Abstract
Epigenetic modifications help orchestrate sweeping developmental, aging, and disease-causing changes in phenotype by altering transcriptional activity in multiple genes spanning multiple biologic pathways. Although previous epigenetic research has focused primarily on dividing cells, particularly in cancer, recent studies have shown rapid, dynamic, and persistent epigenetic modifications in neurons that have significant neuroendocrine, neurophysiologic, and neurodegenerative consequences. Here, we provide a review of the major mechanisms for epigenetic modification and how they are reportedly altered in aging and Alzheimer’s disease (AD). Because of their reach across the genome, epigenetic mechanisms may provide a unique integrative framework for the pathologic diversity and complexity of AD.
Keywords: Epigenetics, DNA methylation, histone acetylation, rDNA, miRNA, genetics, gene expression, amyloid β peptide, inflammation, oxidative stress, cell cycle
1.0 Introduction
Alzheimer’s Disease (AD) is a progressive, irreversible neurodegenerative disorder culminating in dementia. Its etiology and pathogenesis are complex, and encompass many genetic and environmental risk factors, changes in the expression of thousands of genes, and upregulation of multiple pathogenic pathways such as amyloid β peptide (Aβ) deposition, tau hyperphosphorylation, inflammation, oxidative stress, energy metabolism, and aberrant re-entry into the cell cycle/apoptosis. Moreover, with the exception of Aβ-inducing mutations, none of these molecular and genetic factors appears to have absolute penetrance in causing the disorder: many individuals may possess the most salient risk factors for AD, as well as express profuse Aβ and tau pathology, yet never develop the disorder (Lue et al., 1996). Indeed, even monozygotic twins can have dichotomous AD outcomes (Raiha et al., 1997; Mastroeni et al., 2009)
The emerging field of epigenetics has its roots in studies of the structure of chromatin and modifications to the structure of DNA, which extend back half a century or more (Felsenfeld, 2007). Although a unitary definition of epigenetics has yet to be reached, the many definitions that have been suggested all invoke heritability, lack of dependence on DNA sequence, and effects on transcription to produce diverse phenotypes. In particular, epigenetic modifications are capable of altering transcriptional activity in a coherent manner across thousands of genes and dozens of biological pathways, yet can do so differentially in monozygotic twins, the same individual at different developmental stages, or adjacent cells in the same organ, all of which share the same genetic code. Epigenetics also provides a means by which environmental factors such as diet, hazardous exposures, and life events can influence gene expression. As such, epigenetic mechanisms may provide a point of intersection for the diverse risk factors and pathophysiologic processes of AD.
The purpose of this review is to briefly describe the major epigenetic mechanisms, histone acetylation, DNA methylation, ribosomal DNA (rDNA), and microRNA (miRNA), and how they are reportedly altered in aging and AD.
2.0 Epigenetic regulation of gene expression
Epigenetic mechanisms modify heritable and non-heritable traits without necessarily altering the underlying DNA sequence. Thus, through epigenetic modification the diverse cellular phenotypes and functions needed by the body can be achieved using a single genetic code for all cells. These effects are typically accomplished by inhibition of transcriptional access to various genes, leading to their repression or silencing. Conversely, release from normal epigenetic repression can enhance gene expression (Wilson and Jones, 1983; Bandyopadhyay and Medrano, 2003; Liu et al., 2003; Fraga and Esteller, 2007; Poulsen et al., 2007). These modifications can occur at specific gene loci in specific cells to yield specific cellular phenotypes, or can encompass many genes in many cells, an orchestrating mechanism that is widely assumed to help drive such broad biological processes as development and aging (Wilson and Jones, 1983; Bandyopadhyay and Medrano, 2003; Liu et al., 2003; Fraga and Esteller, 2007; Poulsen et al., 2007).
Because phenotype and function are affected and effected by what genes are expressed and what genes are repressed, epigenetically regulated and dysregulated transcription states can give rise not only to different cell types and developmental stages, but also to favorable and unfavorable outcomes for specific cells within the same organ system. Thus, changes in epigenetic regulation can cause some cells to develop structural, physiologic, and metabolic abnormalities, while other cells of the same type remain normal. This is thought to occur, for example, in several hematologic malignancies after aberrant epigenetic silencing of genes that control proliferation (Mund et al., 2006).
In addition to direct regulation of gene expression, epigenetic modifications can mimic, exacerbate, or even cause genetic mutations. For example, epigenetic repression of tumor suppressor genes can mimic loss-of-function mutations of tumor suppressor genes, and both are highly associated with cancer development (Mund et al., 2006). Epigenetic silencing of the alpha subunit of the stimulatory G protein, a signaling peptide essential for the actions of parathyroid hormone, can cause pseudohypoparathyroidism, just as mutations to the alpha subunit do (Bastepe, 2008). Epigenetic modifications can exacerbate the effects of gene mutations, as occurs with Apc gene mutations in colorectal cancers (Colnot et al., 2004), E-cadherin mutations in gastric cancers (Strathdee, 2002), and mitochondrial DNA mutations in Leber’s disease (Johns and Neufeld, 1993). Beyond these interactions, epigenetic repression of DNA repair genes may also induce gene mutations (Jacinto and Esteller, 2007).
Finally, epigenetic mechanisms provide a means by which environmental events can be translated to the cellular and molecular level. For example, ultraviolet ray exposure may induce epigenetic modifications in skin cells that culminate in cutaneous malignancies (Millington, 2008). Likewise, epigenetic changes induced by different environments, or simply by stochastic events, are thought to underlie the subtle phenotypic differences that emerge with time in monozygotic twins (Fraga et al., 2005; Flanagan et al., 2006).
2.1 Histone modifications
Epigenetic mechanisms typically involve changes in the micro- and macro-structure of chromatin, a complex of DNA, chromosome proteins, and histone proteins in which the histone proteins are tethered together in structures around which double-stranded DNA is wound. Conformational changes in histone proteins or modifications of the way in which DNA wraps around the histones may then differentially alter access of the transcriptional machinery to some genes while leaving access to other genes intact (Allfrey, 1966).
Although there are multiple mechanisms by which histones are modified, including methylation, phosphorylation, ubiquitination, sumoylation, citrullination, ADP-ribosylation, and other post-translational modifications of the amino acids that make up histone proteins, histone acetylation is one of the most ubiquitous and well studied. Here, histone acetyltransferases (HATs) catalyze the transfer of an acetyl group from acetyl-coenzyme A to lysine residues on the N-termini of histone proteins (Fig. 1). As a result of acetylation, the positive charge of the histone proteins is neutralized, decreasing interactions of the histone protein tails with negatively-charged phosphate groups of associated DNA. This conformational relaxation of the chromatin permits access to and transcription of genes within the complex. Conversely, the histone deacetylases (HDACs) transfer acetyl groups from acetylated histone proteins back to coenzyme A, producing a more condensed chromatin state and decreased or silenced gene transcription.
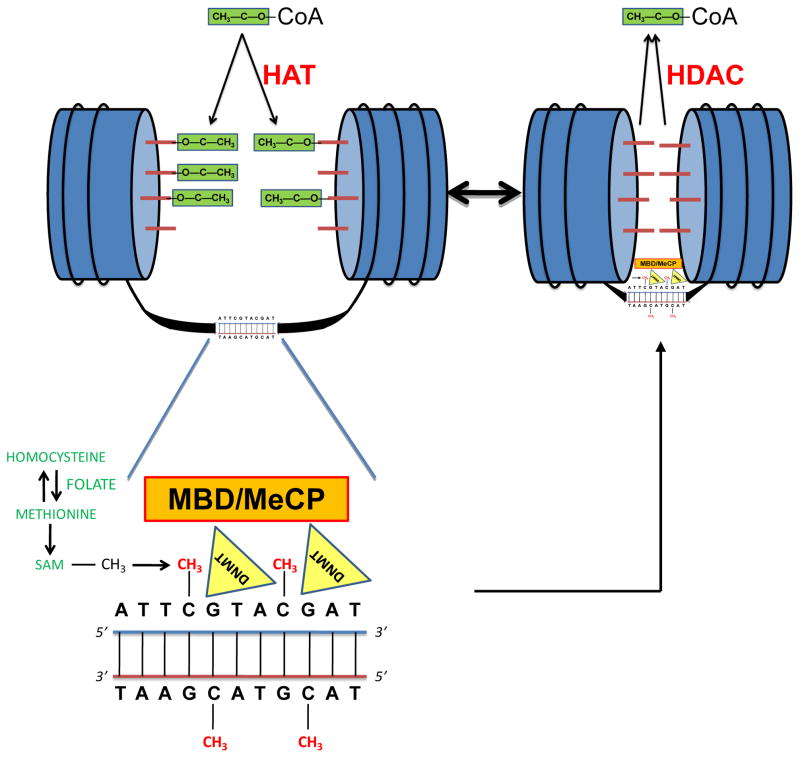
Fig. 1. Simplified schematic of histone acetylation and DNA methylation.
(Upper Left) In transcriptionally active genes the chromatin, made up of histones (blue cylinders) around which DNA is wrapped, is in a relaxed state, permitting transcriptional access to unwound DNA. This relaxed, euchromatin state is, in part, mediated by acetylation of histone tails (red rods) in which acetyl groups (green blocks) are transferred from acetyl-coenzyme A (acetyl-CoA) to the histone tails by histone acetyltransferases (HATs). (Bottom Left) Within the DNA, the cytosines of adjacent C-G/G-C dinucleotides (CpGs) may be methylated. The methyl group ultimately derives from methyltetrahydrofolate in conjunction with the methionine/homocysteine cycle, and is transferred from S-adenosylmethionine (SAM) to the cytosine and incorporated into the genome by DNA methyltransferases (DNMTs). CpG-methyl-binding-domain proteins (MBDs) and methylation complex proteins (MeCPs) (which may contain MBDs) become associated with methylated CpGs, further inhibiting transcriptional access and repression of the gene. (Upper Right). DNA methylation and histone modifications are integrally linked, because MBDs and MeCPs attract histone deacetylases (HDACs) that transfer acetyl groups on the histone tail back to CoA. Histone deacetylation, in turn, promotes the condensed, heterochromatin state characteristic of silenced or repressed genes.
2.2 DNA Methylation
DNA methylation provides a further means for histone modification, and is highly interactive with histone acetylation and the other histone-modifying mechanisms. For this reason, DNA methylation can, to some extent, be taken as a surrogate for histone modification profiles.
Beginning with seminal research in the late 1970s (Bird, 1978), it is now well established that adjacent cytosine-guanine (CpGs) dinucleotides within DNA can be methylated by the actions of the DNA methyltransferases, DNMT1, DNMT2, DNMT3a/b, and DNMT4. In mammals, DNMT1 appears to be primarily involved in maintenance methylation of hemimethylated DNA after DNA replication, whereas DNMT3a and DNMT3b are particularly important for de novo DNA methylation. DNMT2 is typically considered to be an RNA methyltransferase, although it also has 5-cytosine DNA methyltransferase activity (Tang et al., 2003) and forms denaturant-resistant complexes with DNA (Dong et al., 2001). The methyl group that is transferred to cytosine by the DNMTs ultimately derives from methyltetrahydrofolate through its interactions with S-adenosylmethionine (SAM) in the homocysteine-methionine cycle (Fig. 1).
Through these processes, approximately 70% of CpG dinucleotides within the human genome are methylated. Although DNA methylation can take place at any CpG site, whether in coding or non-coding regions, previous studies have often focused on CpG-rich stretches (CpG islands) within the promoter region. Some 50,267 CpG islands exist in the human genome, with 28,890 in simple repeat and low complexity sequences that are masked (Bandyopadhyay and Medrano, 2003; Liu et al., 2003). Because CpG islands contain a high proportion of CpGs (and opportunities for CpG methylation), they are, perforce, among the most highly methylated regions of the genome. However, it has recently been shown that the percentage (rather than the absolute number) of methylated CpGs is relatively low in conventional CpG islands, and is actually higher in promoters with intermediate CpG densities (Eckhardt et al., 2006; Weber et al., 2007). Moreover, tissue-specific methylation patterns appear to be most pronounced not at CpG islands but at “CpG shores”, regions within approximately 2 kb of CpG-enriched sequences. It has been suggested, in fact, that methylation patterns of CpG shores are “sufficiently conserved to completely discriminate tissue types regardless of species of origin” (Irizarry et al., 2009). Differential methylation of CpG shores in brain compared to liver has also been found to be highly correlated with differences in gene expression for these two organs (Irizarry et al., 2009).
Methylation of CpG sequences may alter gene expression by inducing histone modifications that inhibit access of the transcriptional machinery (Bird and Wolffe, 1999; Zhang et al., 2002) (Fig. 1). One would expect, then, that highly methylated genes would be repressed, and that hypomethylation of a gene would lead to enhanced expression or overexpression compared to the normally repressed, methylated state. Some notable exceptions to these expected states have been found, however. For example, the p16INK4a promoter is progressively hypermethylated with age (So et al., 2006), but expression of the p16INK4a gene appears to increase with age (Kim and Sharpless, 2006). More generally, Gius and colleagues (2004) found that chemical hypomethylation of nearly half the genes they surveyed resulted in silencing rather than upregulation. Indeed, altering the methylation status of some CpG sites within a gene can be inconsequential compared to alterations at other sites (c.f., Murgatroyd et al., 2009; Yakovlev et al., 2009). Such findings indicate that it will be important to follow up genome-wide DNA methylation profiling, which can only survey a limited number of CpGs per gene, with more detailed DNA methylation maps that include shores and flanking regions. In addition, even highly-detailed DNA methylation profiles will require validation of functional effects at the gene expression and protein levels.
A second, linked mechanism by which DNA methylation may modify gene expression is through methyl-CpG-binding proteins (MeCPs) such as MeCP2. When bound to methylated DNA, MeCP2 has been shown to recruit HDACs, which, as noted earlier, may then induce a more condensed chromatin state and decreased or silenced gene transcription (Jones et al., 1998; Wade et al., 1998). Although previous studies assumed that MeCP2 required a methylated chromatin substrate for direct binding (Lewis et al., 1992), more recent research has demonstrated that MeCP2 can directly condense chromatin even in the absence of DNA methylation, histone deacetylase activity, or the cooperation of other transcriptional co-repressors such as mSin3A (Wade, 2001). Mutations of the MeCP2 gene cause Rett’s Syndrome, with dysregulation of neural development, mental retardation, and motor dysfunction (Amir et al., 1999).
MeCP1, a macromolecule made up of some 10 different peptides, may also act as a mediator between DNA methylation and histone acetylation, recognizing and binding to CpG dinucleotides, recruiting HDACs, and inducing transcriptional repression (Feng and Zhang, 2001). Unlike MeCP2, however, MeCP1 does not bind directly to methylated DNA, but to a single methyl-CpG-binding domain protein, MBD2. In addition to inducing histone modifications, MBD2-bound MeCP1 helps maintain the DNA methylation status of CpGs by recruiting DNMT1. DNMT1 is then able to recognize and repair CpGs that have lost methyl groups on one DNA strand but not the other.
Recent studies have also revealed a further modification to methylated CpGs that may make them even more inaccessible to transcription. Here, the 5-methylcytosines are hydroxylated (oxidized) to form 5-hydroxymethylcytosine (Valinluck and Sowers, 2007; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Hydroxymethylated DNA has been observed in neurons (Kriaucionis and Heintz, 2009), and may occur as a result of oxidative damage and/or the actions of specific oxidative enzymes, particularly TET1 (Tahiliani et al., 2009). Normally, 5-hydroxymethylcytosines are stripped from genomic DNA by glycosylases of the base excision repair (BER) system (Steen et al., 2008), permitting replacement with unmodified cytosines for subsequent de novo DNA methylation by the DNMTs. When left intact, however, 5-hydroxymethylcytosines reportedly reduce the interaction of DNA with DNA-binding proteins to an even greater extent than do 5-methylcytosines (Valinluck et al., 2004; Valinluck and Sowers, 2007). Because hydroxymethylated CpGs may go undetected by 5-methylcytosine antibodies, hydroxymethylation could lead to under-reporting of DNA methylation, a possibility that the present authors are now investigating. Conversely, bisulphite sequencing may not discriminate normally-methylated from hydroxymethylated CpGs.
2.3 RNA-related mechanisms
Epigenetic regulation also extends to mechanisms involving RNA such as micro-RNAs (miRNAs) and hereditable and cell cycle-maintained silencing of a portion of the ribosomal RNA genes. rRNA transcripts from repeated rRNA genes (rDNA) provide a structure and primary catalytic site for the eukaryotic ribosome, wherein gene expression culminates in protein synthesis. Different cell types exhibit different proportions of active rRNA genes, suggesting that the fraction of rRNA gene copies may be altered in development and differentiation. Epigenetic mechanisms are now known to play important roles in this process by silencing rRNAs, thereby providing a dynamic balance between active and inactive rRNAs. DNA methylation appears to be one of these epigenetic mechanisms, and may be of particular importance given the unusually high frequency of CpG dinucleotides within the rRNA genes and their unusually high states of DNA methylation. For example, two studies have shown a correlation between DNA methylation status and activity of rRNA genes (Bird et al., 1981; Santoro & Grummt, 2001), and treatment with 5-aza-2-deoxycytidine (azacytidine), a hypomethylating agent, enhances expression by rDNA genes (Santoro & Grummt, 2001). For an excellent and comprehensive summary of rDNA methylation, as well as histone modifications and chromatin remodeling, which also play key roles in epigenetic regulation of rDNA, the reader is referred to the recent review of McStay and Grummt (2008).
The study of miRNAs represents an additional, critical area for epigenetics research. Often deriving from their own genes with their own promoter and regulatory elements, approximately 700–800 miRNAs have been identified in the human genome. These small (~22 nucleotide) RNAs regulate gene expression in a post-transcriptional manner by binding to their target mRNAs, inhibiting translation or, less often, inducing cleavage of the mRNAs (reviewed in Yang, 2007).
3.0 Dynamic epigenetic regulation in adult neurons
3.1 Histone modifications
Histone modifications have been implicated in broad neurobiological processes such as development of the CNS (reviewed in MacDonald and Roskams, 2009), post-traumatic stress disorders (Sokolova et al., 2006), childhood abuse/suicide (Meaney et al., 2007; McGowan et al., 2009), memory formation (Gupta et al., 2010), and addiction (Impey, 2007); specific physiologic processes such as neuronal differentiation (Kular et al., 2009), regulation of choline acetyltransferase activity (Aizawa and Yamamuro, 2010), astrocyte GDNF and BDNF transcription (Wu et al., 2008), microglial apoptosis (Chen et al., 2007), and axon pathfinding (Zinovyeva et al., 2006); and various neurologic disorders, including Parkinson’s disease (Chen et al., 2007; Wu et al., 2008), motor neuron disease (reviewed in Echaniz-Laguna et al., 2008), multiple sclerosis (reviewed in Gray and Dangond, 2006), X-linked mental retardation (Tahiliani et al., 2007), and stroke/cerebral palsy (Meisel et al., 2006). All of these histone-related processes occur in the context of the CNS, and many have been reported to function as dynamic regulatory mechanisms in postmitotic neurons.
3.2 miRNA
Like histone modifications, miRNA has been implicated in both broad and specific CNS processes and disorders. For example, miRNA-206 appears to promote neuromuscular synapse regeneration (Williams et al., 2009), and miR-329, miRNA-134 and miRNA-381, which are induced by neuronal activity, have been suggested to be essential for activity-dependent dendritic outgrowth of hippocampal neurons (Khudayberdiev, Fiore, and Schratt, 2009). In turn, these and other miRNA-mediated mechanisms have been investigated in various CNS disorders, including HIV-dementia (Witwer et al., 2010), amyotrophic lateral sclerosis (Williams et al., 2009), Tourette’s syndrome (Abelson et al., 2005), AD (see section 5.2, below), and other neurologic conditions (reviewed in Maes et al., 2009).
Once again, these epigenetic processes have been demonstrated to occur in the context of postmitotic neurons. Indeed, it has been reported that the switch from a replicative state to the postmitotic state of neurons may itself be controlled by miRNA mechanisms (Yoo et al., 2009).
3.3 DNA methylation
Previously, DNA methylation reactions were considered primarily in the context of maintenance of the DNA methylation pattern across cell divisions. Why neurons and other postmitotic cells should express DNA methylation markers such as DNMT1 was therefore unclear. Likewise, how long-term DNA methylation alterations might be accomplished and sustained in postmitotic cells was unknown. Recently, however, new studies have shown that hypo- and hypermethylation are dynamic events that can occur within cells (Kangaspeska et al., 2008; Metivier et al., 2008), including postmitotic neurons (Levenson et al., 2006; Murgatroyd et al., 2009), on the scale of tens of minutes. These findings open the door for long-suspected, dynamic epigenetic mechanisms that may help mediate neuronal and synaptic plasticity (e.g., (Arendt, 2005). For example, gene-specific hypomethylation of hippocampal neurons after DNMT inhibition blocks long-term potentiation (Levenson et al., 2006) and fear conditioning (Miller and Sweatt, 2007). Weaver and co-workers (2005) have suggested that early life events—specifically, maternal care—alter adult stress responses through sustained DNA methylation changes in rat hippocampal neurons.
The role of DNA methylation in dynamic regulation of neuronal activity and function began to emerge with studies by Bredy and colleagues (2003), and has been vividly confirmed by recent work linking early-life stress to enduring molecular, physiologic, memory, and behavioral changes in mice via epigenetic modifications to hypothalamic neurons (Murgatroyd et al., 2009). In particular, early exposure of mice to stress during the first 10 days of life has been found, as much as a year later, to be associated with impaired step-down avoidance learning, sustained hyperactivity of the hypothalamic-pituitary-adrenal axis, and corticosterone and pituitary adrenocorticotropin pro-hormone hypersecretion. These effects, in turn, were elegantly traced to persistent arginine vasopressin (AVP) overexpression by parvocellular neurons of the hypothalamic paraventricular nucleus, and to hypomethylation of specific CpG sites within the AVP gene. Notably, age-dependent increases in AVP gene hypomethylation at multiple CpG sites were observed in control mice in these studies, but hypomethylation of CpGs within a CpG island (CGI3) in the AVP enhancer region approximately 0.5 kb downstream from the AVP gene itself appeared to be the primary determinant of AVP overexpression in early-life stress mice. Further experiments went on to show that CGI3 hypomethylation was specific to the paraventricular nucleus compared to the supraoptic nucleus, and to provide a key mechanism for the epigenetic modifications that were observed: CGI3 CpG sequences serve as preferential and selective DNA-binding sites for MeCP2. Phosphorylation of MeCP2 by calmodulin-dependent protein kinase II as a result of neuronal membrane depolarization decreases MeCP2 occupancy of CGI3 CpGs, thereby enhancing transcription (Murgatroyd et al., 2009). This landmark study therefore shows that dynamic and long-lasting DNA methylation changes can and do occur in postmitotic neurons.
Similarly, Yakovlev et al. (2010) have traced age-dependent downregulation of caspase-3 production in rat brain to significantly lower levels of histone 3 acetylated Lys14 and histone 4 acetylated Lys5, 8, 12, and 16, as well as to differential methylation of specific CpG sites within the caspase-3 promoter. These sites are in a region that is essential for caspase-3 promoter activity, and correspond to predicted binding sites for several transcription factors such as Ets-1 and Ets-2 that are known to help control caspase-3 synthesis and to play critical roles in neuronal differentiation, development, and death. Notably, Ets-1 and Ets-2 themselves did not show age-related decline in the study, highlighting the potential importance of interactions between DNA methylation modifications and transcription factor activity. That is, transcriptional control of a gene by its transcription factors may be significantly altered by differential methylation of the binding sites for those transcription factors.
New research has also shown that, in addition to their roles in DNA methylation, MBD2 and DNMT3a/b may participate in dynamic demethylation processes. It has been reported, for example, that transient co-expression of MBD2 and methylated promoters results in demethylation and activation of gene expression, whereas knockdown of MBD2 inhibits replication-independent, active demethylation by valproate (reviewed in Szyf, 2009). Cyclical methylation/demethylation processes that mediate bouts of active and inactive transcription over periods of tens of minutes have also been demonstrated, and appear to help regulate the expression of multiple genes (Metivier et al., 2008; Kangaspeska et al., 2008). Here, DNMT3a/b, in concert with p68, bind and deaminate selective CpG sites, creating mismatches that are recognized by TDG and repaired by the BER. Because the repaired CpGs are no longer methylated, the local chromatin environment becomes poised for transcription. Following transcription, MBDs, MeCP2, and DNMTs are recruited and remethylate the CpG sites. Transcription is therefore turned off or retarded. The cycle may then begin again with DNMT3-mediated CpG deamination (Metivier et al., 2008). Although these dynamic MBD and DNMT demethylating mechanisms have, as yet, been examined only in non-CNS cells, there is no obvious reason why they should not occur in neurons. Similarly, a recent study has suggested that histone acetylation and deacetylation, like DNA methylation and demethylation, are dynamic, rapid-turnover processes that can poise genes for transcription in peripheral cells (Clayton et al., 2006). As such, all these mechanisms may warrant significant attention in future neuroepigenetics research.
4.0 Epigenetic regulation of aging
Aging is universally considered to be one of the most salient risk factors for AD, with increasing risk for the disorder cumulating until at least the ninth decade of life (Gao et al., 1998; Kukull et al., 2002). Why aging should be a risk factor for AD (and other age-related disorders), however, is not well understood, particularly at a mechanistic level. Potentially deleterious changes in mitochondria/oxidative stress (Crouch et al., 2007), gonadotropins (Fuller et al., 2007), calcium (Thibault et al., 2007), glucocorticoids (Landfield et al., 2007), inflammation (Duenas-Gonzalez et al., 2008), trace metals (Brewer, 2007), insulin (Craft, 2005), cerebrovascular supply (Bailey et al., 2004), the cell cycle (Macaluso et al., 2005), Aβ (Selkoe, 2003), tau (Maeda et al., 2006), and hundreds to thousands of genes (Parachikova et al., 2007; Berchtold et al., 2008) occur both in aging and AD, but a coherent explanation for why they occur and if their co-occurrence in aging and AD is coincidence or meaningful remains elusive.
DNA methylation and histone modifications have been widely implicated in the phenotypic alterations that occur during cellular senescence and the aging of various organisms (reviewed in Wilson and Jones, 1983; Bandyopadhyay and Medrano, 2003; Liu et al., 2003; Fraga and Esteller, 2007; Poulsen et al., 2007), and may provide a link between aging and AD. Histone acetylation mechanisms, particularly those involving the Sir2 family of histone deacetylases, have been linked to aging and senescence in yeast and invertebrates (reviewed in Bandyopadhyay and Medrano, 2003), but have yet to be investigated in mammals. By contrast, many studies have reported a genome-wide tendency to DNA hypomethylation with age in multiple vertebrate organs, including brain, liver, small intestine mucosa, heart, and spleen; multiple cell types, including fibroblasts and T lymphocytes; and multiple vertebrate species, including aging salmon, mice, rats, cows, and humans (Vanyushin et al., 1970; Vanyushin et al., 1973; Romanov and Vaniushin, 1980; Wilson et al., 1987; Golbus et al., 1990; Richardson, 2002). Yu and colleagues (2006) assayed human peripheral blood mononuclear cell DNA for the percentage of methylated to total cytosines and observed a 3% per decade decrease from the first to the tenth decade of life. In vitro, hypomethylation of human and mouse fibroblasts cultured to senescence has also been observed (Wilson and Jones, 1983). Likewise, progressive age-related decline in total genomic methylcytosine has been reported in various organisms (Mays-Hoopes, 1989). Because DNMT1, which is responsible for maintaining DNA methylation of CpG sites, is also progressively lost with age (Lopatina et al., 2002), it has been speculated that progressive, age-related, genome-wide hypomethylation may be due to parallel DNMT1 deficits (Liu et al., 2003), and that the process overall may serve as a counting mechanism that triggers cellular senescence (Neumeister et al., 2002). Age-dependent increases in S-adenosyl-homocysteine (SAH) relative to SAM (Gharib et al., 1982; Varela-Moreiras et al., 1994) might also play a role, since SAH inhibits methylation reactions, including DNA methylation. In turn, overall decline in genomic methylation with age has been linked to specific age-related pathogenic processes such as aberrant cell cycle events (e.g., p-53-dependent apoptosis) (Jackson-Grusby et al., 2001) and the increased inflammatory tone that occurs with advancing age (Wilson, 2008).
Hypomethylation of non-coding regions and other sites also occurs with age and has been suggested to be relevant to the aging process. For example, repetitive sequences (Romanov and Vanyushin, 1981; Mays-Hoopes et al., 1986; Rath and Kanungo, 1989), retrotransposons (Barbot et al., 2002), and endogenous retroviruses (Ono et al., 1989) that are normally repressed by DNA methylation can become hypomethylated with age, potentially promoting chromosome translocations, retrotransposon activation, and retrovirus emergence, respectively (reviewed in Richardson, 2002).
Age-dependent hypomethylation of a number of specific genes related to AD has been reported. For example, methylation of cytosines in the APP promoter, particularly GC-rich elements from approximately −270 to −182, is significantly lower in autopsy cases 70 years old compared to cases <70 years old (Tohgi et al., 1999a). DNA methylation within the tau promoter reportedly declines overall with age, but with interesting variations at different transcription factor binding sites: binding sites for GCF, which represses GC-rich promoters, become hypomethylated with age, whereas binding sites for Sp1, a transcriptional activator, become hypermethylated. These changes might therefore represent a double hit on tau gene transcriptional activity, causing decreased activity overall with age (Tohgi et al., 1999c). Promoter methylation of the receptor for advanced glycation end products (RAGE) gene exhibits similar complexity. Overall methylation of the promoter declines with age, but the change is manifest at cytosine residues other than CpG dinucleotides: CpC, CpA, and CTG sequences within AP2 and SP1 binding sites show significant hypomethylation with age (Tohgi et al., 1999b). Expression of the immune/inflammatory antigen CD11a increases with age (Pallis et al., 1993), an effect that appears to be linked to an age-related hypomethylation of flanking repeats some 1 kb 5′ to the CD11a promoter start site (Zhang et al., 2002).
Despite the trend to genome-wide and gene-specific DNA methylation with age, it should be emphasized that the trend is no more than that, as many genes exhibit age-related decreases in expression rather than the upregulation that is predicted by hypomethylation or histone acetylation. CpG islands on several specific genes undergo age-dependent hypermethylation (e.g., estrogen receptors, insulin-like growth factor 2) (Issa et al., 1994, 1996). Tumor suppressor genes appear particularly apt to show increasing methylation with age, providing a potential link to age-related cancers (Romanov and Vaniushin, 1980; Mays-Hoopes, 1989; Issa et al., 1994; Issa and Baylin, 1996; Neumeister et al., 2002; Richardson, 2002; Liu et al., 2003; Kim and Sharpless, 2006; So et al., 2006; Fraga and Esteller, 2007; Jacinto and Esteller, 2007). Many of these hypermethylation events could be due to age-related increases in DNMT3a/b expression (Lopatina et al., 2002; Liu et al., 2003), as these methyltransferases are responsible for de novo methylation of DNA. Alternatively, they have also been linked to demethylation processes (Metivier et al., 2008; Kangaspeska et al., 2008).
Further adding to the complexity of aging changes in DNA methylation, tissue-specific patterns should also be noted. For example, the tumor suppressor gene c-fos exhibits increasing CpG methylation with age in liver, but not brain or spleen (Ono et al., 1989). In brain, methylation profiles may differ substantially from one region to another (Ladd-Acosta et al., 2007) and even from one subregion to another (e.g., hippocampal dentate gyrus and CA fields) (Brown et al., 2008), underscoring the value of brain regional comparisons in epigenetic studies of aging and AD.
In addition to aging, epigenetics plays a major role in development (reviewed in Reik and Dean, 2001). These mechanisms could be relevant to AD since overt clinical symptoms of the disorder virtually never appear until after the developmental stages of infancy, childhood, and early adolescence have been completed, and this is true not simply in late-onset patients but in patients carrying APP, PS1, or PS2 mutations. Thus, epigenetic changes earlier in life might be a necessary but not sufficient step toward AD in susceptible individuals, a key concept in the “LEARn” (latent early-life associated regulation) model of age-related neurologic disorders (Lahiri et al., 2007; Wu et al., 2008b). Support for this hypothesis comes from APP transgenic mouse research wherein earlier epigenetic manipulations appear to accelerate or delay the expression of Aβ pathology (Fuso et al., 2008). Likewise, early exposure of monkeys to Pb reportedly decreases DNMT activity, increases APP, BACE, and SP1 expression, and alters levels and distribution of Aβ in the animals in late life (Wu et al., 2008a).
5.0 Epigenetic alterations in AD
5.1 Histone modifications
Several reports have demonstrated alterations in histone proteins in AD. Phosphorylation of histone 3, a key step in the activation of the mitotic machinery, is increased to a hyperphosphorylated state in AD hippocampal neurons (Ogawa et al., 2003). A non-nuclear form of histone 1 appears to be upregulated in astrocytes and neurons in brain regions that are rich in AD pathology (Bolton et al., 1999). Linker Histone H1, a vital component of chromatin, has been reported to preferentially bind Aβ-42, as well as Aβ–like structures of numerous proteins (Duce et al., 2006). In addition, the H1 molecule has been shown to be a major target for poly (ADP-ribosyl)ation in areas of AD brain with ischemic brain injury (Love et al., 1999).
Manipulation of histone tail acetylation with HDAC inhibitors has been investigated in several animal models of AD. For example, it has been reported that after fear conditioning training in APP/PS1 mice, levels of hippocampal acetylated histone 4 (H4) were about 50% lower than in wild-type littermates. Treatment with the HDAC inhibitor Trichostatin A increased the levels of acetylated H4 and contextual freezing performance to wild-type values (Francis et al., 2009). Treatment with HDAC inhibitors has also been shown to induce sprouting of dendrites, increase the number of synapses, and reinstate learning behavior and access to long-term memories in CK-p25 transgenics (Fischer et al., 2007). In addition, valproic acid, which has HDAC1 inhibitor activity, has been shown to decrease Aβ production and reduce plaque burden in the brains of PDAPP(APP(V717F)) transgenic mice (Su et al., 2004). Similarly, in the Tg2576 mouse model of AD, a daily dose of phenylbutyrate, another HDAC inhibitor, reversed spatial memory loss and normalized levels of phosphorylated tau in the hippocampus, but failed to alter Aβ levels (Ricobaraza et al., 2009). Conversely, in a cortical neuron culture model, overexpression of APP resulted in a decrease in histone 3 and histone 4 acetylation, as well as a decrease in CREB-binding protein levels (Lonze and Ginty, 2002; Rouaux et al., 2003).
In summary, although it appears that histone modifications occur in AD, AD animal models, and AD culture models, the pattern of changes is complex and could entail both histone acetylation increases and decreases at specific loci that function disjointedly or in concert.
5.2 miRNAs
Global analyses of AD versus normal elderly control brains have revealed changes in the levels of several specific miRNAs that were concordant across two separate studies (Hebert et al., 2008; Nunez-Iglesias et al., 2010). Notably, however, both positive and negative relationships between levels of the miRNAs and levels of their targets were observed, suggesting the operation of upstream factors (Tsang, Zhu, and van Oudenaarden, 2007). A third study has provided the additional caveat that significant AD changes in many miRNAs may be common to both pathologically-vulnerable and pathologically-spared brain regions. Other miRNAs, however, were specific both to AD and to areas of extensive AD pathology, and their targets had resonance with mainstream AD pathologic pathways (Cogswell et al., 2008).
In vitro experiments with HeLa, COS1, and HEK293 cells have shown that luciferase expression controlled by the APP 3′UTR can be regulated by miR-20a, miR-17-5p, and miR-106b miRNAs. Transient transfection of these miRNAs downregulated APP—expression in the case of miR-20a by inhibiting translation rather than by degradation of APP mRNA. Conversely, blocking expression of miR-20a increased endogenous APP levels by some 50%. Subsequent developmental studies of mouse brain revealed dramatic reductions in all three miRNAs that were significantly correlated with increased APP protein expression. The fact that APP mRNA levels remained stable under these conditions again suggested that the miRNAs have their effects on APP by inhibiting translation rather than promoting cleavage of APP mRNA. Finally, human pathology studies have shown that miR-106b is significantly decreased in AD cortex. Although all these findings indicate that APP may be targeted by miRNA mechanisms, levels of miR-20a, miR-17-5p, and miR-106b in AD cortex do not appear to correlate with levels of APP protein (Hebert et al., 2009).
Two studies have used computational analysis methods to reveal miRNA target sites in BACE mRNA that may be functionally relevant to AD pathogenesis. Wang and colleagues (2008) found multiple target sites for miR-107 in the 3′-untranslated region of BACE, and went on to show significant decreases in miR-107 that were apparent even in early AD cases, particularly in the large pyramidal cell cortical layers that may be especially vulnerable to AD pathology (Rogers and Morrison, 1985). Moreover, when assayed in the same cases BACE mRNA expression appeared to be negatively associated with miRNA-107 levels (Wang et al., 2008). An additional miRNA, miR-29a/b-1, also appears to target BACE mRNA and, like miR-107, is decreased in AD and inversely correlated with BACE in this case, BACE protein levels (Hebert et al., 2008).
5.3 DNA methylation
5.3.1 Genome-wide and multi-gene studies
Although an early analysis reported no significant difference in percent CCGG methylation of DNA in AD cortex, a number of caveats were given (Schwob et al., 1990), particularly the fact that CCGG methylation only covers approximately 20–30% of CpG sites in the genome. Methylation status of 12 specific genes that have been implicated in AD pathology has also been reported to exhibit significant “epigenetic drift”, although the manner in which the data were analyzed makes it difficult to determine whether methylation was increased or decreased in AD. The study did note, however, that an age-specific epigenetic drift was observed in some of the CpG sites within the DNMT1 promoter (Wang et al., 2008).
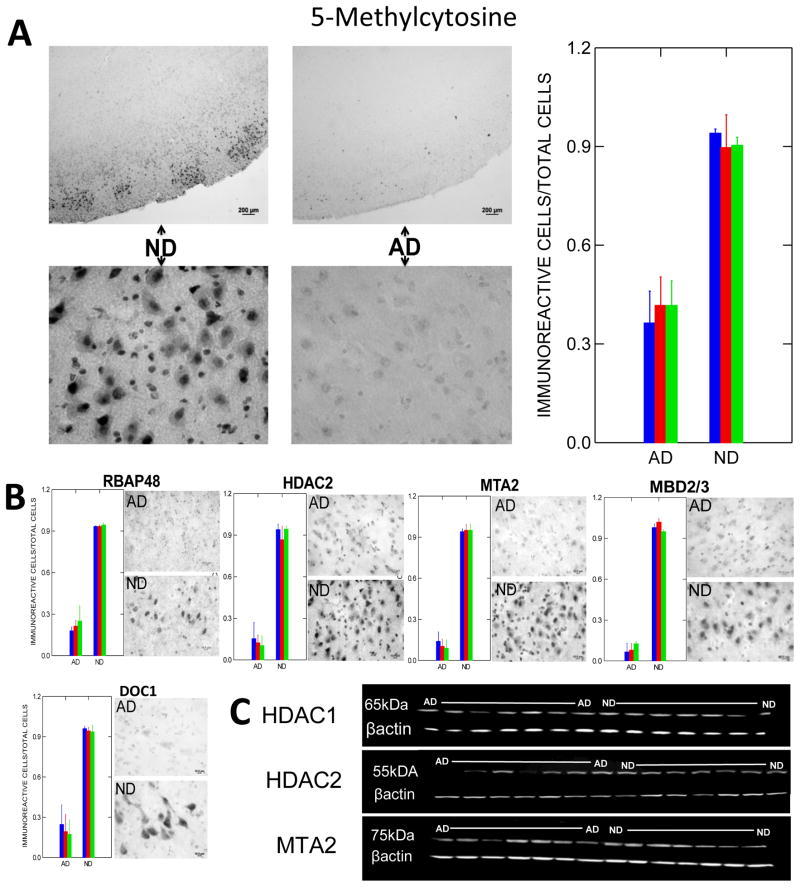
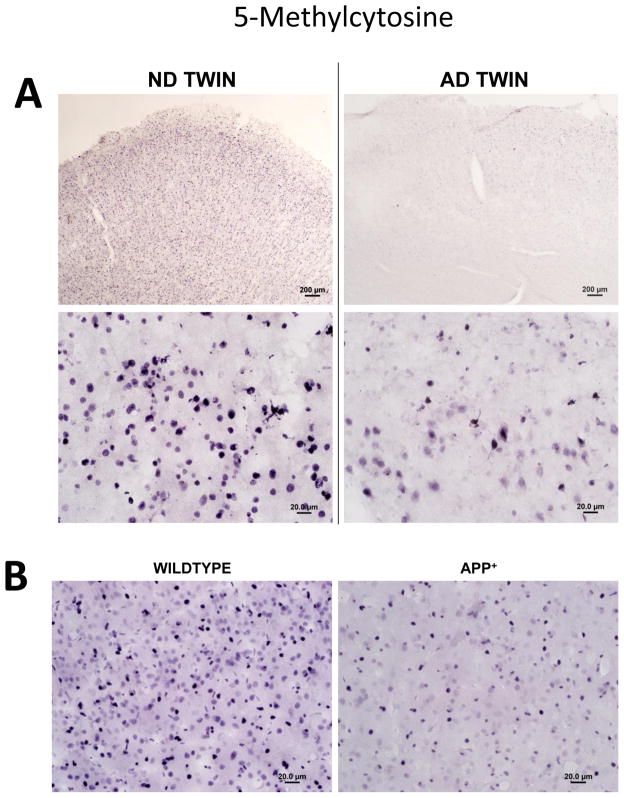
From a genome-wide perspective, our laboratory has reported decreased immunoreactivity for some seven different markers of DNA methylation in AD compared to matched, non-demented elderly control (ND) cortical neurons and glia (Figs. 2, 3), whereas no such changes were observed in cerebellum, a brain region that is relatively spared in AD (Mastroeni et al., 2008). Highly similar results were subsequently obtained in a set of monozygotic twins discordant for AD (Mastroeni et al., 2009) (Fig. 4A), as well as in APP-overexpressing transgenic mice (Fig. 4B).
Fig. 2. DNA methylation markers in AD and ND cortex.
A) Typical immunoreactivity for 5-methylcytosine, a global marker of DNA methylation, in AD and ND entorhinal cortex (from Mastroeni et al., 2008, with permission). Cases were well matched for age, gender, and postmortem intervals, which were all less than 3 hours 15 minutes. Shaded bars represent means for different cases. Although glia and virtually all types of neurons exhibit 5-methylcytosine immunoreactivity, layer II “island” neurons, among the most vulnerable to AD pathology, exhibit particularly intense staining in ND cases, as shown in the upper left micrograph at low power. Such staining is weak to absent in AD cases (upper right micrograph). High power micrographs show the expected nuclear localization of immunoreactivity. Far right panel shows counts of immunoreactive neurons per total neurons per field. Normalizing to total neurons is important, as it helps to demonstrate loss of methylation within cell nuclei rather than loss of the methylated cell population itself. The significant decrement in AD (P < 0.001) is typical of dozens of AD and ND cases examined, with little to no overlap in any case. B) Representative immunoreactivity and cell counts for various MeCP1 components in AD and ND neocortex. Significant AD decrements (P < 0.05) were observed with all the markers—again with little to no overlap. C) Western blots (normalized to β-actin) for these and other methylation markers exhibit immunoreactivity at appropriate molecular weights, with AD/ND differences similar to those observed by immunohistochemistry, suggesting that the latter are not due to cross-reactivity with other antigens.
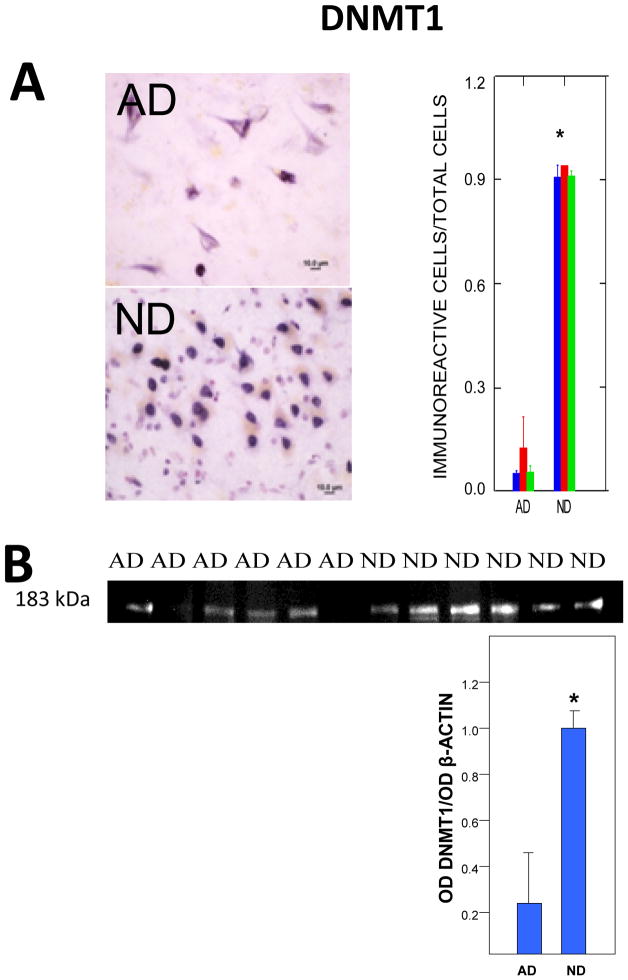
Fig. 3. Immunoreactivity for DNMT1, the most prevalent methyltransferase in adult mammals.
A) Typical DNMT1 immunoreactivity and cell counts (P < 0.001) in AD and ND neocortex. Shaded bars represent means for different cases. B) Western blots (normalized to β-actin standards) show immunoreactive bands at appropriate molecular weights for DNMT1 and a significant (P < 0.01) decrement in AD cases.
Fig. 4. Decreased overall DNA methylation in monozygotic twins discordant for AD.
A) Global hypomethylation (5-methylcytosine immunoreactivity) in an AD monozygotic twin compared to his normal sibling at low (upper micrographs) and high (bottom micrographs) power (Mastroeni et al., 2009) (courtesy of PLoS1). B) Similar findings have recently been observed by our group in APP transgenic mice compared to their wildtype littermates.
As previously noted, folate/methionine/homocysteine metabolism is critically linked to DNA methylation mechanisms. Folate deficiency in humans and in animal models, for example, typically results in global hypomethylation that is at least partially reversible with folate supplementation (reviewed in Choi and Mason, 2002; Choi and Friso, 2005; Fuso et al., 2005, 2008). Deficits in folate and alterations in the methionine/homocysteine cycle have been reported in aging and AD (reviewed in Smith, 2008), and may therefore provide a basis for the tendency to genome-wide hypomethylation summarized earlier in this review. Although one prospective study failed to find an association between dietary folate, vitamin B12, or vitamin B6 with incident AD (Morris et al., 2006), CSF folate has nonetheless been reported to be significantly decreased in AD (Serot et al., 2001), as has CSF and brain SAM and one of its synthesizing enzymes, methionine S-adenosyltransferase (Bottiglieri et al., 1990; Morrison et al., 1996). Increases in brain SAH (Kennedy et al., 2004) and plasma homocysteine (Clarke et al., 1998; Smith et al., 2008), which inhibit DNA methylation, have also been observed. Elevated plasma homocysteine has been reported to be a significant risk factor for AD in dementia-free cohorts of both the Framingham Study of Aging (Seshadri et al., 2002) and the Conselice Study of Brain Aging (Ravaglia et al., 2005). In fact, dietary effects of folate and homocysteine manipulation have been implicated in cognitive impairment generally, and in a wide range of neurologic conditions, including AD, Parkinson’s disease, depression, cortico-basal degeneration, multiple sclerosis, and fronto-temporal dementia (Obeid et al., 2007).
5.3.2 Aβ-related genes
Epigenetic influences on Aβ-producing mutations have long been suspected based on the heterogeneity of clinical presentation by patients who share mutations in the same APP, BACE, or PS1 genes—sometimes in identical promoter sites (Larner and Doran, 2006). Consistent with this notion, the APP (Davidson et al., 1992; West et al., 1995; Mani & Thakur, 2006), BACE, and PS1 (Fuso et al., 2005) genes all contain manipulable, methylated CpG sites. A case study has reported complete demethylation of the APP gene in an AD postmortem cortical sample, but not in similar samples from a normal control or Pick’s disease patient (West et al., 1995). In vitro, expression of the BACE and PS1 genes is enhanced after folate deprivation-induced hypomethylation, and restored to normal when folate deprivation is accompanied by SAM supplementation. Expression of TACE, ADAM10, and APP was unaffected by these manipulations (Fuso et al., 2005). In vivo, exposure of APP-overexpressing transgenic mice to a folate/B12/B6-deficient diet is associated with enhanced SAH relative to SAM, PS1 and BACE upregulation, enhanced Aβ deposition, and an accelerated appearance of intraneuronal Aβ and cognitive deficits (although the latter was quite modest) (Fuso et al., 2008).
Finally, it has been demonstrated that Aβ itself may induce genome-wide hypomethylation in murine cerebral endothelial cell cultures while, at the same time, causing specific hypermethylation and repression of the gene for neprilysin, an Aβ degrading enzyme (Chen et al., 2009). Our laboratory has replicated the global hypomethylating effects of Aβ in human SK-N-BE2 neuroblastoma cells, and extended the results to show hypomethylation (as well as several instances of hypermethylation) of specific CpG islands in the BACE (Fig. 5) and caspase-3 genes (Grover et al., unpublished). Together with hypermethylation of neprilysin, these effects suggest the potential for a vicious cycle in which Aβ-induced methylation changes feed back to enhance Aβ production, further methylation changes, and further Aβ production. Moreover, if the overall trend to hypomethylation after Aβ exposure were to functionally impact other key AD genes, additional synergisms might occur. For example, TNF-α (Wilson, 2008) and caspase-3 are upregulated when hypomethylated (Muerkoster et al., 2008), and increased levels of TNF-α (Janelsins et al., 2008; McAlpine et al., 2009; Sommer et al., 2009) and caspases (Xie et al., 2008; Xiong et al., 2008) enhance Aβ expression, potentially generating new vicious cycles.
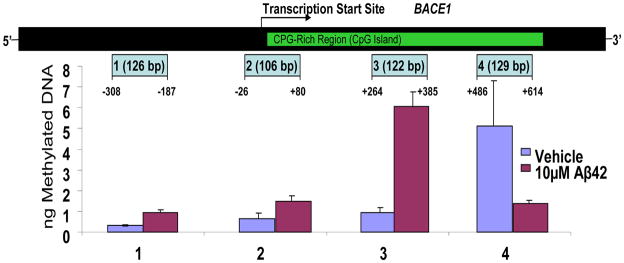
Fig. 5. MethylMiner methylation profile of selected flanking and initial BACE promoter sites after exposure of differentiated SK-N-BE(2) neuron-like cultures to 10 μM Aμ42.
MethylMiner Methylated DNA Enrichment Kits (Invitrogen, Carlsbad, CA) were employed to enrich and fractionate double-stranded DNA based on CpG methylation density. The highly methylated region showed significant hypomethylation while poorly methylated regions exhibited hypermethylation (Grover et al., unpublished). Correlations with Aβ production can help establish the functional relevance of methylation modifications at these and other CpG sites within the BACE gene.
5.3.3 Tau
Methylation mechanisms with respect to tau and neurofibrillary tangle formation have also been explored. As previously noted, in normal adults the AP2 binding site of the tau promoter is not methylated, but the SP1 and GCF binding sites are. SP1, a transcriptional activator site, is increasingly methylated and GCF, a promoter repressor site, is increasingly demethylated with age, suggesting an overall downregulation of tau gene expression (Tohgi et al., 1999c). Although a corresponding age-related decrease in normal tau protein, particularly in frontal cortex and hippocampus, has been reported, there was no correlation with the modest neurofibrillary tangle pathology in the same subjects (Mukaetova-Ladinska et al., 1996).
Tau phosphorylation mechanisms are, however, subject to cytoplasmic methylation reactions, and have been the subject of several recent reports. Our studies, for example, revealed co-localized immunoreactivity for the methyl binding complex component p66α (as well as HDAC1) with PHF1-positive neurofibrillary tangles (Mastroeni et al., 2008). PP2A is an enzyme that can dephosphorylate phosphorylated tau, an action that may be potently activated by methylation of the PP2A catalytic subunit at its L309 site. In N2a cultures carrying the APP Swedish mutation (APPswe) and in APPswe/PS1 transgenic mice, levels of demethylated PP2A at L309 were significantly increased, corresponding with increases in tau phosphorylation at the Tau-1 and PHF-1 sites. Treatment with Aβ25–35 led to demethylation and enhanced tau phosphorylation (Zhou et al., 2008). Like treatment with Aβ25–35, exposure of rodent primary neuron cultures to methotrexate, a folate antagonist, also has been reported to result in demethylation of PP2A, with attendant enhancement of tau phosphorylation (as well as upregulation of APP and BACE) (Yoon et al., 2007). Consistent with all these findings, injections of homocysteine into rats for two weeks yielded decreased PP2A L309 methylation and PP2A activity, effects that were reversed by simultaneous administration of folate and vitamin B12. Hippocampal samples from the rats and from AD patients exhibited immunohistochemical co-localization of demethylated, but not methylated PP2A with hyperphosphorylated tau (Zhang et al., 2008).
5.3.4 Aberrant cell cycle/apoptosis
A wide range of evidence suggests that attempted or aberrant re-entry into the cell cycle and/or apoptosis of neurons may be a common neurodegenerative mechanism in AD (reviewed in Neve and McPhie, 2006). Many of the critical components of the cell cycle and apoptosis pathways are upregulated in degenerating AD neurons, and are subject to regulation by DNA methylation, including the P16, P21, P27, P53, RB1 (Moreira et al., 2009), cyclin B2 (Tschop and Engeland, 2007), ARF (Robertson and Jones, 1998), caspase 1 (Jee et al., 2005), caspase 3, caspase 7, caspase 8, and caspase 9 (Muerkoster et al., 2008) genes. Hypomethylation of these genes would be expected to promote aberrant cell cycle events, and global hypomethylation has been reported to occur in cells as they move from the G(0) stage characteristic of postmitotic neurons to the G(1) stage characteristic of cell cycle re-entry (Brown et al., 2007). Indirect support for the role of hypomethylation in aberrant cell cycle events is provided by studies demonstrating apoptosis of cultured neurons when exposed to high homocysteine levels (Ho et al., 2002). Although many other biologic effects are possible, such treatment is known to hypomethylate DNA (Reynolds, 2006), and concurrent treatment with SAM antagonized the apoptotic effect (Ho et al., 2002). Interactions of the histone acetyltransferase Tip60 with the γ-secretase-generated APP C-terminal fragment APP-CT58, which translocates to the nucleus, also lead to apoptosis of human H4 neuroglioma cells (Kinoshita et al., 2002).
5.3.5 Inflammation
Key genes at almost every level of the inflammatory response appear to be subject to DNA methylation influences. A highly abbreviated list of examples includes complement C3, factor B, IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-13, TNF-α, TNFRI, INF-γ, SOCS, S100A2, the chemokine receptors CXCR4 and CCR7, clusterin (apoJ), and iNOS (Mejia et al., 1995; Mori et al., 2005; Buslei et al., 2006; Suuronen et al., 2007; Mi and Zeng, 2008; Nile et al., 2008; Parker et al., 2008; Pieper et al., 2008; van Panhuys et al., 2008; van Rietschoten et al., 2008). The TNF-α promoter, for example, contains 12 CpG-rich sites. Two of those sites (−304 and −205) are hypomethylated on exposure of macrophages to the classic inflammatory stimulus lipopolysaccharide, and their extent of hypomethylation is correlated with increasing expression of TNF-α (Wilson, 2008). Likewise, a single CpG site in the IL-6 promoter (−1,181) is significantly hypomethylated in rheumatoid arthritis and this hypomethylation is correlated with IL-6 mRNA levels (Nile et al., 2008). Although virtually none of the many factors in inflammation has been investigated with respect to its DNA methylation status in AD, this could be a fertile area for investigation given the heightened expression of these molecules in the disorder. A recent Parkinson’s disease study, for example, has demonstrated that the TNF-α promoter is hypomethylated in the substantia nigra relative to several other brain regions. Since DNA methylation of CpG sites in the TNF-α promoter inhibits SP1 and AP-2 transcription factor binding and decreases TNF-α expression, it has been speculated that nigral TNF-α hypomethylation might explain the heightened vulnerability of nigral dopamine neurons to TNF-α-mediated inflammatory reactions (Pieper et al., 2008).
5.3.6. Apolipoproteins
Again, no specific studies have been done on the methylation status of apolipoprotein E (ApoE) in AD. Notably, however, Wang and colleagues (2008) have reported that although the ApoE promoter is poorly methylated, the ApoE ε4 allele contains methylated CpG sequences that are not extant in the ε2 or ε3 alleles. Because not all ε4 carriers develop AD, it would be of great interest to know if methylation status at ε4 CpG sites is altered in ε4 carriers who develop (or do not develop) AD. Similarly, it might be useful to determine whether the significant but relatively low penetrance of ApoJ (clusterin) SNPs to AD risk (Harold et al., 2009; Lambert et al., 2009) might be explained by methylation changes, since the ApoJ promoter is rich in CpG sites and expression is increased on hypomethylation (Suuronen et al., 2007).
6.0 Conclusions
Global epigenetic changes, acting on a wide range of genes and biological pathways, appear to help orchestrate the cellular alterations that drive development, aging, and, in some cases, disease. Likewise, global epigenetic changes have been observed in pathologically-vulnerable regions of the AD brain, and key genes in virtually every mainstream pathologic pathway in AD are known to be labile to such changes. The ability of epigenetic mechanisms to initiate an extremely wide range of pathogenic responses—an orchestrating capacity perhaps equaled only by transcription factors (which themselves often work together with epigenetic mechanisms to direct expression in specific sets of genes) (e.g., Agrawal et al., 2007; Ivascu et al., 2007, Yakovlev et al., 2010)—provides a relatively unique integrative framework for the diverse genetic factors and multifactorial pathology of AD, including Aβ, tau, inflammation, mitochondrial metabolism, oxidative stress, and aberrant cell cycle/apoptosis events. Moreover, the epigenetic modifications that have been reported in AD, particularly with respect to DNA methylation, typically resonate with similar trends in aging, and may therefore help explain not only the pathologic complexity of AD, but also the particular salience of aging as an AD risk factor.
Finally, whereas the field of epigenetics has previously emphasized mechanisms for preserving epigenetic profiles across generations of dividing cells, there is now ample precedent for active, dynamic epigenetic alterations in postmitotic cells, including neurons, that play important roles in neuroendocrine, learning and memory, and apoptotic processes. These latter, landmark studies provide the tools for subsequent explorations of how the epigenetic modifications that have already been reported in AD occur, as well as a mechanistic underpinning for the AD genome-wide methylation profiling that is now in progress.
7.0 Future Directions
At the level of basic research, DNA methylation profiling of aging and AD subjects is eagerly awaited in order to develop a better portrait of the normal methylation status of all genes across the AD genome, how that status may change in AD, and whether or not such changes implicate AD-related proteins and pathogenetic processes. These studies would be especially significant if they were conducted in tandem with genome-wide gene expression arrays because the experiments would then provide validation of the functional effects of DNA methylation changes on gene expression. In addition, knowing the methylation states of genes containing putative AD SNPs could be useful. Many such SNPs, for example, remain controversial because their odds ratios for disease risk consistently hover at the statistical edges of significance or they fail to replicate in some studies but not others. Epigenetic regulation of the SNP genes could account for this variability. For example, a gain or decrease of function SNP could be compensated for by epigenetic downregulation or upregulation, respectively, of the gene’s expression, so that some carriers might in effect possess the SNP with relative impunity. Finally, as valuable as large-scale epigenetic profiling of the AD genome will be, it will still not tell us why or how the profiles were altered, nor will it give us a detailed profile of each gene. Genome-wide methylation profiling is presently only able to sample a few of the CpG-rich sites within each gene, although the technology is rapidly expanding. Because both hyper- and hypomethylation can occur at different CpG sites in the same gene, with one but not the other causing functional changes in gene expression (e.g., Murgatroyd et al., 2009), follow-up studies giving detailed methylation maps of AD-relevant genes and concomitant changes in their expression will be essential. These same considerations may also apply to other neurologic conditions such as schizophrenia and bipolar disorders, where epigenetic mechanisms are being pursued (reviewed in Pidsley and Mill, 2010).
Of course, to hypothesize that epigenetic changes play a role in brain aging, AD, and other neurologic disorders still begs the question of what causes the epigenetic changes? The environment that cells and organisms are exposed to can have a profound influence on epigenetic mechanisms (Waterland and Michels., 2007; Smith and Kim, 2008), but simple stochastic processes may do so as well (Jaenisch and Bird, 2003). Whether as environmentally-driven or randomly-occurring events, however, the probability of epigenetic modifications must logically increase with time, and increased time is precisely what the advanced ages reached by many human beings may afford. Some of these modifications may be inconsequential, depending on the CpG site, the gene, or the organ. For example, inadvertent upregulation of an Aβ-synthesizing gene might have little to no impact on a muscle cell, whereas it could be highly significant in a pyramidal neuron. Similarly, the brain lacks several major defenses against inflammatory attack (Gasque et al., 2002; Zanjani et al., 2005), so that the inadvertent upregulation of a pro-inflammatory gene might be uniquely problematic there. Thus, the origin and organ-specific consequences of epigenetic modifications are important considerations for AD epigenetic studies, and will continue to be critical targets for AD basic research into epigenetic mechanisms.
At the clinical level, the initiation of AD trials with folate/B vitamins/SAM may constitute one means of testing an epigenetic orchestration hypothesis of AD, although it is becoming increasingly evident that reversing the course of human AD with any treatment may be an over-ambitious goal. For example, in a recent trial B vitamin treatment significantly slowed cognitive decline in mild AD, but was without effect in more advanced cases (Aisen et al., 2008). A trial in mild cognitive impairment patients might therefore be of great interest. A second impediment to succesful treatment of epigenetic defects may be achieving sufficiently high levels of epigenetic therapeutics not only within cells, but within neuronal nuclei. Moreover, the many different biochemical pathways impacted by epigenetic mechanisms may make targeting specific disease processes difficult. Beyond folate and other such approaches, cancer chemotherapeutics that are directed at epigenetic mechanisms are available, but would need to be considered carefully. DNA demethylating agents such as 5-azacytidine and decitabine, for example, might actually prove harmful in AD given the profound global hypomethylation of AD neurons (Mastroeni et al., 2008, 2009). HDAC inhibitors such as valproate, by contrast, might counter many of the epigenetic changes that have been reported in AD, and have, in fact, been used successfully to improve cognitive outcome measures in AD transgenic mouse models (Su et al., 2004; Fischer et al., 2007; Francis et al., 2009; Ricobaraza et al., 2009; Guan et al., 2009). Treatment trials in human AD patients, however, have not been particularly encouraging, perhaps due to somnolence, agitation, and other side effects of the drug (Profenno et al., 2005; Herrman et al., 2007) or to lack of specificity of the drug to epigenetic mechanisms alone.
With respect to the development of new treatments for AD, a direct role for epigenetics would be the design and application of epigenetic therapeutics that have appropriate effects on specific epigenetic mechanisms in specific genes or sets of genes. As we have emphasized throughout, however, one of the defining hallmarks of epigenetic mechanisms is their ability to exert effects over many genes, and, accordingly, virtually all present epigenetic therapeutics also exert their effects over many genes. Unless a broad modifier such as an HDAC inhibitor can be found that just happens to impact the right genes, while sparing significant effects in others, the specificity requirements of an AD epigenetic therapeutic will be challenging. Nonetheless, elucidating specific genes that undergo significant epigenetic alterations in AD—as is now in progress in our laboratory and elsewhere—could, at the very least, help direct our attention to the most salient pathogenic elements of the disorder and to more conventional (e.g., agonist/antagonist) approaches to the protein products of the epigenetically-modified genes.
Acknowledgments
Preparation of this review was supported by NIA AGO-7367-19 (JR), NIA AG 036400 (PC), and the Arizona Alzheimer’s Disease Consortium (JR).
Glossary
- HAT
Histone acetyltransferases
- HDAC
Histone deacetylase
- CpG
Cytosine-phosphate-guanine
- DNMT
DNA methyltransferase
- MECP
Methyl CpG binding protein
- SAM
S-adenosylmethionine
- SAH
S-adenosyl-homocysteine
- rDNA
Ribosomal DNA
- miRNA
Micro-RNA
- BACE
Beta secretase
- PS1
Presenillin 1
- PS2
Presenillin 2
- APP
Amyloid precursor protein
Footnotes
Disclosure
The authors state that they have no actual or potential conflict of interest that could inappropriately influence this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin M-R, Gunel M, Spertus JA, Leckman JF, Dure LS, Kurlan R, Singer HS, Gilbert DL, Farhi A, Louvi A, Lifton RP, Sestan N, State MW. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Murphy RF, Agrawal DK. DNA methylation in breast and colorectal cancers. Mod Pathol. 2007;20:711–721. doi: 10.1038/modpathol.3800822. [DOI] [PubMed] [Google Scholar]
- Aisen PS, Schneider LS, Sano m, Diaz-Arrastia R, Van Dyck CH, Weiner MF, Bottiglieri T, Jin S, Stokes KT, Thomas RG, Thal LJ. High-dose B vitamin supplementation and cognitive decline in Alzheimer disease: a randomized controlled trial. JAMA. 2008;300:1774–1783. doi: 10.1001/jama.300.15.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa S, Yamamuro Y. Involvement of histone acetylation in the regulation of choline acetyltransferase gene in NG108-15 neuronal cells. Neurochem Int. 2010 doi: 10.1016/j.neuint.2010.01.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Allfrey VG. Structural modifications of histones and their possible role in the regulation of ribonucleic acid synthesis. Proc Can Cancer Conf. 1966;6:313–335. [PubMed] [Google Scholar]
- Amir RE, Van Den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arendt T. Alzheimer’s disease as a disorder of dynamic brain self-organization. Prog Brain Res. 2005;147:355–378. doi: 10.1016/S0079-6123(04)47025-3. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Rivara CB, Rocher AB, Hof PR. The nature and effects of cortical microvascular pathology in aging and Alzheimer’s disease. Neurol Res. 2004;26:573–578. doi: 10.1179/016164104225016272. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay D, Medrano EE. The emerging role of epigenetics in cellular and organismal aging. Exp Gerontol. 2003;38:1299–1307. doi: 10.1016/j.exger.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv Exp Med Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. The occurrence and transmission of a pattern of DNA methylation in Xenopus laevis ribosomal DNA. Philos Trans R Soc Lond B Biol Sci. 1978;283:325–327. doi: 10.1098/rstb.1978.0032. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Bolton SJ, Russelakis-Carneiro M, Betmouni S, Perry VH. Non-nuclear histone H1 is upregulated in neurones and astrocytes in prion and Alzheimer’s diseases but not in acute neurodegeneration. Neuropathol Appl Neurobiol. 1999;25:425–432. doi: 10.1046/j.1365-2990.1999.00171.x. [DOI] [PubMed] [Google Scholar]
- Bottiglieri T, Godfrey P, Flynn T, Carney MW, Toone BK, Reynolds EH. Cerebrospinal fluid S-adenosylmethionine in depression and dementia: effects of treatment with parenteral and oral S-adenosylmethionine. J Neurol Neurosurg Psychiatry. 1990;53:1096–1098. doi: 10.1136/jnnp.53.12.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy T, Grant R, Champagne D, Meaney M. Maternal care Influences neuronal survival in the hippocampus of the rat. European journal Neurosci. 2003;10:2903–2909. doi: 10.1111/j.1460-9568.2003.02965.x. [DOI] [PubMed] [Google Scholar]
- Brewer GJ. Iron and copper toxicity in diseases of aging, particularly atherosclerosis and Alzheimer’s disease. Exp Biol Med. 2007;232:323–335. [PubMed] [Google Scholar]
- Brown SE, Fraga MF, Weaver IC, Berdasco M, Szyf M. Variations in DNA methylation patterns during the cell cycle of HeLa cells. Epigenetics. 2007;2:54–65. doi: 10.4161/epi.2.1.3880. [DOI] [PubMed] [Google Scholar]
- Brown SE, Weaver IC, Meaney MJ, Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Buslei R, Kreutzer J, Hofmann B, Schmidt V, Siebzehnrubl F, Hahnen E, Eyupoglu IY, Fahlbusch R, Blumcke I. Abundant hypermethylation of SOCS-1 in clinically silent pituitary adenomas. Acta Neuropathol. 2006;111:264–271. doi: 10.1007/s00401-005-0009-9. [DOI] [PubMed] [Google Scholar]
- Chen KL, Wang SS, Yang YY, Yuan RY, Chen RM, Hu CJ. The epigenetic effects of amyloid-beta(1–40) on global DNA and neprilysin genes in murine cerebral endothelial cells. Biochem Biophys Res Commun. 2009;378:57–61. doi: 10.1016/j.bbrc.2008.10.173. [DOI] [PubMed] [Google Scholar]
- Chen PS, Wang CC, Bortner CD, Peng GS, Wu X, Pang H, Lu RB, Gean PW, Chuang DM, Hong JS. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience. 2007;149:203–212. doi: 10.1016/j.neuroscience.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Friso S. Interactions between folate and aging for carcinogenesis. Clin Chem Lab Med. 2005;43:1151–1157. doi: 10.1515/CCLM.2005.200. [DOI] [PubMed] [Google Scholar]
- Choi SW, Mason JB. Folate status: effects on pathways of colorectal carcinogenesis. J Nutr. 2002;132:2413S–2418S. doi: 10.1093/jn/132.8.2413S. [DOI] [PubMed] [Google Scholar]
- Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitamin B12, and serum total homocysteine levels in confirmed Alzheimer disease. Arch Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Colnot S, Niwa-Kawakita M, Hamard G, Godard C, Le Plenier S, Houbron C, Romagnolo B, Berrebi D, Giovannini M, Perret C. Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Lab Invest. 2004;84:1619–1630. doi: 10.1038/labinvest.3700180. [DOI] [PubMed] [Google Scholar]
- Craft S. Insulin resistance syndrome and Alzheimer’s disease: age- and obesity-related effects on memory, amyloid, and inflammation. Neurobiol Aging. 2005;26(Suppl 1):65–69. doi: 10.1016/j.neurobiolaging.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Crouch PJ, Cimdins K, Duce JA, Bush AI, Trounce IA. Mitochondria in aging and Alzheimer’s disease. Rejuvenation Res. 2007;10:349–357. doi: 10.1089/rej.2007.0592. [DOI] [PubMed] [Google Scholar]
- Davidson JS, West RL, Kotikalapudi P, Maroun LE. Sequence and methylation in the beta/A4 region of the rabbit amyloid precursor protein gene. Biochem Biophys Res Commun. 1992;188:905–911. doi: 10.1016/0006-291x(92)91141-c. [DOI] [PubMed] [Google Scholar]
- Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duce JA, Smith DP, Blake RE, Crouch PJ, Li QX, Masters CL, Trounce IA. Linker histone H1 binds to disease associated amyloid-like fibrils. J Mol Biol. 2006;361:493–505. doi: 10.1016/j.jmb.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Duenas-Gonzalez A, Candelaria M, Perez-Plascencia C, Perez-Cardenas E, De La Cruz-Hernandez E, Herrera LA. Valproic acid as epigenetic cancer drug: preclinical, clinical and transcriptional effects on solid tumors. Cancer Treat Rev. 2008;34:206–222. doi: 10.1016/j.ctrv.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Bousiges O, Loeffler JP, Boutillier AL. Histone deacetylase inhibitors: therapeutic agents and research tools for deciphering motor neuron diseases. Curr Med Chem. 2008;15:1263–1273. doi: 10.2174/092986708784534974. [DOI] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. A brief history of epigenetics. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2007. pp. 15–22. [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, Petronis A. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Francis YI, Fa M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, Arancio O. Dysregulation of Histone Acetylation in the APP/PS1 Mouse Model of Alzheimer’s Disease. J Alzheimers Dis. 2009;18:131–139. doi: 10.3233/JAD-2009-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller SJ, Tan RS, Martins RN. Androgens in the etiology of Alzheimer’s disease in aging men and possible therapeutic interventions. J Alzheimers Dis. 2007;12:129–142. doi: 10.3233/jad-2007-12202. [DOI] [PubMed] [Google Scholar]
- Fuso A, Nicolia V, Cavallaro RA, Ricceri L, D’anselmi F, Coluccia P, Calamandrei G, Scarpa S. B-vitamin deprivation induces hyperhomocysteinemia and brain S-adenosylhomocysteine, depletes brain S-adenosylmethionine, and enhances PS1 and BACE expression and amyloid-beta deposition in mice. Mol Cell Neurosci. 2008;37:731–746. doi: 10.1016/j.mcn.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Fuso A, Seminara L, Cavallaro RA, D’anselmi F, Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol Cell Neurosci. 2005;28:195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Gao S, Hendrie HC, Hall KS, Hui S. The relationships between age, sex, and the incidence of dementia and Alzheimer disease: a meta-analysis. Arch Gen Psychiatry. 1998;55:809–815. doi: 10.1001/archpsyc.55.9.809. [DOI] [PubMed] [Google Scholar]
- Gasque P, Neal JW, Singhrao SK, Mcgreal EP, Dean YD, Van BJ, Morgan BP. Roles of the complement system in human neurodegenerative disorders: pro-inflammatory and tissue remodeling activities. Mol Neurobiol. 2002;25:1–17. doi: 10.1385/mn:25:1:001. [DOI] [PubMed] [Google Scholar]
- Gharib A, Sarda N, Chambannes B, Cronenberger L, Pacheco H. The regional concentration of S-andenosyl-L-methionine, S-adenosyl-L-homocysteine, and adenosine in rat brain. J Neurochem. 1982 doi: 10.1111/j.1471-4159.1982.tb08702.x. [DOI] [PubMed] [Google Scholar]
- Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, Young L, Brandenburg SA, Hu Y, Bisht KS, Ho AS, Mattson D, Sun L, Munson PJ, Chuang EY, Mitchell JB, Feinberg AP. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell. 2004;6:361–371. doi: 10.1016/j.ccr.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–1872. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- Gottfries CG, Lehmann W, Regland B. Early diagnosis of cognitive impairment in the elderly with the focus on Alzheimer’s disease. J Neural Transm. 1998;105:773–786. doi: 10.1007/s007020050094. [DOI] [PubMed] [Google Scholar]
- Gray SG, Dangond F. Rationale for the use of histone deacetylase inhibitors as a dual therapeutic modality in multiple sclerosis. Epigenetics. 2006;1:67–75. doi: 10.4161/epi.1.2.2678. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–99. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, Mcguinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Van Den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, Mcquillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrman N, Lanctot KL, Rothenburg LS, Eryavec G. A placebo-controlled trial of valproate for agitation and aggression in Alzheimer’s disease. Dement Geriatr Cogn Disord. 2007;23:116–9. doi: 10.1159/000097757. [DOI] [PubMed] [Google Scholar]
- Ho PI, Ortiz D, Rogers E, Shea TB. Multiple aspects of homocysteine neurotoxicity: glutamate excitotoxicity, kinase hyperactivation and DNA damage. J Neurosci Res. 2002;70:694–702. doi: 10.1002/jnr.10416. [DOI] [PubMed] [Google Scholar]
- Impey S. A histone deacetylase regulates addiction. Neuron. 2007;56:415–417. doi: 10.1016/j.neuron.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP, Baylin SB. Epigenetics and human disease. Nat Med. 1996;2:281–282. doi: 10.1038/nm0396-281. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivascu C, Wasserkort R, Lesche R, Dong J, Stein H, Thiel A, Eckhardt F. DNA methylation profiling of transcription factor genes in normal lymphocyte development and lymphomas. Int J Biochem Cell Biol. 2007;39:1523–1538. doi: 10.1016/j.biocel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Jacinto FV, Esteller M. Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis. 2007;22:247–253. doi: 10.1093/mutage/gem009. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Janelsins MC, Mastrangelo MA, Park KM, Sudol KL, Narrow WC, Oddo S, Laferla FM, Callahan LM, Federoff HJ, Bowers WJ. Chronic neuron-specific tumor necrosis factor-alpha expression enhances the local inflammatory environment ultimately leading to neuronal death in 3xTg-AD mice. Am J Pathol. 2008;173:1768–1782. doi: 10.2353/ajpath.2008.080528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee CD, Lee HS, Bae SI, Yang HK, Lee YM, Rho MS, Kim WH. Loss of caspase-1 gene expression in human gastric carcinomas and cell lines. Int J Oncol. 2005;26:1265–1271. [PubMed] [Google Scholar]
- Johns DR, Neufeld MJ. Pitfalls in the molecular genetic diagnosis of Leber hereditary optic neuropathy (LHON) Am J Hum Genet. 1993;53:916–920. [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Bottiglieri T, Arning E, Ziegler MG, Hansen LA, Masliah E. Elevated S-adenosylhomocysteine in Alzheimer Brain: influence on methyltransferases and cognitive funtion. J Neural Transm. 2004;111:547–567. doi: 10.1007/s00702-003-0096-5. [DOI] [PubMed] [Google Scholar]
- Khudayberdiev S, Fiore R, Schratt G. MicroRNA as modulators of neuronal responses. Commun Integr Biol. 2009;2:411–413. doi: 10.4161/cib.2.5.8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Whelan CM, Berezovska O, Hyman BT. The gamma secretase-generated carboxyl-terminal domain of the amyloid precursor protein induces apoptosis via Tip60 in H4 cells. J Biol Chem. 2002;277:28530–28536. doi: 10.1074/jbc.M203372200. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukull WA, Higdon R, Bowen JD, Mccormick WC, Teri L, Schellenberg GD, Van Belle G, Jolley L, Larson EB. Dementia and Alzheimer disease incidence: a prospective cohort study. Arch Neurol. 2002;59:1737–1746. doi: 10.1001/archneur.59.11.1737. [DOI] [PubMed] [Google Scholar]
- Kular RK, Cvetanovic M, Siferd S, Kini AR, Opal P. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J Biol Chem. 2009;284:7783–7792. doi: 10.1074/jbc.M806150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, Callinan PA, Fan JB, Potash JB, Feinberg AP. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Basha MR, Ge YW, Zawia NH. How and when environmental agents and dietary factors affect the course of Alzheimer’s disease: the “LEARn” model (latent early-life associated regulation) may explain the triggering of AD. Curr Alzheimer Res. 2007;4:219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, De Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Landfield PW, Blalock EM, Chen KC, Porter NM. A new glucocorticoid hypothesis of brain aging: implications for Alzheimer’s disease. Curr Alzheimer Res. 2007;4:205–212. doi: 10.2174/156720507780362083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner AJ, Doran M. Clinical phenotypic heterogeneity of Alzheimer’s disease associated with mutations of the presenilin-1 gene. J Neurol. 2006;253:139–158. doi: 10.1007/s00415-005-0019-5. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- Love S, Barber R, Wilcock GK. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer’s disease. Brain. 1999;122 (Pt 2):247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- Lue LF, Brachova L, Civin WH, Rogers J. Inflammation, A beta deposition, and neurofibrillary tangle formation as correlates of Alzheimer’s disease neurodegeneration. J Neuropathol Exp Neurol. 1996;55:1083–1088. [PubMed] [Google Scholar]
- Macaluso M, Montanari M, Cinti C, Giordano A. Modulation of cell cycle components by epigenetic and genetic events. Semin Oncol. 2005;32:452–457. doi: 10.1053/j.seminoncol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- MacDonald JL, Roskams AJ. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog Neurobiol. 2009;88:170–183. doi: 10.1016/j.pneurobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Maeda S, Sahara N, Saito Y, Murayama S, Ikai A, Takashima A. Increased levels of granular tau oligomers: an early sign of brain aging and Alzheimer’s disease. Neurosci Res. 2006;54:197–201. doi: 10.1016/j.neures.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani ST, Thakur MK. In the cerebral cortex of female and male mice, amyloid precursor protein (APP) promoter methylation is higher in females and differentially regulated by sex steroids. Brain Res. 2006;1067:43–47. doi: 10.1016/j.brainres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.12.005. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Mckee A, Grover A, Rogers J, Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays-Hoopes L, Chao W, Butcher HC, Huang RC. Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Dev Genet. 1986;7:65–73. doi: 10.1002/dvg.1020070202. [DOI] [PubMed] [Google Scholar]
- Mays-Hoopes LL. DNA methylation in aging and cancer. J Gerontol. 1989;44:35–36. doi: 10.1093/geronj/44.6.35. [DOI] [PubMed] [Google Scholar]
- Mcalpine FE, Lee JK, Harms AS, Ruhn KA, Blurton-Jones M, Hong J, Das P, Golde TE, Laferla FM, Oddo S, Blesch A, Tansey MG. Inhibition of soluble TNF signaling in a mouse model of Alzheimer’s disease prevents pre-plaque amyloid-associated neuropathology. Neurobiol Dis. 2009;34:163–177. doi: 10.1016/j.nbd.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan P, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):241–243. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M, Seckl JR. Epigenetic mechanisms of perinatal programming of hypothalamic-pituitary-adrenal function and health. Trends Mol Med. 2007;13(7):269–277. doi: 10.1016/j.molmed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Meisel A, Harms C, Yildirim F, Bösel J, Kronenberg G, Harms U, Fink KB, Endres M. Inhibition of histone deacetylation protects wild-type but not gelsolin-deficient neurons from oxygen/glucose deprivation. J Neurochem. 2006;98:1019–1031. doi: 10.1111/j.1471-4159.2006.04016.x. [DOI] [PubMed] [Google Scholar]
- Mejia JE, Jahn I, Hauptmann G. DNA methylation and the origin of complement factor B polymorphism. Hum Immunol. 1995;42:241–244. doi: 10.1016/0198-8859(94)00109-4. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Mi XB, Zeng FQ. Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. Acta Pharmacol Sin. 2008;29:105–112. doi: 10.1111/j.1745-7254.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Millington GW. Epigenetics and dermatological disease. Pharmacogenomics. 2008;9:1835–1850. doi: 10.2217/14622416.9.12.1835. [DOI] [PubMed] [Google Scholar]
- Moreira PR, Guimaraes MM, Guimaraes AL, Diniz MG, Gomes CC, Brito JA, Gomez RS. Methylation of P16, P21, P27, RB1 and P53 genes in odontogenic keratocysts. J Oral Pathol Med. 2009;38:99–103. doi: 10.1111/j.1600-0714.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- Mori T, Kim J, Yamano T, Takeuchi H, Huang S, Umetani N, Koyanagi K, Hoon DS. Epigenetic up-regulation of C-C chemokine receptor 7 and C-X-C chemokine receptor 4 expression in melanoma cells. Cancer Res. 2005;65:1800–1807. doi: 10.1158/0008-5472.CAN-04-3531. [DOI] [PubMed] [Google Scholar]
- Morris MC, Evans DA, Schneider JA, Tangney CC, Bienias JL, Aggarwal NT. Dietary folate and vitamins B-12 and B-6 not associated with incident Alzheimer’s disease. J Alzheimers Dis. 2006;9:435–443. doi: 10.3233/jad-2006-9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison LD, Smith DD, Kish SJ. Brain S-adenosylmethionine levels are severely decreased in Alzheimer’s disease. J Neurochem. 1996;67:1328–1331. doi: 10.1046/j.1471-4159.1996.67031328.x. [DOI] [PubMed] [Google Scholar]
- Muerkoster SS, Werbing V, Koch D, Sipos B, Ammerpohl O, Kalthoff H, Tsao MS, Folsch UR, Schafer H. Role of myofibroblasts in innate chemoresistance of pancreatic carcinoma--epigenetic downregulation of caspases. Int J Cancer. 2008;123:1751–1760. doi: 10.1002/ijc.23703. [DOI] [PubMed] [Google Scholar]
- Mund C, Brueckner B, Lyko F. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics. 2006;1:7–13. doi: 10.4161/epi.1.1.2375. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Mukaetova-Ladinska EB, Harrington CR, Roth M, Wischik CM. Alterations in tau protein metabolism during normal aging. Dementia. 1996;7:95–103. doi: 10.1159/000106861. [DOI] [PubMed] [Google Scholar]
- Neumeister P, Albanese C, Balent B, Greally J, Pestell RG. Senescence and epigenetic dysregulation in cancer. Int J Biochem Cell Biol. 2002;34:1475–1490. doi: 10.1016/s1357-2725(02)00079-1. [DOI] [PubMed] [Google Scholar]
- Neve RL, Mcphie DL. The cell cycle as a therapeutic target for Alzheimer’s disease. Pharmacol Ther. 2006;111:99–113. doi: 10.1016/j.pharmthera.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS One. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeid R, Mccaddon A, Herrmann W. The role of hyperhomocysteinemia and B-vitamin deficiency in neurological and psychiatric diseases. Clin Chem Lab Med. 2007;45:1590–1606. doi: 10.1515/CCLM.2007.356. [DOI] [PubMed] [Google Scholar]
- Ogawa O, Zhu X, Lee HG, Raina A, Obrenovich ME, Bowser R, Ghanbari HA, Castellani RJ, Perry G, Smith MA. Ectopic localization of phosphorylated histone H3 in Alzheimer’s disease: a mitotic catastrophe? Acta Neuropathol. 2003;105:524–528. doi: 10.1007/s00401-003-0684-3. [DOI] [PubMed] [Google Scholar]
- Ono T, Shinya K, Uehara Y, Okada S. Endogenous virus genomes become hypomethylated tissue--specifically during aging process of C57BL mice. Mech Ageing Dev. 1989;50:27–36. doi: 10.1016/0047-6374(89)90056-0. [DOI] [PubMed] [Google Scholar]
- Pallis M, Robins A, Powell R. Quantitative analysis of lymphocyte CD11a using standardized flow cytometry. Scand J Immunol. 1993;38:559–564. doi: 10.1111/j.1365-3083.1993.tb03241.x. [DOI] [PubMed] [Google Scholar]
- Parachikova A, Agadjanyan MG, Cribbs DH, Blurton-Jones M, Perreau V, Rogers J, Beach TG, Cotman CW. Inflammatory changes parallel the early stages of Alzheimer disease. Neurobiol Aging. 2007;28:1821–1833. doi: 10.1016/j.neurobiolaging.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Chambers PA, Lodge JP, Pratt JR. Ischemia- reperfusion injury and its influence on the epigenetic modification of the donor kidney genome. Transplantation. 2008;86:1818–1823. doi: 10.1097/TP.0b013e31818fe8f9. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Mill J. Epigenetic Studies of Psychosis: Current Findings, Methodological Approaches, and Implications for Postmortem Research. Biol Psychiatry. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Pieper HC, Evert BO, Kaut O, Riederer PF, Waha A, Wullner U. Different methylation of the TNF-alpha promoter in cortex and substantia nigra: Implications for selective neuronal vulnerability. Neurobiol Dis. 2008;32:521–527. doi: 10.1016/j.nbd.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Esteller M, Vaag A, Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- Profenno LA, Jakimovich L, Holt CJ, Porsteinsson A, Tariot PN. A randomized, double-blind, placebo-controlled pilot trial of safety and tolerability of two doses of divalproex sodium in outpatients with probable Alzheimer’s disease. Curr Alzheimer Res. 2005;2:553–8. doi: 10.2174/156720505774932205. [DOI] [PubMed] [Google Scholar]
- Raiha I, Kaprio J, Koskenvuo M, Rajala T, Sourander L. Alzheimer’s disease in twins. Biomed Pharmacother. 1997;51:101–104. doi: 10.1016/s0753-3322(97)86906-5. [DOI] [PubMed] [Google Scholar]
- Rath PC, Kanungo MS. Methylation of repetitive DNA sequences in the brain during aging of the rat. FEBS Lett. 1989;244:193–198. doi: 10.1016/0014-5793(89)81191-3. [DOI] [PubMed] [Google Scholar]
- Ravaglia G, Forti P, Maioli F, Martelli M, Servadei L, Brunetti N, Porcellini E, Licastro F. Homocysteine and folate as risk factors for dementia and Alzheimer disease. Am J Clin Nutr. 2005;82:636–643. doi: 10.1093/ajcn.82.3.636. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22:2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Reynolds E. Vitamin B12, folic acid, and the nervous system. lancet Neuro. 2006;5:949–960. doi: 10.1016/S1474-4422(06)70598-1. [DOI] [PubMed] [Google Scholar]
- Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132:2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Morrison JH. Quantitative morphology and regional and laminar distributions of senile plaques in Alzheimer’s disease. J Neurosci. 1985;5:2801–2808. doi: 10.1523/JNEUROSCI.05-10-02801.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Vaniushin BF. Intragenomic specificity of DNA methylation in animals. Qualitative differences in tissues and changes in methylation of repeating sequences during aging, carcinogenesis and hormonal induction. Mol Biol (Mosk) 1980;14:357–368. [PubMed] [Google Scholar]
- Romanov GA, Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981;653:204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 2003;22:6537–6549. doi: 10.1093/emboj/cdg615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob NG, Nalbantoglu J, Hastings KE, Mikkelsen T, Cashman NR. DNA cytosine methylation in brain of patients with Alzheimer’s disease. Ann Neurol. 1990;28:91–94. doi: 10.1002/ana.410280117. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Aging, amyloid, and Alzheimer’s disease: a perspective in honor of Carl Cotman. Neurochem Res. 2003;28:1705–1713. doi: 10.1023/a:1026065122854. [DOI] [PubMed] [Google Scholar]
- Serot JM, Christmann D, Dubost T, Bene MC, Faure GC. CSF-folate levels are decreased in late-onset AD patients. J Neural Transm. 2001;108:93–99. doi: 10.1007/s007020170100. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’agostino RB, Wilson PW, Wolf PA. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N Engl J Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- Smith AD. The worldwide challenge of the dementias: a role for B vitamins and homocysteine? Food Nutr Bull. 2008;29:s143–s172. doi: 10.1177/15648265080292S119. [DOI] [PubMed] [Google Scholar]
- Smith AD, Kim YI, Refsum H. Is folic acid good for everyone? Am J Clin Nutr. 2008;87(3):517–33. doi: 10.1093/ajcn/87.3.517. [DOI] [PubMed] [Google Scholar]
- So K, Tamura G, Honda T, Homma N, Waki T, Togawa N, Nishizuka S, Motoyama T. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci. 2006;97:1155–1158. doi: 10.1111/j.1349-7006.2006.00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova NE, Shiryaeva NV, Dyuzhikova NA, Savenko YN, Vaido AI. Effect of long-term mental and pain stress on the dynamics of H4 histone acetylation in hippocampal neurons of rats with different levels of nervous system excitability. Bull Exp Biol Med. 2006;142:341–343. doi: 10.1007/s10517-006-0361-3. [DOI] [PubMed] [Google Scholar]
- Sommer G, Kralisch S, Lipfert J, Weise S, Krause K, Jessnitzer B, Lossner U, Bluher M, Stumvoll M, Fasshauer M. Amyloid precursor protein expression is induced by tumor necrosis factor alpha in 3T3-L1 adipocytes. J Cell Biochem. 2009;108:1418–1422. doi: 10.1002/jcb.22382. [DOI] [PubMed] [Google Scholar]
- Strathdee G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Semin Cancer Biol. 2002;12:373–379. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- Su Y, Ryder J, Li B, Wu X, Fox N, Solenberg P, Brune K, Paul S, Zhou Y, Liu F, Ni B. Lithium, a common drug for bipolar disorder treatment, regulates amyloid-beta precursor protein processing. Biochemistry (Mosc) 2004;43:6899–6908. doi: 10.1021/bi035627j. [DOI] [PubMed] [Google Scholar]
- Suuronen T, Nuutinen T, Ryhanen T, Kaarniranta K, Salminen A. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357:397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tang LY, Reddy MN, Rasheva V, Lee TL, Lin MJ, Hung MS, Shen CK. The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J Biol Chem. 2003;278:33613–33616. doi: 10.1074/jbc.C300255200. [DOI] [PubMed] [Google Scholar]
- Thibault O, Gant JC, Landfield PW. Expansion of the calcium hypothesis of brain aging and Alzheimer’s disease: minding the store. Aging Cell. 2007;6:307–317. doi: 10.1111/j.1474-9726.2007.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region −224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res. 1999a;70:288–292. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Ukitsu M, Genda Y. Decrease with age in methylcytosines in the promoter region of receptor for advanced glycated end products (RAGE) gene in autopsy human cortex. Brain Res Mol Brain Res. 1999b;65:124–128. doi: 10.1016/s0169-328x(98)00351-9. [DOI] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Ukitsu M, Genda Y. The methylation status of cytosines in a tau gene promoter region alters with age to downregulate transcriptional activity in human cerebral cortex. Neurosci Lett. 1999c;275:89–92. doi: 10.1016/s0304-3940(99)00731-4. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop K, Engeland K. Cell cycle-dependent transcription of cyclin B2 is influenced by DNA methylation but is independent of methylation in the CDE and CHR elements. Febs J. 2007;274:5235–5249. doi: 10.1111/j.1742-4658.2007.06045.x. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Panhuys N, Le Gros G, Mcconnell MJ. Epigenetic regulation of Th2 cytokine expression in atopic diseases. Tissue Antigens. 2008;72:91–97. doi: 10.1111/j.1399-0039.2008.01068.x. [DOI] [PubMed] [Google Scholar]
- Van Rietschoten JG, Gal-Yam EN, Jeong S, Cortez CC, Verweij CL, Jones PA. Epigenetic regulation and nucleosome positioning in the human TATA-less IL-1 alpha promoter. Genes Immun. 2008;9:582–590. doi: 10.1038/gene.2008.53. [DOI] [PubMed] [Google Scholar]
- Vanyushin BF, Nemirovsky LE, Klimenko VV, Vasiliev VK, Belozersky AN. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia. 1973;19:138–152. [PubMed] [Google Scholar]
- Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animal DNA. Nature. 1970;225:948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Varela-Moreiras G, Perez-Olleros L, Garcia-Cuevas M, Ruiz-Roso B. Effects of ageing on folate metabolism in rats fed a long-term folate deficient diet. Int J Vitam Nutr Res. 1994;64:294–299. [PubMed] [Google Scholar]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Veenstra GJ, Imhof A, Sera T, Tse C, Ge H, Shi YB, Hansen JC, Wolffe AP. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harb Symp Quant Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS ONE. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;87:517–533. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39(4):457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79:1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-{beta} protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184:2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Brock B, Cox DP, Cardozo-Pelaez F, Mcpherson CA, Harry J, Rice DC, Maloney B, Chen D, Lahiri DK, Zawia NH. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J Neurosci. 2008a;28:3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Basha MR, Zawia NH. The environment, epigenetics and amyloidogenesis. J Mol Neurosci. 2008b;34:1–7. doi: 10.1007/s12031-007-0009-4. [DOI] [PubMed] [Google Scholar]
- Wu X, Chen PS, Dallas S, Wilson B, Block ML, Wang CC, Kinyamu H, Lu N, Gao X, Leng Y, Chuang DM, Zhang W, Lu RB, Hong JS. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int J Neuropsychopharmacol. 2008;11:1123–1134. doi: 10.1017/S1461145708009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Vilkaitis G, Yu B, Klimasauskas S, Chen X. Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymol. 2007;427:139–154. doi: 10.1016/S0076-6879(07)27008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Culley DJ, Dong Y, Zhang G, Zhang B, Moir RD, Frosch MP, Crosby G, Tanzi RE. The common inhalation anesthetic isoflurane induces caspase activation and increases amyloid beta-protein level in vivo. Ann Neurol. 2008;64:618–627. doi: 10.1002/ana.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M, Zhang T, Zhang LM, Lu SD, Huang YL, Sun FY. Caspase inhibition attenuates accumulation of beta-amyloid by reducing beta-secretase production and activity in rat brains after stroke. Neurobiol Dis. 2008;32:433–441. doi: 10.1016/j.nbd.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Yakovlev A, Khafizova M, Abdullaev Z, Loukinov B, Kondratyev A. Epigenetic regulation of caspase-3 gene expression in rat brain development. Gene. 2010;450:103–108. doi: 10.1016/j.gene.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Vilkaitis G, Yu B, Klimasauskas S, Chen X. Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymology. 2007;427:139–154. doi: 10.1016/S0076-6879(07)27008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Choi HI, Choi JE, Sul CA, Choi JM, Kim DH. Methotrexate decreases PP2A methylation and increases tau phosphorylation in neuron. Biochem Biophys Res Commun. 2007;363:811–816. doi: 10.1016/j.bbrc.2007.09.060. [DOI] [PubMed] [Google Scholar]
- Yu MK. Epigenetics and chronic lymphocytic leukemia. Am J Hematol. 2006;81:864–869. doi: 10.1002/ajh.20718. [DOI] [PubMed] [Google Scholar]
- Zanjani H, Finch CE, Kemper C, Atkinson J, Mckeel D, Morris JC, Price JL. Complement activation in very early Alzheimer disease. Alzheimer Dis Assoc Disord. 2005;19:55–66. doi: 10.1097/01.wad.0000165506.60370.94. [DOI] [PubMed] [Google Scholar]
- Zhang CE, Tian Q, Wei W, Peng JH, Liu GP, Zhou XW, Wang Q, Wang DW, Wang JZ. Homocysteine induces tau phosphorylation by inactivating protein phosphatase 2A in rat hippocampus. Neurobiol Aging. 2008;29:1654–1665. doi: 10.1016/j.neurobiolaging.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Deng C, Lu Q, Richardson B. Age-dependent DNA methylation changes in the ITGAL (CD11a) promoter. Mech Ageing Dev. 2002;123:1257–1268. doi: 10.1016/s0047-6374(02)00014-3. [DOI] [PubMed] [Google Scholar]
- Zhou XW, Gustafsson JA, Tanila H, Bjorkdahl C, Liu R, Winblad B, Pei JJ. Tau hyperphosphorylation correlates with reduced methylation of protein phosphatase 2A. Neurobiol Dis. 2008 doi: 10.1016/j.nbd.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Zinovyeva AY, Graham SM, Cloud VJ, Forrester WC. The C. elegans histone deacetylase HDA-1 is required for cell migration and axon pathfinding. Dev Biol. 2006;289:229–242. doi: 10.1016/j.ydbio.2005.10.033. [DOI] [PubMed] [Google Scholar]