R2 − R1 is 8, 6, and 4 vl. When only one voxel separates the structures, R2 − R1 = 1 vl, relaxation alone will not suffice: the images must be zoomed, i.e., more degrees of freedom are needed, and then several relaxation steps performed. In the last three rows, note that zooming with only one relaxation step is already a significant improvement on one relaxation step with no zooming. The row R2 − R1 = 4 vl shows that zooming is unnecessary when enough degrees of freedom are present, so that 5 relaxation steps with or without zooming gets the correct result.
Application to Subject Data
For each subject in the ADNI database, followup images were nonlinearly registered to the baseline and the volume-change field calculated from the displacement field. Example results for a subject, initially diagnosed with MCI, are shown in Fig. 3: B is the baseline, F is the two-year followup affine registered to baseline, and F’ is the followup nonlinearly registered to baseline; F-B is the followup minus baseline subtraction image, and F’-B the subtraction image for the nonlinearly registered followup. Though far more informative than “eyeballing” affine registered serial scans (let alone unregistered scans, as is more the norm in radiological practice), interpretation of the monophasic and biphasic signals in F-B images in terms of shifts, expansions, and contractions can be subtle due to the 3D nature of the images—see Bydder (1995) for a detailed discussion. The Ux, Uy, and Uz panes in Fig. 3 are the x-, y-, and z-displacement fields that register F to B. The dVol pane shows the corresponding volume-change field overlaid on the baseline image; the color scale ranges from about 20% shrinkage (blue) to 60% expansion (yellow) (this is reproduced in SI Fig. S3 which also provides the color-scale bar).
After baseline, this subject had scans at six-month intervals through two years. (SI Figs. S1 and S2 are similar to Fig. 3 but for change at 6-months and 1-year from baseline, respectively). SI Fig. S4 shows the automatic cortical and subcortical tissue segmentation at baseline (ROIs are defined using FreeSurfer version 3.02 (Fischl et al., 2002)), and the volume-change field, coronally and on the left hemispheric cortical surface, for 6- and 12-month followups. The left lateral ventricle (subject’s left is on the right of the brain image), for example, expanded by 10%, 20%, 25% and (Fig. 3) 33%, at 6, 12, 18, and 24 months, respectively. For the left hippocampus, the decrease in volume at six month intervals was 3.4%, 3.9%, 5.5%, and 6.3%. Note in particular the localized cortical gray matter shrinkage—up to about 20% at 24 months—on the lateral and inferior temporal lobes, quite distinct from the adjacent and less pronounced white matter shrinkage.
Comparison with Standard Methods
To assess the performance on anatomical images of Quarc vis-à-vis standard methodologies, we analyzed disease-specific e ect sizes, using Cohen’s d, on publicly available data sets from research groups funded by ADNI (Jack et al., 2010), downloaded from www.loni.ucla.edu/ADNI /Data through 09/22/2010. Along with Quarc, these data sets comprise measures derived from longitudinal structural MRI processed with: (1) standard FreeSurfer v4.3 (Dale et al., 1999; Fischl et al., 1999, 2002; Desikan et al., 2006; Tosun et al., 2010); (2) Boundary Shift Integral (BSI) (Freeborough and Fox, 1997; Leung et al., 2010); (3) Tensor Based Morphometry (TBM) (Hua et al., 2008a,b, 2009, 2010); and (4) Voxel Based Morphometry (VBM) (Ashburner and Friston, 2000; Tzourio-Mazoyer et al., 2002; Alexander and Chen, 2010). The measures in these data sets are for various ROIs, both pre-defined tissue regions and data-driven regions, at baseline and followup (generally 6-months apart, as described in the Methods section).
Pairwise head-to-head comparisons with Quarc were performed for each of the four other methodologies, using only measures for baseline and 12-month followup. Publicly available FreeSurfer Longitudinal (v4.4) baseline and 12-month data for any subject implicitly involve the subject’s full available data set, up to 3 years, while baseline and 12-month data for all other methodologies involve those two time-points only; with this caveat, however, results for FreeSurfer Longitudinal are in SI. Explicit quality control (QC) information was provided for FreeSurfer, BSI, and Quarc data, and used for filtering out subject visits that did not have values as follows: FreeSurfer QVERALLQC=“Pass” or “Partial”; BSI KMNREGRATING≤3; and Quarc QCPASS=1; for TBM and VBM, QC was implicit in that only subject-visits that passed QC were publicly available. The total numbers of remaining subjects in common with Quarc for FreeSurfer, BSI, TBM, and VBM, individually, are shown in Table 2. For agiven methodology and ROI, the disease-specific effect size was defined as
| (6) |
also known as Glass’s, where μAD is the average annual change in the AD cohort, μHC is the average annual change in the healthy control (HC) cohort, and σAD is the standard deviation in the AD cohort. A posteriori distributions for the AD and HC means can be built from sampling Student’s t distributions, and an a posteriori distribution for the AD standard deviation can be built from sampling a chi-square distribution for the variance (Rosner, 2006). An a posteriori distribution for d can then be built from the ratio of the a posteriori distribution for the difference in AD and HC means, and the a posteriori distribution for the standard deviation. The 95% confidence interval on d can then be calculated from the cumulative distribution for d. Effect sizes from Quarc with 95% confidence intervals for a global measure (whole brain), a cortical measure (entorhinal), and a subcortical measure (hippocampus) that are important biomarkers for AD (Holland et al., 2009) are shown in Fig. 4; numerical values are in Table 3, and results for FreeSurfer Longitudinal are in SI Table 1. Of these three ROIs, whole brain was available for KN-BSI, and hippocampus was available for VBM; all three were available for FreeSurfer; for TBM, data were available only for the statistically-defined temporal lobe “Stat-ROI”—the optimal ROI for TBM (Hua et al., 2010). Statistical comparisons of Cohen’s d effect sizes were performed using 107 samples drawn from the a posteriori distributions of means and standard deviations. P-values are provided in Table 3 for same-ROI effect size comparisons of methodologies with Quarc; for TBM, where there was no tissue ROI in common with Quarc, the p-value is for a comparison of the Stat-ROI effect size with the Quarc whole brain effect size.
Table 2.
Number of subjects for head-to-head comparisons of Quarc with four standard methodologies used in ADNI (Jack et al., 2010).
| Methodology | AD | HC |
|---|---|---|
| FreeSurfer | 96 | 140 |
| KN-BSI | 87 | 127 |
| TBM | 57 | 108 |
| VBM | 88 | 134 |
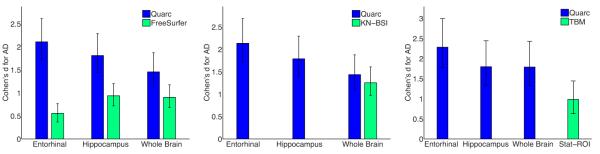
Figure 4.
Head-to-head comparison of Quarc with FreeSurfer, KN-BSI, and TBM, using (absolute) Cohen’s d effect size estimates for annual change in AD controlled for normal aging (Eq. 6), with 95% confidence intervals. Numerical values, including p-values, are in Table 3; numbers of subjects are in Table 2. Stat-ROI is the optimal ROI for TBM.
Table 3.
Cohen’s d effect size estimates for annual regional change in AD controlled for normal aging (Eq. 6), from a head-to-head comparison of Quarc with FreeSurfer, KN-BSI, TBM, and VBM, using data for baseline and 12-months only. Values in brackets are 95% confidence intervals.
| ROI | Quarc | FreeSurfer | p |
|---|---|---|---|
| Entorhinal | −2.11 [ −2.56 −1.75 ] | −0.56 [ −0.78 −0.35 ] | ≤1.0e-6 |
| Hippocampus | −1.81 [ −2.20 −1.50 ] | −0.94 [ −1.19 −0.72 ] | 1.6e-5 |
| Whole Brain | −1.46 [ −1.81 −1.17 ] | −0.90 [ −1.16 −0.68 ] | 0.0051 |
| ROI | Quarc | KN-BSI | p |
|---|---|---|---|
| Entorhinal | −2.13 [ −2.61 −1.76 ] | ||
| Hippocampus | −1.79 [ −2.20 −1.47 ] | ||
| Whole Brain | −1.44 [ −1.80 −1.14 ] | −1.25 [ −1.53 −1.02 ] | 0.38 |
| ROI | Quarc | TBM | p† |
|---|---|---|---|
| Entorhinal | −2.28 [ −2.92 −1.82 ] | ||
| Hippocampus | −1.80 [ −2.32 −1.41 ] | ||
| Whole Brain | −1.79 [ −2.32 −1.39 ] | ||
| Stat-ROI | −0.98 [ −1.29 −0.72 ] | 0.0016 |
| ROI | Quarc | VBM† | p |
|---|---|---|---|
| Entorhinal | −2.09 [ −2.56 −1.72 ] | ||
| Hippocampus | −1.82 [ −2.23 −1.50 ] | 0.32 [ 0.07 0.59 ] | ≤1.0e-6 |
| Whole Brain | −1.40 [ −1.76 −1.10 ] |
p-value for Stat-ROI compares the best TBM measure (Hua et al., 2010) with the Quarc whole brain measure.
For VBM, HC change more than AD, giving positive d.
The Quarc measures present a significant improvement over those of FreeSurfer, particularly the entorhinal cortex (p≤1.0e-6). The KN-BSI effect size for whole brain, though lower, is not significantly different from the Quarc whole brain effect size (p=0.38). The TBM Stat-ROI has a significantly smaller effect size that the Quarc whole brain (p=0.0016). For VBM, hippocampal data were not meaningful: HCs changed slightly more than AD subjects. It should be noted that official ADNI data from VBM and FreeSurfer were provided by groups that were not developers of these methods, and thus may not accurately reflect the capability of these methods.
Conclusion
The nonlinear registration method presented here enables very precise quantification of large whole brain deformations, cortical surface shrinkage, and subtle sub-cortical changes in small ROIs, based on serial MRI scans of an individual. The method’s precision, however, relies on tissue intensity signatures not changing over time. Therefore, application of the method, Quarc, to quantifying structural development in neonatal brains, or to progression of diseases such as multiple sclerosis, will require modeling of associated MRI contrast changes (Studholme et al., 2006).
Though presented here with a focus on the brain, Quarc is applicable to deformation and structural change in organs generally. For example, imaging livers requires breath-holding, but from breath-hold to breath-hold the organ will be non-affinely deformed, necessitating nonlinear transformation to a chosen target. As another example, when tumor image contrast remains fairly constant over time, tumor growth or shrinkage detectable on T1- or T2-weighted scans could accurately be tracked (Wang et al., 2009). As a visual aid in detecting organ changes over time, the method should greatly assist clinical assessment of subjects. Moreover, its quantitative precision should facilitate early clinical diagnosis, accurate treatment monitoring, and assessing efficacy in clinical trials. Indeed, as demonstrated for AD, in locating potential surrogate, complementary, disease-specific biomarkers (Holland et al., 2009; Jack et al., 2010), the precision will allow for smaller sample sizes in AD clinical trials, and/or shorter trials. The ability to track subtle changes over time throughout an organ may enable determination of disease-specific deformation patterns, and given a pair of serial scans for a subject, perhaps along with genetic information or other biomarkers, assess where the subject is on a disease trajectory.
Supplementary Material
Acknowledgments
Thanks to Graeme Bydder, Yoon Chung, Matt Erhart, Donald J. Hagler, Robin Jennings, Alan Koyama, and Chris Pung. This research was supported by National Institute of Health Grants R01AG031224, R01AG22381, U54NS056883, P50-NS22343, and P50MH081755 (to A.M.D.). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc, F. Ho man-La Roche, Schering-Plough, Synarc, Inc., as well as non-profit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement Anders M. Dale is a founder and holds equity in CorTechs Labs, Inc, and also serves on the Scientific Advisory Board. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
References
- Alexander GE, Chen K. 2010 adni_uaspmvbm_2010-05-23.csv. http://www.loni.ucla.edu/
- Ashburner J, Andersson J, Friston K. High-dimensional image registration using symmetric priors. Neuroimage. 1999 Jun;9:619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Andersson JL, Friston KJ. Image registration using a symmetric prior–in three dimensions. Hum Brain Mapp. 2000 Apr;9:212–225. doi: 10.1002/(SICI)1097-0193(200004)9:4<212::AID-HBM3>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry–the methods. Neuroimage. 2000 Jun;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Neelin P, Collins D, Evans A, Friston K. Incorporating prior knowledge into image registration. Neuroimage. 1997 Nov;6:344–352. doi: 10.1006/nimg.1997.0299. [DOI] [PubMed] [Google Scholar]
- Atiya M, Hyman B, Albert M, Killiany R. Structural magnetic resonance imaging in established and prodromal Alzheimer disease: a review. Alzheimer Dis Assoc Disord. 2003;17:177–195. doi: 10.1097/00002093-200307000-00010. [DOI] [PubMed] [Google Scholar]
- Bobinski M, de Leon MJ, Wegiel J, Desanti S, Convit A, Saint Louis LA, Rusinek H, Wisniewski HM. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer’s disease. Neuroscience. 2000;95:721–725. doi: 10.1016/s0306-4522(99)00476-5. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Bradley WG, Bydder GM. Advanced MR Imaging Techniques. Informa Healthcare. 1997 January; [Google Scholar]
- Bydder GM. The Mackenzie Davidson Memorial Lecture: detection of small changes to the brain with serial magnetic resonance imaging. Br J Radiol. 1995 Dec;68:1271–1295. doi: 10.1259/0007-1285-68-816-1271. [DOI] [PubMed] [Google Scholar]
- Christensen GE. Information Processing in Medical Imaging. Vol. 1613. Springer-Verlag; Berlin, Germany: 1999. Ch. Consistent linear-elastic transformations for image matching; pp. 224–237. [Google Scholar]
- Christensen GE, Johnson HJ. Consistent image registration. IEEE Trans Med Imaging. 2001 Jul;20:568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Rabbit RD, Miller MI. Deformable templates using large deformation kinematics. IEEE T Image Process. 1996;5:1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999 Feb;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Davies DE, Salmond DJ. Calculation of the volume of a general hexahedron for flow predictions. AIAA J. 1985 Jun.23:954–956. [Google Scholar]
- DeKosky S, Marek K. Looking backward to move forward: early detection of neurodegenerative disorders. Science. 2003 Oct;302:830–834. doi: 10.1126/science.1090349. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006 Jul;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002 Jan;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999 Feb;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Blennow K, Brewer JB, Dale AM. Brain Atrophy in Healthy Aging Is Related to CSF Levels of Abeta1-42. Cereb Cortex. 2010a Jan; doi: 10.1093/cercor/bhp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM. One-year brain atrophy evident in healthy aging. J. Neurosci. 2009 Dec;29:15223–15231. doi: 10.1523/JNEUROSCI.3252-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM. CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. J. Neurosci. 2010b Feb;30:2088–2101. doi: 10.1523/JNEUROSCI.3785-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Espeseth T, Reinvang I, Dale AM, Holland D, Walhovd KB. Cortical gray matter atrophy in healthy aging cannot be explained by undetected incipient cognitive disorders: a comment on Burgmans et al. (2009) Neuropsychology. 2010c Mar;24:258–263. doi: 10.1037/a0018827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frackowiak RS, Ashburner JT, Penny WD, Zeki S, Friston KJ, Frith CD, Dolan RJ, Price CJ, editors. Human Brain Function. 2nd Edn Elsevier Academic Press; Amsterdam ; Boston: 2004a. Ch. Rigid Body Registration; pp. 635–653. [Google Scholar]
- Frackowiak RS, Ashburner JT, Penny WD, Zeki S, Friston KJ, Frith CD, Dolan RJ, Price CJ, editors. Human Brain Function. 2nd Edn Elsevier Academic Press; Amsterdam ; Boston: 2004b. Ch. High-Dimensional Image Warping; pp. 673–694. [Google Scholar]
- Freeborough PA, Fox NC. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging. 1997 Oct;16:623–629. doi: 10.1109/42.640753. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. Modeling brain deformations in alzheimer disease by fluid registration of serial 3d mr images. J Comput Assist Tomo. 1998;22:838–843. doi: 10.1097/00004728-199809000-00031. [DOI] [PubMed] [Google Scholar]
- Garg VK. Applied Computational Fluid Dynamics. Marcel Dekker, Inc.; New York: 1998. Ch. 2. Numerical Techniques. [Google Scholar]
- Gershenfeld N. The Nature of mathematical Modeling. Cambridge University Press; 1999. [Google Scholar]
- Grandy J. Efficient computation of volume of hexahedral cells. Lawrence Livermore National Laboratory; Oct, 1997. Informal report UCRL-ID-128886. [Google Scholar]
- Gudbjartsson H, Patz S. The rician distribution of noisy MRI data. Magn Reson Med. 1995;34:910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal JV, Saeed N, Soar EJ, Oatridge A, Young IR, Bydder GM. A registration and interpolation procedure for subvoxel matching of serially acquired MR images. J Comput Assist Tomo. 1995 March/April;12:289–96. doi: 10.1097/00004728-199503000-00022. [DOI] [PubMed] [Google Scholar]
- Holland D, Brewer JB, Hagler DJ, Fennema-Notestine C, Dale AM, the Alzheimer’s Disease Neuroimaging Initiative Subregional neuroanatomical change as a biomarker for Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 2009 Dec;106:20954–20959. doi: 10.1073/pnas.0906053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Hibar DP, Yanovsky I, Leow AD, Toga AW, Jack CR, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schu N, Alexander GE, Weiner MW, Thompson PM. Mapping Alzheimer’s disease progression in 1309 MRI scans: power estimates for different inter-scan intervals. Neuroimage. 2010 May;51:63–75. doi: 10.1016/j.neuroimage.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Lee S, Yanovsky I, Leow AD, Chou YY, Ho AJ, Gutman B, Toga AW, Jack CR, Bernstein MA, Reiman EM, Harvey DJ, Kornak J, Schu N, Alexander GE, Weiner MW, Thompson PM. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009 Dec;48:668–681. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow A, Lee S, Klunder A, Toga A, Lepore N, Chou Y, Brun C, Chiang M, Barysheva M, Jack C, Jr., Bernstein M, Britson P, Ward C, Whitwell J, Borowski B, Fleisher A, Fox N, Boyes R, Barnes J, Harvey D, Kornak J, Schu N, Boreta L, Alexander G, Weiner M, Thompson P. 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage. 2008a May;41:19–34. doi: 10.1016/j.neuroimage.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Leow AD, Parikshak N, Lee S, Chiang MC, Toga AW, Jack CR, Weiner MW, Thompson PM. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage. 2008b Nov;43:458–469. doi: 10.1016/j.neuroimage.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, Schu N, Krueger G, Killiany RJ, Decarli CS, Dale AM, Carmichael OW, Tosun D, Weiner MW. Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimers Dement. 2010 May;6:212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, MacFall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. NeuroImage. 2006 April;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Chiang MC, Lee AD, Klunder AD, Lu A, Becker JT, Davis SW, Toga AW, Thompson PM. Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans Med Imaging. 2007 Jun;26:822–832. doi: 10.1109/TMI.2007.892646. [DOI] [PubMed] [Google Scholar]
- Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR, Weiner MW, Fox NC, Ourselin S. Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage. 2010 Apr;50:516–523. doi: 10.1016/j.neuroimage.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB, Fox NC. Correction of differential intensity inhomogeneity in longitudinal MR images. Neuroimage. 2004 Sep;23:75–83. doi: 10.1016/j.neuroimage.2004.04.030. [DOI] [PubMed] [Google Scholar]
- Liang Z-P, Lauterbur PC. Principles of Magnetic Resonance Imaging: a signal processing perspective. SPIE Optical Engineering Press; IEEE Press; Bellingham, Washington: New York: 2000. [Google Scholar]
- McDonald CR, Gharapetian L, McEvoy LK, Fennema-Notestine C, Hagler DJ, Holland D, Dale AM. Relationship between regional atrophy rates and cognitive decline in mild cognitive impairment. Neurobiol Aging. 2010 May; doi: 10.1016/j.neurobiolaging.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CR, McEvoy LK, Gharapetian L, Fennema-Notestine C, Hagler DJ, Holland D, Koyama A, Brewer JB, Dale AM. Regional rates of neocortical atrophy from normal aging to early Alzheimer disease. Neurology. 2009 Aug;73:457–465. doi: 10.1212/WNL.0b013e3181b16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Edland SD, Holland D, Hagler DJ, Roddey JC, Fennema-Notestine C, Salmon DP, Koyama AK, Aisen PS, Brewer JB, Dale AM. Neuroimaging enrichment strategy for secondary prevention trials in Alzheimer disease. Alzheimer Dis Assoc Disord. 2010;24:269–277. doi: 10.1097/WAD.0b013e3181d1b814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009 Apr;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Holland D, Fennema-Notestine C, Hagler DJ, Brewer JB, Dale AM. Baseline and longitudinal structural MRI measures improve predictive prognosis. Radiology. 2011 doi: 10.1148/radiol.11101975. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Christensen GE, Amit Y, Grenander U. Mathematical textbook of deformable neuroanatomies. P Natl Acad Sci USA. 1993;90:11944–8. doi: 10.1073/pnas.90.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EA, Holland D, Donohue M, McEvoy LK, Hagler DJ, Dale AM, Brewer JB. Six-month atrophy in MTL structures is associated with subsequent memory decline in elderly controls. Neuroimage. 2010 Dec;53:1310–1317. doi: 10.1016/j.neuroimage.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannalal B. Modern Digital and Analog Communication Systems. 3rd Edn Oxford University Press; New York: 1998. pp. 520–521. [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010 Jan;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A, Jensen J, Helpern J. Quantitative MR imaging in Alzheimer disease. Radiology. 2006 Oct;241:26–44. doi: 10.1148/radiol.2411050628. [DOI] [PubMed] [Google Scholar]
- Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. Neuroimage. 2010 Dec;53:1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner B. Fundamentals of Biostatistics. 6th Edn Duxbury Press; 2006. Ch. 6 / Estimation. [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998 Feb;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Studholme C, Drapaca C, Iordanova B, Cardenas V. Deformation-based mapping of volume change from serial brain MRI in the presence of local tissue contrast change. IEEE Trans Med Imaging. 2006 May;25:626–639. doi: 10.1109/TMI.2006.872745. [DOI] [PubMed] [Google Scholar]
- Thompson WK, Holland D. Bias in Tensor Based Morphometry Stat-ROI measures may result in unrealistic power estimates. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2010.11.092. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosun D, Schu N, Truran-Sacrey D, Shaw LM, Trojanowski JQ, Aisen P, Peterson R, Weiner MW. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiol. Aging. 2010 Aug;31:1340–1354. doi: 10.1016/j.neurobiolaging.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 Jan;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van der Vorst HA. A fast and smoothly converging variant of Bi-CG for the solution of nonsymmetric linear systems. SIAM J Sci Stat Comp. 1992;13:631–644. [Google Scholar]
- van der Vorst HA. Iterative Krylov Methods for Large Linear Systems. Cambridge University Press; Cambridge: Apr, 2003. [Google Scholar]
- Wald LL, Schmitt F, Dale A. Systematic spatial distortion in MRI due to gradient nonlinearities. NeuroImage. 2001;13:S50. [Google Scholar]
- Wang CH, Rockhill JK, Mrugala M, Peacock DL, Lai A, Jusenius K, Wardlaw JM, Cloughesy T, Spence AM, Rockne R, Alvord EC, Swanson KR. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel biomathematical model. Cancer Res. 2009 Dec;69:9133–9140. doi: 10.1158/0008-5472.CAN-08-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky I, Leow AD, Lee S, Osher SJ, Thompson PM. Comparing registration methods for mapping brain change using tensor-based morphometry. Med Image Anal. 2009 Oct;13:679–700. doi: 10.1016/j.media.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Avants BB, Das SR, Pluta J, Altinay M, Craige C. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: an illustration in ADNI 3 T MRI data. Neuroimage. 2010 Apr;50:434–445. doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.