Abstract
Recent advances in DNA sequencing methodology have enabled studies of human skin microbes that circumvent difficulties in isolating and characterizing fastidious microbes. Sequence-based approaches have identified greater diversity of cutaneous bacteria than studies using traditional cultivation techniques. However, improved sequencing technologies and analytical methods are needed to study all skin microbes, including bacteria, archaea, fungi, viruses, and mites, and how they interact with each other and their human hosts. This review discusses current skin microbiome research, with a primary focus on bacteria, and the challenges facing investigators striving to understand how skin micro-organisms contribute to health and disease.
Keywords: skin, microbiome, bacteria, genomic, metagenomics, 16S rRNA, cutaneous, dermatology
Skin microbiome research
The human skin microbiome refers to the entire collection of microbes – bacteria, archaea, fungi, viruses, and mites – that reside in and on human skin. High-throughput sequencing technologies have facilitated studies of the human body’s complex microbial inhabitants by allowing more comprehensive identification of microbes than traditional culture methods. [1-2]. Research efforts such as the National Institutes of Health’s Human Microbiome Project [1, 2] utilize genomic methods to characterize the diversity of microbial communities present at specific body sites, including skin. A more thorough understanding of skin microbial inhabitants (commensals and symbionts) and pathogens provides a foundation for future research regarding the interactions between microbes and humans as well as among the community of microbes. Skin microbes include bacteria, fungi, viruses, archaea, and mites. Most early human microbiome research has focused on bacteria with the ultimate goal of using genomic methods to also study non-bacterial microbes.
Skin serves as a protective barrier and houses large numbers of colonizing bacteria. Based on research and clinical observation, generations of dermatologists have asserted that microbes influence the natural courses of several skin diseases. Staphylococcus epidermidis, for instance, is frequently cultured from healthy skin and may protect humans from pathogenic bacteria [3]. Acne, a common condition often affecting teenagers, is strongly associated with the presence of Propionibacterium acnes. Children with atopic dermatitis (AD; see Glossary), an itchy chronic skin disease that can be disabling, commonly have Staphylococcus aureus infections. Furthermore, severe AD is a risk factor for potentially life-threatening dissemination of herpes viral infections. Questions about the potentially beneficial role of normal resident microorganisms and about the many pathogenic microbes that cause both cutaneous and systemic diseases have long stimulated skin microbial studies. Development of antimicrobials and resistant strains of bacteria have spurred additional cutaneous bacteria research.
Culture-based methods have been used frequently to isolate, identify, and study cutaneous microbes like bacteria, and continue to be essential. However, some bacteria have fastidious growth requirements and are difficult to isolate. In a given clinical sample, some bacterial species (such as staphylococci) grow readily in the laboratory and overcrowd more fastidious bacteria. Differential recovery of various skin bacteria can lead to misinterpretations regarding the true relative abundances of bacterial species that exist and predominate in a clinical sample. Skin microbial research has often also focused on investigating a small number of individual bacterial species which are typically associated with common skin disorders. In contrast, contemporary human microbiome research examines the entire complex microbial community and uses culture-independent methods to reduce potential experimental bias. This review will provide background on the various genomic technologies that are used to identify microbes, primarily bacteria, and will discuss work that has utilized these approaches in healthy skin and skin disorders to gain insight into the effects of the bacterial microbiome on human health.
Microbial genomics
16S rRNA-based sequencing and analysis
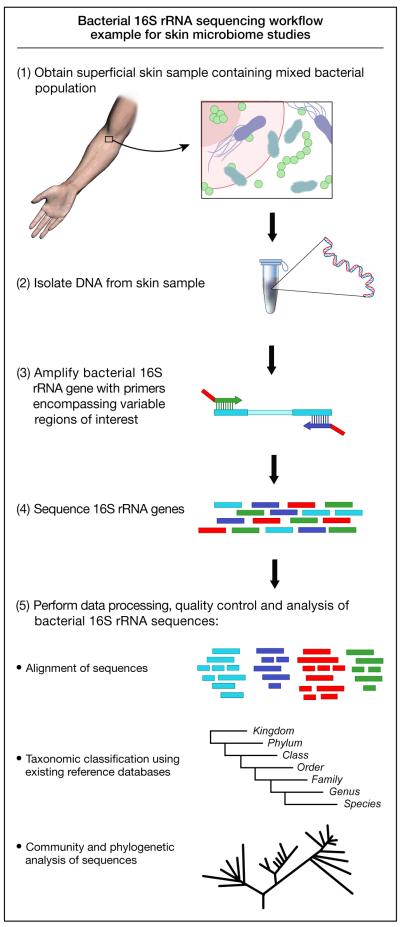
Microbiome analyses take advantage of the universal presence of the small-subunit (16S) ribosomal RNA gene in all prokaryotes (Figure 1). The 16S rRNA gene contains highly conserved regions, facilitating PCR, while hypervariable regions enable phylogenetic categorization. Users can query candidate sequences against online databases, such as the Ribosomal Database Project (RDP) [4], which catalog hundreds of thousands of validated 16S rRNA gene sequences. Thus, sequencing the 16S rRNA gene (Box 1) allows identification of bacteria present in a given sample.
Figure 1.
Schematic view of 16S ribosomal RNA gene-based bacterial sequencing workflow. The 16S ribosomal RNA gene is universal among prokaryotes and can be used for phylotyping bacterial sequences. This diagram shows the process from collecting a skin sample to DNA sequencing and analysis used in many skin microbiome studies.
Box 1. Sequencing strategies used in skin microbiome studies.
Sanger sequencing
This process has been the standard sequencing method and enabled sequencing of the human genome. The Sanger method requires a PCR-amplified or cloned target and incorporates radioactive or fluorescent dideoxynucleotide triphosphates leading to premature chain termination. The resulting fragments of varying lengths are separated in a capillary-based polymer gel. Different methods exist to detect the order of each incorporated nucleotide to determine the DNA sequence. Since this method is more labor-intensive, the cost per kilobase is higher than more recent technologies. Average read length is 800-1000 base pairs.
Next-generation sequencing
This term is used to refer to sequencing methods that parallelize sequencing resulting in millions of short sequences from amplified DNA clones. These high-throughput platforms have reduced the cost of DNA sequencing. The methods have been developed since 2005, and include 454/Roche pyrosequencing, Illumina GA, and SOLiD platforms.
Metagenomics
This term describes the use of cultivation-independent genomics-based investigation to characterize complex microbial communities and has been described in detail by others [60]. Metagenomics can move beyond fundamental phylotyping of the many bacteria present in a given sample by sequencing entire genomes of all microbes to reveal functional information about the microbial communities. Advances in computing and software will be required to fully analyze data resulting from metagenomics studies.
Whole genome shotgun sequencing
This method randomly shears the DNA in a given sample into fragments followed by sequencing of these smaller DNA pieces resulting in short sequences. These numerous reads are assembled into a continuous sequence based on a reference or through overlapping sequences to form larger contigs.
Sanger sequencing provides the benefit of full-length sequencing of the 16S rRNA gene. However, with human microbiome analyses demonstrating a large number of rare species, Sanger sequencing has proven to be too slow with low throughput to fully explore bacterial diversity. Pyrosequencing, one of several next-generation high-throughput sequencing technologies, utilizes detection of pyrophosphate release upon nucleotide incorporation and allows for multiple reactions simultaneously with subsequent faster sequencing. This method produces a significantly higher number of reads (up to a million reads per run), but with shorter read lengths (~400bp). An advantage of pyrosequencing is the ability to sequence multiple samples in a single run by tagging PCR products with error-correcting barcodes, which are unique labels enabling one to multiplex and associate individual PCR products with a specific sample [5]. Neither Sanger nor next-generation platforms are able to circumvent potential biases introduced by PCR, such as primer bias for certain phylotypes and the generation of chimeras. However, algorithms to analyze sequences and identify putative chimeras are under continuous development. Next-generation sequencing technologies have errors in individual reads and costs are non-trivial, yet these 16S rRNA gene-based methods continue to evolve to provide less expensive, longer, and more accurate sequences.
There are several issues to consider when using 16S rRNA gene-based technologies to identify bacteria [6-7]. Sequencing results are generally only semi-quantitative [8], sequencing cannot distinguish between living or dead bacteria, and some phylotypes cannot be differentiated at a species level. Moreover, clinical microbiologists and health care workers generally associate clinical relevance of microorganisms with genus and species level identification. Another challenge in identifying bacteria is that the definition of a bacterial species has traditionally been based on phenotypic characteristics. However, bacterial strains that share a phenotype, such as specific growth requirements, may have significant genomic differences. This raises the issue of defining “species.” Additional issues relate to the bioinformatics required to store and process vast amounts of sequence data, the need for chimera-checking, and the accuracy of 16S rRNA gene databases. Existing and ongoing development of analysis tools that assist in phylotyping 16S rRNA gene sequences include, but are not limited to, UniFrac [9], mothur [10], Ribosomal Database Project [4], and QIIME [11]. Despite the challenges discussed, the use of 16S rRNA gene-based methods has already increased our understanding of the diversity of skin microorganisms.
Sequencing other microorganisms
The association between microorganisms and human skin extends beyond bacteria. Fungi and viruses are well known human pathogens that cause diseases such as athlete’s foot and chickenpox. However, the ability to perform large-scale genomic analyses to survey fungal microorganisms on human skin in a manner similar to bacteria is complicated by the use of different conserved regions, such as the 18S rRNA, 28S rRNA, and the internal transcribed spacer (ITS) regions, and the limited number of comprehensive analytical tools to survey existing reference databases. Large databases for fungal sequences exist for the internal transcribed spacer (ITS) region of fungal rRNA genes, but the software tools to analyze the “mycobiome” [12] have not been developed at the same rapid pace as compared to the bacterial microbiome. Whole-genome shotgun sequencing will provide a growing resource of fungal sequences [13] that may facilitate the development of necessary analytical software tools.
Viruses are important vectors for lateral gene transfer that contributes to a significant amount of genetic variation, such as bacterial diversity mediated by bacteriophage (bacterial virus) insertion. Investigating the viral microbiome will facilitate understanding of the complex interactions among the different types of microbes. For example, bacteriophages are considered an important source of genetic material encoding antibiotic-resistance and virulence [14]. However, the immense diversity (double- and single-stranded DNA and RNA viruses) and rapid evolution of viruses complicates the development of a library of viral genomes. Viruses also do not have conserved regions analogous to bacterial 16S or fungal ITS to generate amplicons (pieces of DNA) for sequencing surveys. Therefore, development of widely available software tools to analyze viral sequences will require more comprehensive databases. Technologies that do not rely on the use of known viral sequences to sequence clinical and environmental samples may allow viral discovery [15-17]. Fungal and viral [18] human microbiome research continues to progress and will likely provide valuable information on the role of these microorganisms in the balance between human health and disease in the future.
Metagenomics
Metagenomics refers to the genetic material in a given community and their aggregate functions [19]. While 16S rRNA gene sequencing enables phylogenetic categorization of bacterial community members, whole genome shotgun sequencing-based methods randomly shear and sequence the entire genomic DNA in a given sample. Sequences are then analyzed for phylogenetics (identification of the microorganisms) and for functional activity such that the biological functions of the entire community is known. This approach has resulted in insights into human health issues, such as gut microbiome research demonstrating that gut bacteria in obese animals and humans have greater energy harvesting efficiency [20-25]. However, metagenomics requires a sufficient quantity of DNA, therefore, the low biomass (small amount of DNA) yielded from skin samples currently limits the ability to perform skin metagenomics. Metagenomics also brings additional computational challenges related to data management for extensive amounts of sequence data, limited availability of computational analysis tools, and incomplete reference genomes of the various microbial populations. The research community is currently confronting the challenges introduced by metagenomics, including the linkage of databases with bacterial genomic sequences and metabolic functions. These advanced sequencing approaches are being used to study the bacterial populations living in the unique environment of human skin.
Skin as a diverse habitat
Human skin can be an inhospitable environment that is characterized by large desiccated regions, acidic pH, and continual shedding of superficial skin cells. Host skin defense includes molecules such as proteases, lysozymes, and antimicrobial peptides [26]. Despite these protective mechanisms, microorganisms survive and thrive.
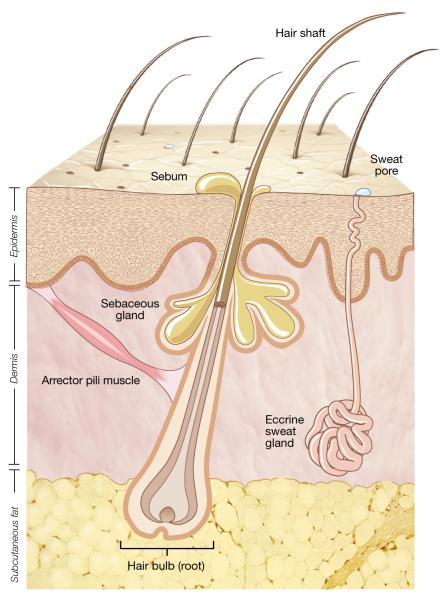
The topography of human skin varies at a microscopic as well as a macroscopic level (Figure 2). The skin surface is uneven with lines, ridges, and invaginations from skin appendages. Appendageal structures such as hair follicles, sebaceous glands, nails, and apocrine and eccrine sweat glands result in skin infoldings of differing depths beneath the top skin layer. Sebaceous glands produce sebum that spills into hair follicles and coats hair and adjacent skin. Apocrine sweat glands are localized to the scalp, armpits, and groin and excrete a fatty substance that is broken down by bacteria to cause an odor. Eccrine sweats glands are ubiquitous with especially high density on the scalp, palms, and soles. Eccrine sweat is a salty fluid that upon evaporation helps cool the body. Each invagination provides a distinct reservoir where bacteria flourish and repopulate the skin surface.
Figure 2.
Representation of human skin. The topographical surface of human skin is irregular and consists of multiple skin appendageal structures including hair follicles, sebaceous (oil) glands, and sweat glands and ducts. Each of the skin appendages provides a potential niche for skin bacteria. The external communication of the skin with the terrestrial environment and the heterogeneity of the appendageal invaginations contribute to the diversity of the skin microbiome.
On a macroscopic level, the human skin surface is comprised of a variety of unique regions: plateaus such as forearm and back, prominences including elbow and shoulder, and crevices (e.g. ear canal and toe web). Additionally, the distribution of skin appendages is non-uniform over the skin surface. For example, there is variable hair density on the scalp, nose, and forearms; greater humidity in the armpit; and increased sebaceous (oily) skin on the face, chest, and upper back. The thickness of the epidermis and dermis also varies over the body surface and affects skin texture and pliability. The irregular topography and additional different skin site characteristics provide distinctive habitats for bacteria.
Many other elements potentially contribute to the composition of the microbial communities residing on and in skin. External factors such as ambient humidity, seasonal weather conditions, prior antibiotic treatments, clothing types, use of lotions/creams, cleansers, or deodorants/antiperspirants, hygiene frequency, and other environmental surfaces [27] interact with and can influence cutaneous bacteria. Intrinsic factors such as age, genetic makeup, and host immune system also influence the composition of skin microorganisms. These factors may result in interindividual skin microbiome differences. A challenge for metagenomics will be to determine how much interpersonal variation of skin microbiomes differentially impacts human hosts. A portion of the variance observed in skin microbiomes may result from the presence of different species of skin microorganisms that exhibit functional redundancy.
Bacterial skin populations can be categorized as transient (contaminant, non-reproducing), temporary resident (not typically resident, yet can colonize), and resident (growing, reproducing) flora [28-29]. The ‘normal’ resident skin flora includes Propionibacteriumacnes, Staphylococcus epidermis, Staphylococcus aureus, Corynebacterium diphtheria, Corynebacterium jeikeium, and Pseudomonas aeruginosa, as determined by traditional cultivation methods [26, 30]. The roles of resident bacteria on human skin are highly varied and incompletely understood.
Resident skin bacteria can be beneficial to humans; some are thought to defend against pathogenic bacteria [30]. Staphylococcus aureus is a common pathogen -- approximately 20-30% of healthy individuals are asymptomatic nasal carriers [31-33] -- that can cause both localized and systemic infections. Some bacteria, including Staphylococcus epidermis [34] and Corynebacterium spp., can inhibit or reverse S. aureus colonization of human nares. A subset of S. epidermis, which secrete the serine protease Esp, inhibited S. aureus when introduced into the nasal cavities of carriers [3]. Furthermore, a small molecule produced by S. epidermidis induced antimicrobial peptide expression in mice with associated reduction in susceptibility to group A Streptococcus skin infections [35].
In addition to protecting human hosts, microbes have been implicated in the pathogenesis and/or clinical course of several skin diseases, including seborrheic dermatitis (Malassezia spp.) and atopic dermatitis (Staphylococcus aureus). Although not considered skin infections, clinical management of such disorders routinely includes the use of antimicrobial agents. Skin disease in general may result from a resident microorganism becoming pathogenic under particular conditions or from newly established colonization by a typically pathogenic microbe. Cutaneous disorders often present in a specific distribution pattern which can aid in diagnosis. For example, seborrheic dermatitis is characterized as greasy white-yellow scales affecting oily areas such as the scalp, creases of the nose, and the external ear canal. In contrast, atopic dermatitis typically presents with scaly and itchy red rashes on the antecubital (inner bend of the arm) and popliteal (behind the knees) creases as well as the folds behind the ears. It is possible that skin diseases presenting at particular anatomical sites are related to the skin physiologic characteristics, such as whether it is oily or moist, or even the bacteria that preferentially reside at those specific skin sites. Understanding how microbes contribute to skin diseases requires a greater understanding of the human skin microbiome, and much recent work has been done in the area of the bacterial microbiome.
Skin microbiome - bacterial
An important goal of human skin microbiome research is to understand the role of bacteria in the pathogenesis of human skin diseases. However, the skin microbiome of disease states must be studied in the context of the healthy or “normal” skin microbiome. Several groups have demonstrated the potential power of direct 16S rRNA gene-based sequencing in assessing the bacterial composition of healthy individual skin sites, including the external ear canal [36], forehead [37], hands [38], front of the elbow [39], and inner forearm [40] (Table 1). This work suggests that anatomically distinct skin sites harbor different bacterial communities. 16S rRNA gene-based methods confirmed the presence of Corynebacterium spp., Propionibacterium spp., and Staphylococcus spp. known to exist on human skin [37-43] and showed greater bacterial skin diversity when compared to concurrent cultures [37] or to existing culture-based knowledge of skin bacteria. In addition, sequencing methods also have identified novel species [36-37, 40].
Table 1.
Summary of recent 16S rRNA sequencing-based human skin bacterial microbiome studies in healthy subjects and in patients with skin disorders
| Health status | Skin site sampled | Study Conclusions | Reference |
|---|---|---|---|
| Healthy | Outer ear canal | 24 subjects (adult, children from 3 families). Sanger sequencing of representative RFLP types. Sex and age affected bacterial diversity. Two most prevalent sequences were bacteria known to cause middle ear infections, suggesting outer ear canal is possible reservoir. |
[36] |
| Healthy | Forehead | 5 subjects. Sanger sequencing with concurrent bacterial cultures. Sequencing showed greater bacterial diversity than cultures and identified novel bacteria. |
[37] |
| Healthy | Forearms | 6 subjects (4 re-sampled 8-10 months later). Sanger sequencing with concurrent bacterial cultures. 16% of species identified via sequencing were cultured from same specimens. Right/left samples were more similar than follow-up samples from same individual. |
[39] |
| Healthy | Volar (inner) forearms, antecubital fossa (front of elbows) |
5 subjects. Sanger sequencing. 3 sampling methods (swab, scrape, punch biopsy) showed dominant bacterial phyla similarly represented by all methods. |
[38] |
| Healthy | Palms | 51 subjects. Pyrosequencing. Bacterial species on right/left palms on same and different individuals had low similarity. Handedness, time from washing, and an individual’s sex were associated with similarly structured bacterial communities. |
[37] |
| Healthy | Glabella (forehead), alar crease (side of nose), external auditory canal (outer ear canal), occiput (back of scalp), manubrium (upper chest), back, nares (anterior nose), axilla (armpit), antecubital fossae (front of elbow), fingerweb, inguinal crease (side of groin), gluteal crease (uppermost part of fold between buttocks), popliteal crease (back of knee), heel, umbilicus (navel), forearm, palm, buttock |
10 subjects (5 resampled). Sanger sequencing. 20 sites selected based on sites of predilection of skin diseases. Skin physiological characteristics (oily, moist, dry) affected composition of bacterial communities. Interpersonal variation and stability over time varied by site. |
[43] |
| Healthy | Index fingers, palms, volar (inner) forearm, forehead, external nose, external ears (pinna), hair, armpits (axilla), soles of feet, navel (umbilicus), backs of knees (popliteal fossae), nostrils (nares), external auditory canal (outer ear canal) |
9 subjects (resampled up to 4 times). Pyrosequencing. Up to 18 skin sites sampled along with gut and oral sites. Bacterial composition depended on site and varied by individual sampled. |
[41] |
| Healthy | Fingers, axilla (armpits) | Up to 9 subjects (and personal computer equipment) Pyrosequencing. Bacterial composition of each subject was similar to that found on personal computer equipment. |
[27] |
| Healthy | Ventral forearms (newborns and mothers), forehead (newborns) |
10 newborns, 9 mothers (4 vaginal deliveries, 5 Cesarean sections). Pyrosequencing. Newborn bacterial communities were homogeneous across skin sites and determined by mode of delivery |
[42] |
| Acne | Facial hair follicles (patients and controls), skin (2 patients) |
5 patients, 3 controls. Sanger sequencing. Hair follicle bacterial composition showed limited diversity in patients and only one species in controls |
[44] |
| Chronic wounds | Decubitus/pressure (7), neuropathic/decreased nerve sensation (7), venous stasis (3), post-surgical (3), other (4) ulcers |
24 patients (12 with diabetes, 14 received antibiotics). Wound bed curette samples. Pyrosequencing with concurrent cultures. Bacterial composition was more diverse with sequencing than cultures and influenced by antibiotic exposure. |
[49] |
| Chronic wounds | Venous leg ulcers | 8 ulcers (40 total samples). Wound bed curette samples. Pyrosequencing. Bacterial composition showed topographical differences within large ulcers. |
[58] |
| Chronic wounds | Diabetic wounds, intact skin | 23 paired samples. Debridement and skin swab samples. Pyrosequencing. Intact skin had greater bacterial diversity yet lower levels of known opportunistic wound pathogens than wound samples. |
[56] |
| Psoriasis | Psoriasis plaques (affected, unaffected) |
6 patients. Sanger sequencing. Greater bacterial diversity observed in psoriasis lesions than unaffected skin in patients and controls. |
[47] |
Given the irregular topography of the skin surface, the impact of different sampling methods (skin swabs, noninvasive scrapes, and punch biopsies) from the antecubital fossae (inner bend of the arm) has been evaluated [39]. All three methods identified similar dominant bacterial groups. Less common species were differentially identified with the individual sampling methods, which were sequenced using 16S rRNA gene-based Sanger method. Using methods that increase the depth of sequencing (produce a greater number of sequences per sample) will help determine if the different abundances among the rare species are related to technical limitations of Sanger sequencing. It may appear counterintuitive that superficial sampling methods are comparable to more invasive methods. One explanation is that excretion of sweat gland contents and outward migration of differentiating skin cells likely transport living and dead microorganisms that dwell within appendageal structures and in deeper layers of the skin to the skin surface.
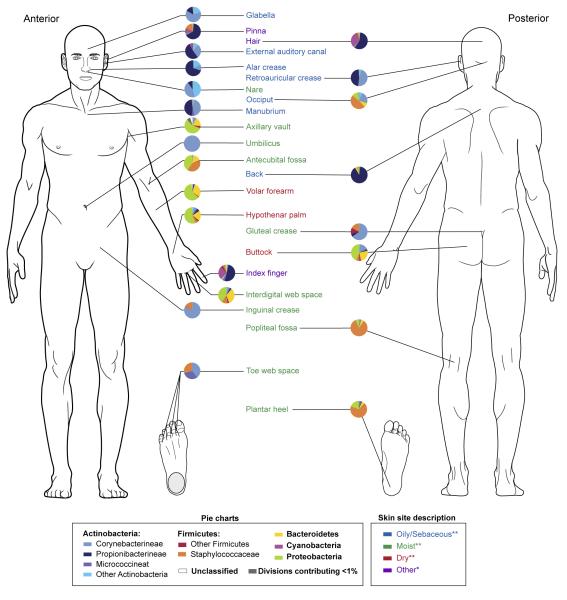
Bacterial diversity across multiple skin sites has also been assessed in a survey of healthy volunteers [41, 43] (Figure 3). Twenty specific sites were selected based on the predilection for particular dermatologic disorders to manifest at certain body sites that exhibit unique microenvironments. In addition to confirming significant bacterial diversity at the 20 different skin sites, similar bacterial communities were identified at skin sites with similar physiological characteristics: sebaceous, moist, and dry [43]. Corynebacterium spp. predominated at moist sites for instance, which corresponds with culture-based observations indicating that these organisms appear to favor skin with high humidity.
Figure 3.
Heterogeneity and pattern of distribution of the skin microbiome for over 20 skin sites. Each pie chart represents the relative abundance of different bacterial phyla at the indicated sites. Bacterial diversity varies among the many skin sites. Sites sharing physiologic features such as oily skin have greater similarities in the bacterial composition. Data derived primarily from [43]** with additional data from [41]*.
Several investigators have examined the variability of the skin bacterial microbiome in individual subjects, between individuals, and over time. When comparing sites with bilateral symmetry (left and right), healthy subjects had low intraindividual variability [40-41, 43], except for hands [38]. These studies suggest that, for most skin sites, a contralateral site could be used as a potential control. Certain body sites consistently exhibited high bacterial diversity across multiple individuals [38, 40-41, 43]. In contrast, several skin sites, particularly sebaceous sites, housed similar bacterial species across individuals [41, 43]. The high intrapersonal and interpersonal variation observed for hands is likely related to many factors, including frequent handwashing and potential contact with numerous environmental elements. The extent to which the skin bacterial microbiome fluctuates over weeks to months differed between individuals [38, 40-41, 43], but was influenced by the particular skin site studied [43]. In a small cohort of eight individuals [41], the skin bacterial microbiome was compared to non-cutaneous sites, and exhibited the greatest diversity of phylotypes and the highest degree of variability over time. Investigators also showed differential ability of distinct skin sites (forehead and forearm) to retain native bacterial communities after exposure to swabs from other skin and body sites [38]. This and other work [36-37, 40, 43] emphasizes the importance of careful skin site selection when studying the skin bacterial microbiome and the range of variability encountered when investigating the skin microbiome.
Skin characteristics also change as people age. Age-related alterations in skin bacterial communities have been examined in detail using culture-based techniques [28-29]. Before birth, neonates are sterile and the role of delivery in establishing the skin microbiota in newborns has been studied [42]. Unlike the bacterial site-specificity observed in adults, the bacterial communities from different anatomical sites of newborn skin were similar. Furthermore, infants delivered vaginally harbored bacteria similar to maternal vaginal bacteria, while newborns delivered by Cesarean section had bacterial skin microbiomes similar to maternal skin bacteria. Longitudinal studies examining fluctuations in skin microbiota may provide additional insight into the establishment and stability of the skin microbiome.
Bacterial microbiome in skin diseases
To understand the influence of bacteria on several dermatologic diseases, investigators have begun to use 16S rRNA-gene sequencing to analyze the skin bacterial microbiome in several skin disease states, including psoriasis, skin ulcers, and acne [44-50]. Culture-based studies showed that S. aureus, particularly toxigenic strains, was significantly increased in lesional skin and nares of psoriasis patients compared to healthy controls [51]. In a sequencing-based comparison of bacteria from the skin of healthy controls and lesional and nonlesional skin of psoriasis patients [47], the bacterial diversity observed from skin lesions of psoriasis patients was greater than the other skin samples, including increased Streptococcus and significantly less Propionibacterium acnes [47]. The authors suggested that more extensive clinical phenotyping may increase the accuracy of identifying microbial associations with disease states and that understanding the over- and underrepresentation of certain bacteria may assist in diagnosis and treatment. Although a subset of psoriasis (guttate) has been associated with streptococcal infections, anti-streptococcal treatment does not ameliorate psoriasis [52]. Therefore, the disease course of psoriasis may not be directly affected by skin microbiome fluctuations.
Slow-healing wounds cause significant morbidity and economic burden and disproportionately affect immobilized, elderly, and diabetic individuals. Chronic wounds such as venous leg ulcers are often treated with local compression, and reliable evidence for the use of systemic and topical antibiotics – in the absence of infection – is insufficient [53-54]. Alternatively, a staphylococcal product appears to suppress antimicrobial peptide-induced inflammation in an acute wound model in mice [55]. Since the role of the skin microbiome in chronic wound healing is unclear, there is interest in understanding the impact of bacteria in the pathogenesis and treatment of chronic skin ulcers [46, 48-49, 56-58]. As in healthy skin, 16S rRNA gene sequence analysis identifies greater microbial diversity in chronic wounds than is found by routine cultures [46], though neither method has identified a specific microorganism as causative and/or associated. Because wound etiology impacts the clinical features and natural history of lesions, the bacterial microbiota of the wound may be dependent on the underlying cause (i.e. diabetes, venous disease, pressure). Studying defined clinical phenotypes based on etiologies of the wounds in conjunction with a greater understanding of wound bacterial microbiota may identify key roles for bacteria in the development, course, or treatment of chronic wounds. For instance, a significant increase in Pseudomonadaceae was observed after antibiotic therapy in a small subset of chronic wound patients and increased prevalence of Streptococcaceae seen in diabetic compared to non-diabetic wounds [49]. A logical application of microbial genomics research is dissection of host-microbe interactions. A longitudinal selective shift of bacterial microbiota from slow-healing diabetic mouse wounds correlates with aberrant expression of skin defense response genes [48]. Future skin microbiome research may help reveal such complex interactions.
Acne has long been associated with the bacteria P. acnes. More recently, however, investigators have used 16S-based sequencing to identify uncultivatable bacteria in acne patients [44]. This study showed that, in fact, healthy follicles are exclusively colonized by P. acnes, whereas acne lesions contain a mixture of S. epidermidis, Corynebacterium spp., and dominant P. acnes [44].
These studies have expanded our knowledge of bacterial microbiota beyond what was known from traditional cultivation techniques and will stimulate further investigation into the complexities of skin microbes. Investigations of the skin microbiome in cutaneous diseases have the potential to identify biomarkers to distinguish disease subtypes and guide development of treatments.
Concluding remarks
The use of advanced sequencing technologies has revolutionized human microbial research. Recent skin microbiome investigations have provided insights into the diverse bacterial microbiota inhabiting distinct skin regions. Although variability exists among individuals, commonalities of the skin microbiome have been identified, particularly in skin sites sharing physiologic characteristics. Early research has begun to examine the relevance of the human microbiome to skin diseases such as acne, chronic wounds, and psoriasis. Much more investigation is needed to identify bacterial microbiota of different skin sites, ascertain the function of these skin microbes in maintaining or advancing the health of the human host, determine how the host immune system interacts with cutaneous microbial communities [59], and examine the possible causative effects of bacteria on skin diseases (Box 2). Such investigations will require: i) continued advances in sequencing technology and development of analyses methods and software; ii) standardization of optimal sample processing and analytical approaches; iii) additional studies of skin diseases incorporating longitudinal design, consistent sampling methods, well-selected controls, and relevant clinical phenotyping and metadata; and iv) perturbation of the microbiota in animal models and human hosts to analyze the function of various microbes. Harnessing the power of genomics, bioinformatics, infectious diseases, clinical microbiology, microbial ecology, and dermatology will fuel ongoing efforts to understand the skin microbiome and its impact on human health and disease.
Box 2. Outstanding questions.
Skin microbes have evolved to become symbionts with their hosts. How do these microorganisms interact with and benefit the human hosts?
Skin microbiome studies have shown the immense diversity of bacteria residing on human skin. How do the myriad of microbes (bacteria, fungi, viruses) interact with each other?
The interindividual variability of the bacterial skin microbiome is relatively high at certain skin sites. Do these groups of highly variable bacteria share redundant functions or do different bacterial species contribute to an individual’s unique degree of skin health?
Standardized nomenclature for bacteria provides a common language, yet often assumes that members of a bacterial species are identical. Bacteria that are considered to represent a single strain often share a phenotype, such as specific growth requirements, but may have significant genomic differences. How will increasing knowledge of genomic differences among bacteria redefine the term “species” and change bacterial nomenclature?
How can we use characterization of the skin microbiome to better understand dermatoses pathophysiology and to ultimately develop better diagnostic tools and novel therapies?
Acknowledgements
The author thanks Julia A. Segre for her close collaboration; Elizabeth A Grice and Mark C. Udey for discussions; and Maria L. Turner for guidance and support. The author has no conflict of interest. This work was supported by the NCI Intramural Research Program and NIH Common Fund AR057504.
Glossary
- 16S rRNA gene
The 16S rRNA gene is universal among prokaryotes. Sequencing relies on sufficient conservation of this gene to allow sequence alignment and adequate variability to enable bacterial identification.
- Atopic dermatitis (AD)
A common, chronic, and itchy skin condition -- commonly referred to as atopic eczema -- that is characterized by scaly red skin. Patients are at risk for skin infections with Staphylococcus aureus. The disease occurs most frequently in children and has a strong genetic predisposition. Individuals may have associated asthma and/or allergic rhinitis as well as elevated serum IgE levels.
- Chimera
A hybrid DNA sequence derived from multiple parent sequences that can result from PCR amplification and sequencing errors. Software tools have been, and continue to be, developed to identify these spurious sequences.
- Psoriasis
A chronic skin condition characterized by thick red skin with silvery scales. The most common form of psoriasis, plaque psoriasis, characteristically affects the elbows, knees, and back of the scalp. There is a genetic predisposition associated with this skin disease.
- Seborrheic dermatitis
A very common, chronic inflammatory skin condition that results in white to yellow greasy flaky skin in areas known to have oily skin (ear canals and scalp). It is more commonly known as dandruff.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwase T, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 4.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamady M, et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Z, et al. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol. 2010;48:3575–3581. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuomo CA, Birren BW. The fungal genome initiative and lessons learned from genome sequencing. Methods Enzymol. 2010;470:833–855. doi: 10.1016/S0076-6879(10)70034-3. [DOI] [PubMed] [Google Scholar]
- 14.Ochman H, et al. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 15.Lipkin WI. Microbe hunting. Microbiol Mol Biol Rev. 2010;74:363–377. doi: 10.1128/MMBR.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finkbeiner SR, et al. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palacios G, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 18.Schowalter RM, et al. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugenholtz P, Tyson GW. Microbiology: metagenomics. Nature. 2008;455:481–483. doi: 10.1038/455481a. [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley RE, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 24.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogen AL, et al. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierer N, et al. Forensic identification using skin bacterial communities. Proc Natl Acad Sci U S A. 2010;107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble WC, Somerville DA, editors. Microbiology of Human Skin. W.B. Saunders Company Ltd; 1974. [Google Scholar]
- 29.Marples M. The Ecology of the Human Skin. Charles C Thomas, Bannerstone House; 1965. [Google Scholar]
- 30.Chiller K, et al. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 31.Frank DN, et al. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuehnert MJ, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J Infect Dis. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 33.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 34.Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai Y, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank DN, et al. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J Clin Microbiol. 2003;41:295–303. doi: 10.1128/JCM.41.1.295-303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekio I, et al. Detection of potentially novel bacterial components of the human skin microbiota using culture-independent molecular profiling. J Med Microbiol. 2005;54:1231–1238. doi: 10.1099/jmm.0.46075-0. [DOI] [PubMed] [Google Scholar]
- 38.Fierer N, et al. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Z, et al. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bek-Thomsen M, et al. Acne is not associated with yet-uncultured bacteria. J Clin Microbiol. 2008;46:3355–3360. doi: 10.1128/JCM.00799-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dekio I, et al. Characterization of skin microbiota in patients with atopic dermatitis and in normal subjects using 16S rRNA gene-based comprehensive analysis. J Med Microbiol. 2007;56:1675–1683. doi: 10.1099/jmm.0.47268-0. [DOI] [PubMed] [Google Scholar]
- 46.Frank DN, et al. Microbial diversity in chronic open wounds. Wound Repair Regen. 2009;17:163–172. doi: 10.1111/j.1524-475X.2009.00472.x. [DOI] [PubMed] [Google Scholar]
- 47.Gao Z, et al. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PLoS ONE. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grice EA, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci U S A. 2010;107:14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price LB, et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS One. 2009;4:e6462. doi: 10.1371/journal.pone.0006462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharschmidt TC, et al. Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. J Invest Dermatol. 2009;129:2435–2442. doi: 10.1038/jid.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balci DD, et al. High prevalence of Staphylococcus aureus cultivation and superantigen production in patients with psoriasis. Eur J Dermatol. 2009;19:238–242. doi: 10.1684/ejd.2009.0663. [DOI] [PubMed] [Google Scholar]
- 52.Owen CM, et al. Antistreptococcal interventions for guttate and chronic plaque psoriasis. Cochrane Database Syst Rev. 2000:CD001976. doi: 10.1002/14651858.CD001976. [DOI] [PubMed] [Google Scholar]
- 53.O’Meara S, et al. Antibiotics and antiseptics for venous leg ulcers. Cochrane Database Syst Rev. 2010:CD003557. doi: 10.1002/14651858.CD003557.pub2. [DOI] [PubMed] [Google Scholar]
- 54.Kantor J, Margolis DJ. Management of leg ulcers. Semin Cutan Med Surg. 2003;22:212–221. doi: 10.1016/S1085-5629(03)00043-9. [DOI] [PubMed] [Google Scholar]
- 55.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gontcharova V, et al. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolcott RD, et al. Bacterial diversity in surgical site infections: not just aerobic cocci any more. J Wound Care. 2009;18:317–323. doi: 10.12968/jowc.2009.18.8.43630. [DOI] [PubMed] [Google Scholar]
- 58.Wolcott RD, et al. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol. 2009;9:226. doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wanke I, et al. Skin Commensals Amplify the Innate Immune Response to Pathogens by Activation of Distinct Signaling Pathways. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.328. [DOI] [PubMed] [Google Scholar]
- 60.National Research Council (U.S.) Committee on Metagenomics: Challenges and Functional Applications. and National Academies Press (U.S.) The new science of metagenomics : revealing the secrets of our microbial planet. National Academies Press; 2007. [PubMed] [Google Scholar]