Abstract
The Rho family of GTPases represents a class of Ras-related signaling molecules often deregulated in cancer. Rho GTPases switch from a GDP-bound, inactive state to a GTP-bound, active state in response to extracellular stimuli such as mitogens and extracellular matrix. In addition, Rho GTPase signaling can be altered in response to cell intrinsic factors such as changes in oncogenic or tumor suppressor signaling. In their active form, these proteins bind to a number of effector molecules, activating signaling cascades which regulate a variety of cellular processes including cytoskeletal reorganization, cell cycle progression, cell polarity and transcription. Here, we focus on one Rho family member, Cdc42, which is overexpressed in a number of human cancers. Consistent with a role in the promotion of tumorigenesis, activating mutations in Cdc42 and guanine nucleotide exchange factors are transforming, while inhibition of Cdc42 activity can impinge on cellular transformation following the activation of oncoproteins or loss of tumor suppressor function. Furthermore, Cdc42 activity has been implicated in the invasive phenotype which characterizes tumor metastasis, further suggesting that Cdc42 may be a useful target for therapeutic intervention. However, several recent studies in mice have unveiled a putative tumor suppressor function of Cdc42 in several tissue types which may involve cell polarity maintenance, suggesting that the role of Cdc42 in cancer development is complex and may be cell type specific.
Keywords: Rho GTPases, Cdc42, transformation, tumorigenesis, cell migration, invasion, polarity
1. Introduction
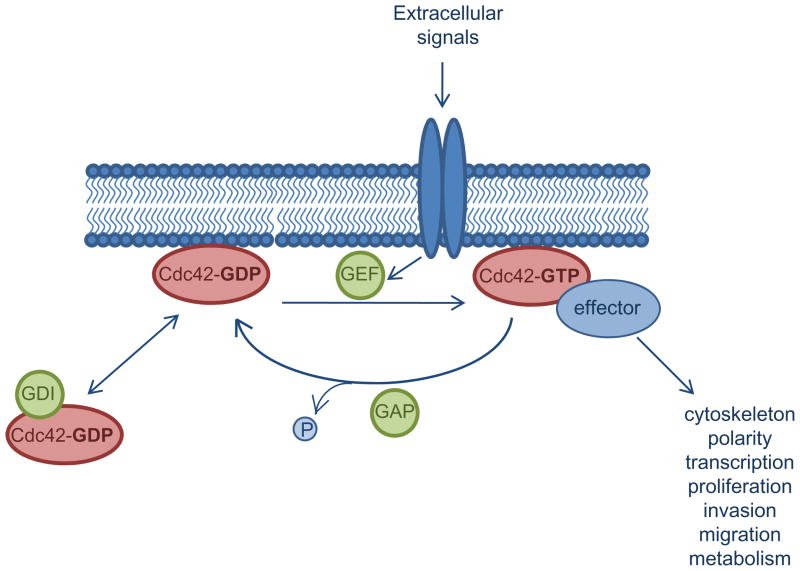
Cdc42 is a member of the Rho family of small GTPases, which function as molecular switches, transitioning from an inactive GDP-bound form to an active GTP-bound form in response to diverse signals [1]. The activation status of Cdc42 is tightly regulated by a variety of interacting proteins including guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs) and guanine nucleotide-dissociation inhibitors (GDIs) (fig. 1). Upon stimulation of cell surface molecules such as receptor tyrosine kinases, G-protein coupled receptors, cytokine receptors and integrins, GEFs mediate the exchange of GDP for GTP resulting in Cdc42 activation [1]. Once activated, Cdc42 can interact with a variety of downstream effectors leading to their activation, usually through induction of conformational changes which alleviate autoinhibitory interactions within the effector molecules [1–3]. The ability of Cdc42 to interact with and stimulate effector signaling is terminated when the bound GTP is hydrolyzed to GDP. Whereas the intrinisic GTPase activity of Cdc42 is low, Rho GAPs recognize the GTP-bound form of Cdc42 and enhance its intrinsic GTPase activity, facilitating the return Cdc42 to its inactive form [4]. Separately, Rho GDIs may function to sequester GDP-bound Cdc42 and extract it from membrane organelles, thereby preventing its activation [5]. Collectively, the Rho GEF, GAP, and GDI families of regulators control the spatially and temporally dynamic process of Cdc42 activation by regulating GDP-GTP exchange, GTP hydrolysis and subcellular localization.
Figure 1. Biochemical regulation of Cdc42 activity by three classes of regulators.
In response to stimulatory signals, activated GEFs catalyze the dissociation of GDP and binding of GTP to Cdc42. In its active, GTP-bound form, Cdc42 binds to effector molecules, leading to the activation of a variety of signaling cascades regulating cellular processes such as proliferation, survival, invasion and migration. GAP proteins enhance the intrinsic GTPase activity of Cdc42, resulting in GTP hydrolysis and inactivation of Cdc42. Cdc42 activity is further inhibited by GDIs which sequester Cdc42 away from cell membranes, thus preventing its activation.
Cdc42 was originally identified in S. cerevisiae as a mediator of cell division through the regulation of bud site selection and subsequent bud formation [6, 7]. Since its original discovery, Cdc42 has been found to influence a variety of signaling events and cellular processes in a myriad of organisms from yeast to mammals [8] (table 1). Efforts to identify downstream effectors of Cdc42 have revealed a number of kinases such as PAKs, MLKs, and MRCKs as well as scaffolding proteins such as Par6, WASP, and IQGAP that are directly regulated by Cdc42 activity [1, 9]. The plethora of effectors activated downstream of Cdc42 set in motion a variety of signaling cascades initiating changes in cellular processes including cell polarity, cytoskeletal remodeling, proliferation, migration, adhesion, membrane trafficking, and transcription [9, 10]. It is clear that tight regulation of these cellular processes by Cdc42 is important for normal cell function and their deregulation has been associated with a number of pathological conditions [11, 12]. Recent gene targeting studies in mouse models have revealed complex signaling roles of Cdc42 that are likely stimuli- and tissue cell type-specific under physiological conditions [13].
Table 1.
| Species | Physiological role of Cdc42 |
|---|---|
| S. cerevisiae | Polarized cell growth; polarized organization of the actin cytoskeleton at sites of bud emergence [6,7] |
| S. pombe | Polarized cell growth [8] |
| C. elegans | Embryonic body elongation [8] |
| D. melanogaster | |
| M. musculus |
|
| H. sapiens |
|
Loss or gain of Cdc42 in certain tissue cell types in mice can manifest pathologic phenotypes mimicking disease states such as holoprosencephaly, myeloid proliferative disease, lymphopenia, anemia, and premature aging (table 1)[13]. Likewise, deregulation of Cdc42 has been associated with several human disease states and/or developmental disorders. For example, faciogenital dysplasia is an inherited, X-linked disease characterized by short stature and craniofacial abnormalities, which results from mutations of the Cdc42 GEF, FGD1 [14]. In Fanconi Anemia, a genetic disorder characterized by a number of hematological abnormalities, defects in patient hematopoietic stem/progenitor cell adhesion and homing are attributed, at least in part, to deregulation of Cdc42 activity [15]. Significantly, accumulating evidence has implicated deregulated Cdc42 activity in cell transformation, cancer development and metastasis. Of the multitude of pathologic conditions in which Cdc42 signaling may be involved, the current review will focus on the involvement of Cdc42 in cancer.
2. Cdc42 as a putative oncoprotein
2.1 Cdc42 activation in cell transformation
Activation of Cdc42 results in the stimulation of a variety of signaling cascades altering cellular processes such as cytoskeletal remodeling, establishment of cell polarity, regulation of cell migration and cell proliferation, as well as modification of transcriptional programs. As such, it is not surprising that inappropriate activation of Cdc42 has been shown to be oncogenic [16, 17]. Several mutations in the Cdc42 protein including Cdc42Q61L and Cdc42G12V, which represent constitutively activative mutations, and Cdc42F28L, which exhibits spontaneous and accelerated cycling between the GDP- and GTP-bound states, are capable of inducing foci formation and/or anchorage-independent growth in immortalized fibroblasts [16, 17]. Interestingly, while the fast-cycling Cdc42F28L mutant readily transformed NIH 3T3 cells [16, 17], constitutively active mutants, which are defective in GTP hydrolysis and effectively locked in the active state, appeared to hinder cell growth [18] but promote foci-formation on soft agar [18]. It is important to note that, unlike the Ras oncoprotein, to date no activating Cdc42 mutations have been detected in human cancer [19]. This is consistent with the observation that Cdc42G12V overexpression actually confers a growth disadvantage and suggests that, unlike Ras, the transforming potential of Cdc42 is dependent on its ability to dynamically cycle between GDP- and GTP-bound states. Alternatively, it may suggest that requirements for Cdc42 activity may fluctuate throughout the process of tumor progression and that constitutively elevated Cdc42 signaling can be advantageous to the tumor cell growth at one stage yet disadvantageous at another.
While mutations in the cdc42 gene have not been detected in human cancers, Cdc42 has been reported to be overexpressed in several types of human cancers including non-small cell lung cancer[20], colorectal adenocarcinoma [21], melanoma [22], breast cancer [23, 24] and testicular cancer [25]. One study in melanoma reported that overexpression of Cdc42 positively correlated with prognostic indicators, suggesting that elevated Cdc42 level may be detrimental to patient survival [22]. While these initial studies seem to implicate elevation of Cdc42 in a variety of human cancers, further supporting the notion that Cdc42 acts as an oncogene, more studies are needed to determine the feasibility and therapeutic benefit of targeting Cdc42 in human disease.
The mechanisms by which Cdc42 activation contributes to oncogenesis, including what signaling pathways activated downstream of Cdc42 are essential for the induction of cellular transformation, have been under active investigation. In yeast, Cdc42 is known to be required for cell division. Similarly, Cdc42 has been shown to be involved in mammalian cell cycle progression. In fact, Cdc42 seems to be required for both G1-S phase progression and mitosis [26–28]. In addition to promoting cell proliferation, Cdc42 has been implicated in regulating cancer cell survival, as well as modulating the transcription factors such as SRF [29], STAT3 [30] and NFkB [31], which are further involved in the cell growth and survival. These proliferative and survival roles of Cdc42 in cancer biology have been extensively reviewed elsewhere [10–12, 32].
One recent, intriguing study implicated the activity of Cdc42, as well as other Rho proteins, in the promotion of cellular transformation by altering cell metabolism [33]. It has long been appreciated that cancer cells exhibit abnormal metabolic features when compared to normal cells. In particular, it is well established that cancer cells consume higher levels of glucose and secrete increased amount of lactate in the presence of oxygen, a phenomenon known as the Warburg effect [34]. In addition to glucose consumption, tumor cells also consume large amounts of the amino acid, glutamine [35]. The TCA cycle is crucial in proliferating cells, because it provides key precursors necessary for macromolecule biosynthesis. High glutamine consumption fosters proliferation by replenishing TCA cycle intermediates allowing for sustained biogenesis of lipids, proteins and nucleic acids [34, 35]. Compared to untransformed cells, mitochondria from the activating Cdc42F28L mutant-transformed fibroblasts exhibit elevated glutaminase activity that is critical for sustained glutamine metabolism. Furthermore, suppression of glutaminase activity by small molecule inhibitors deminished the growth and transformation of Cdc42F28L cells without significantly impacting the growth of control cells, suggesting that the ability of Cdc42 to induce transformation is associated with its ability to enhance the metabolism of cancer cells [33]. Therefore, alteration of cell metabolism to support elevated proliferation of cancer cells may be a novel mechanism by which Cdc42 contributes to cell transformation.
2.2 Deregulation of Cdc42 regulators in transformation
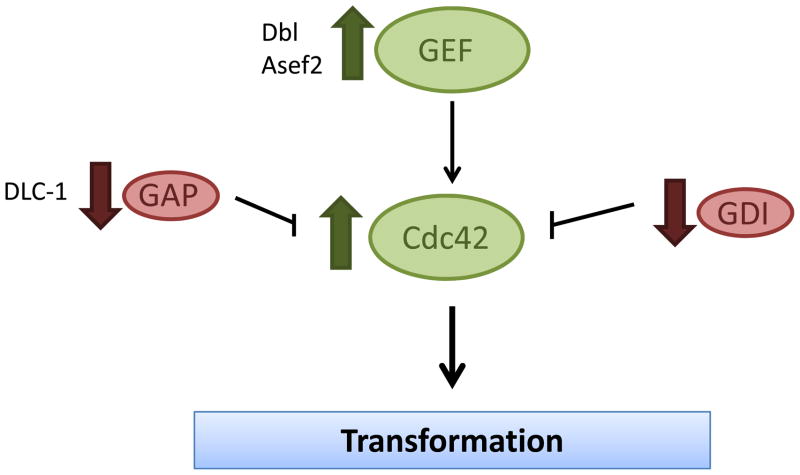
Given that the cycling of Cdc42 between active and inactive states is important for its transforming potential, it seems likely that perturbation of regulators of Cdc42 activity may potentiate oncogenesis caused (fig. 2). Cdc42 is positively regulated by a large number of GEFs which stimulate the dissociation of GDP and subsequent GTP binding. Early evidence of cellular transformation following aberrant activation of GEF activity comes from studies of the oncoprotein, Dbl. Dbl was first identified as a transforming gene product following isolation from a human diffuse B-cell lymphoma. Upon further examination, it was determined that oncogenic Dbl is derived from a chromosomal rearrangement resulting in the deletion of the amino terminus of the Dbl proto-oncogene [36, 37]. Studies in NIH 3T3 cells reveal that oncogenic Dbl is highly transforming [38]. Subsequent studies have demonstrated that the transforming activity of oncogenic Dbl results from its ability to inappropriately activate Rho GTPases, including Cdc42 [39]. Another example of deregulated GEF activity found in cancer is the Cdc42-specific exchange factor, Asef2 [40]. Asef2 activity is regulated through binding to the tumor suppressor, APC. Either Asef2 overexpression or mutations in APC that affect Asef2 binding, which are commonly observed in colorectal cancer, can lead to an inappropriate activation of Cdc42 [41]. Consistently, Asef2 loss of function significantly impairs adenoma formation in APC(min/+) mice [42].
Figure 2. Cdc42 and its regulators are deregulated in cancer cells.
While fast-cycling Cdc42 mutant can transform NIH 3T3 cells, Cdc42 is found overexpressed in a variety of human tumors, consistent with its role as an oncoprotein. Although no activating mutation of Cdc42 has been detected in primary tumors, inappropriate activation of GEFs, which activate Cdc42, is associated with tumorigenesis, while negative regulators of Cdc42 activity, GAPs and GDIs, can suppress tumorigenesis.
While GEFs act as positive regulators of Cdc42 activity, GAPs act as negative regulators through enhancement of the intrinsic GTPase activity of Cdc42 [4]. Therefore, while high GEF activity results in increased Cdc42 activation, loss of GAP activity could similarly result in the inappropriate activation of Cdc42. In fact, loss of function or downregulation of several Rho GAPs have been observed in a variety of cancer types, suggesting that while GEFs may play an oncogenic role through enhancing Rho GTPase activation, GAPs may play a tumor suppressor role by keeping the GTPase activity in check [4]. Consistent with this idea, DLC-1, a Rho and Cdc42-specific GAP, has been shown to be down regulated in a variety of human cancers including HCC, breast and lung cancers [43]. Furthermore, restoration of DLC-1 expression in human cancer cell lines lacking the endogenous protein resulted in reduced proliferation and colony formation as well as reduced tumor formation in xenograft models, suggesting that DLC-1 can in fact act as a tumor suppressor [43, 44]. It is important to note, however, that while this tumor suppressor function was attributed at least in part to the ability of DLC-1 to inhibit Rho GTPase signaling, to what extent its regulation of Cdc42 contributes to the tumor suppressive phenotype has not been thoroughly explored.
Finally, Cdc42 activity can be further regulated through the activity of Rho GDIs. Like GAPs, these proteins are traditionally thought to inhibit Cdc42 signaling; however, they do so by inhibiting GDP dissociation from Cdc42 and further sequestering the Cdc42 protein away from the cell membranes at which it signals. Thus, Rho GDIs may not simply function to inhibit Rho protein activity, but may also be responsible for shuttling the GTPases between different cellular membranes and thereby altering their signaling [9]. Currently, the correlation between GDI function and oncogenesis is somewhat convoluted. In some instances, GDIs seem to function as tumor suppressors, with a number of reports suggesting that GDIs can suppress tumor cell metastasis [45–48]. One study observed that GDI expression is reduced in hepatacellular carcinoma and correlates with an increased invasive phenotype and activation of Rho GTPases, including Cdc42 [49]. However, other studies have found that high levels of GDIs correlate with an invasive phenotype [48, 50]. These discrepancies may represent tumor and tissue-specific differences, and may also reflect the complexity of Rho GTPase regulation by GDIs.
3. Cdc42 as a mediator/modulator of oncogenic transformation
3.1. Cdc42 in oncogenic Ras induced transformation
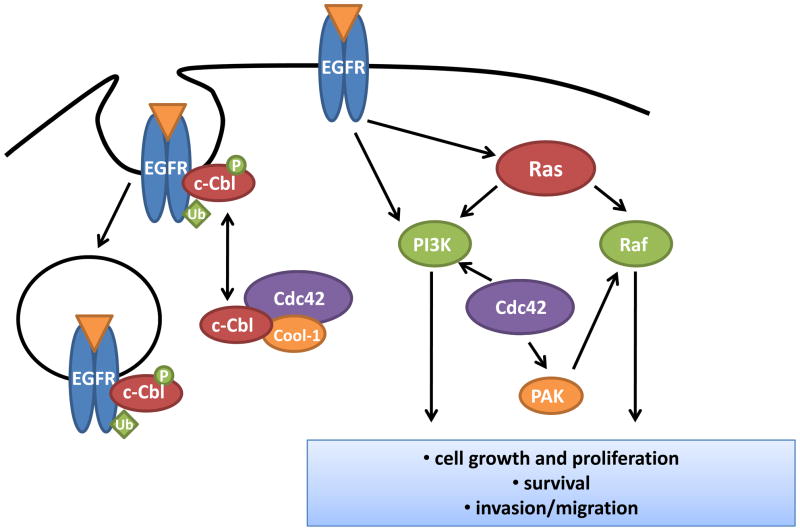
While enhanced activation of Cdc42 has been shown to elicit transformation, Cdc42 has also been shown to modify the ability of other oncoproteins, including Ras and EGFR, to induce cellular transformation. The ras proto-oncogene is a central component of mitogenic signaling and is mutated in approximately 30% of human tumors [51]. Gain-of-function ras mutations can result in the inappropriate activation of signaling pathways, particularly Raf-MEK-ERK and PI3K-Akt, leading to tumor initiation and progression [51]. Early studies suggesting that Cdc42 is involved in transformation by oncogenic Ras came from experiments in Rat1 fibroblasts. Expression of a dominant negative Cdc42 mutant prevented HRasV12 from inducing colony formation in soft agar [52]. Subsequently, multiple studies have reported that Cdc42 becomes activated upon expression of oncogenic Ras, and it can impinge on Ras-induced signaling pathways [53, 54]. For instance, Cdc42 has been shown to bind and activate PI3K [53], while one of the Cdc42 effectors, PAK, has been shown to phosphorylate both Raf1 and MEK, thus enhancing signaling through ERK [54]. In addition to studies carried out in rodent cells, transformation of human fibroblasts with HRasV12 is also inhibited upon dominant negative Cdc42 expression. Interestingly, expression of angiogenic factors was downregulated upon Cdc42 inhibition in this model [55]. However, while this effect may account for differences in tumorigenesis in vivo, it does not explain in vitro changes in cell growth. One intriguing study recently identified two separate cellular pools of HRasV12, which associate with distinct signaling complexes at either the plasma membrane or endomembrane. Signaling at the plasma membrane, which is sufficient to activate Raf1, is only weakly transforming, whereas activation of Cdc42 by Ras at the endomembrane is required to achieve full transformation [56]. These studies suggest that Ras signaling can be characterized by the cellular compartment where it occurs, that signaling effects at these various compartments are distinct, and that Cdc42 is vital for the transforming Ras signal emanating from endomembranes. Future studies must address how the Ras-Cdc42 signaling module at endomembrane promotes transformation, and what signaling pathways are involved.
3.2. Cdc42 in EGFR signaling
In addition to Ras-mediated transformation, Cdc42 has also been shown to modify cellular transformation resulting from aberrant EGFR signaling [9, 57]. The epidermal growth factor receptor (EGFR) is an oncoprotein which has been found to be overexpressed, mutated, and/or inappropriately activated in a variety of human tumors. EGFR contributes to tumor development and progression through activation of mitogenic signaling pathways. As such, the activity of EGFR is tightly regulated, not only by ligand-mediated receptor activation, but also through a course of receptor endocytosis, degradation and recycling [9]. Maintenance of normal EGFR degradation is critical for prevention of aberrant and sustained mitogenic signal. One function of Cdc42 in this process is as a modulator of EGFR degradation. Through interaction with its effector, Cool-1/β-Pix, Cdc42 associates with the ubiquitin ligase, c-Cbl, which is involved in the initiation of EGFR degradation. Activation of Cdc42 resulted in c-Cbl sequestration and prevention of ubiquitin-mediated EGFR degradation, leading to sustained EGFR signaling and ultimately cellular transformation [57]. In fact, treatment of Cdc42F28L transformed cells with an EGFR inhibitor caused a significant reduction in cell growth [57]. The contribution of Cdc42 to EGFR-mediated transformation is also observed in human cancer models. In breast cancer cells overexpressing EGFR, Cdc42 depletion resulted in a c-Cbl-dependent reduction in EGFR protein, and a consequent reduction in cell growth and migration [58]. In addition to mediating EGFR degradation through the regulation of c-Cbl, active Cdc42 has also been shown to interact with the γCOP subunit of the coatomer protein complex, suggesting that Cdc42 may further regulate EGFR signaling by altering its cellular trafficking [9, 59]. However, the mechanism and extent to which Cdc42-mediated EGFR trafficking may alter EGFR signaling and oncogenesis in primary tumor cells remain unclear. Additionally, whether Cdc42 involvement in vesicle trafficking affects the abundance and/or localization of other cell surface receptors associated with tumorigenesis awaits further examination.
3.3. Cdc42 as a mediator of tumor suppressor signaling
In addition to modulation of the transforming activities of oncogenes, Cdc42 can signal downstream of tumor suppressors such as p53, PTEN or NF1 to affect cell transformation elicited following tumor suppressor loss of function. Primary fibroblasts deficient for p53 or p19ARF exhibit increased cell growth, which is partially dependent on Cdc42 activity. This ability of Cc42 to influence cell growth upon p53 loss is related to its role in the regulation of NFκB and cyclin D1 activities [60]. Further, Cdc42 has been implicated in p53-mediated changes in cell morphology and apoptosis [61, 62], whereas activation of Cdc42 following Cdc42GAP deletion resulted in p53-dependent induction of cellular senescence [63]. Parallel to effects on p53 signaling, Cdc42 has also been shown to modulate PTEN and NF1 tumor suppressor signaling. Cdc42, in conjunction with RhoA, has been shown to regulate PTEN localization at the cell surface during chemotaxis [64], suggesting that Cdc42 contributes to PTEN-mediated tumor suppression by regulating its localization and activity at the cell membrane. Activation of the Cdc42 effector, PAK, has been shown to be necessary for transformation upon NF1 loss. The loss of NF1 results in the deregulation of Ras activity, and the contribution of PAK to NF1-mediated transformation is thought to be dependent on its ability to modulate the activation of ERK downstream of Ras [65]. These studies further propose that, in addition to its ability to mediate transformation by oncoproteins such as Ras and EGFR, changes in Cdc42 signaling can modify transformation associated with the loss of tumor suppressor function.
4. Cdc42 in cancer cell metastasis
The acquisition of an invasive and motile phenotype is critical for tumor progression and mestastasis. Metastasis is a complex process in which cancer cells must migrate out of the primary tumor site, invade surrounding tissue, intravasate into the blood or lymphatic system, survive while in circulation, and extravasate and initiate metastatic outgrowth at distant organ sites [66]. A number of studies have implicated Cdc42 function in the regulation of multiple steps of this process. Consistently, studies in human cancer models have identified Cdc42 as an important regulator of metastasis [67–69].
4.1. Cdc42 and invadopodia formation
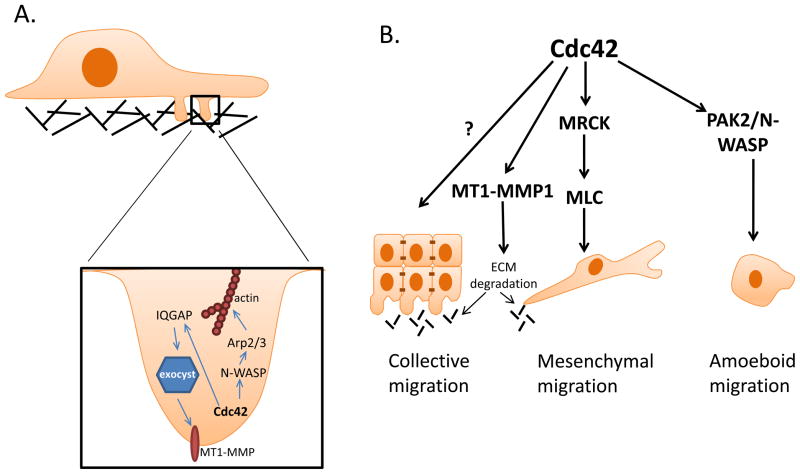
When invasive cancer cells are cultured on an extracellular matrix (ECM) substratum, such as fibronectin or collagen, they extrude actin-rich protrusions from their ventral surface into the matrix. These protrusions, termed invadopodia, provide focused regions of proteolytic matrix degradation by matrix metalloproteinases (MMPs) [70, 71]. There is evidence to suggest that Cdc42 is involved in both the formation of invadopodia structures as well as the production and/or activation of MMPs responsible for the ECM digestion necessary for tumor cell invasion (figure 4A). In rat mammary adenocarcinoma cells, Cdc42 activation upon EGF stimulation is required for invadopodia formation [72]. Activation of Cdc42 led to N-WASP and Arp2/3-mediated actin polymerization resulting in the protrusion of invadopodia structures into the matrix [72]. Furthermore, while activation of N-WASP and downstream Arp2/3 complex are required for invadopodia formation, stabilization of the invadopodia structure requires cofilin activity, which may also be modulated by Cdc42 signaling. Additionally, the highly invasive human breast cancer cell line, MDA-MB-231, exhibits high levels of invadopodia activity, and studies in this cell line further implicate Cdc42 activity in the initiation of invadopodia formation through enhanced signaling of a Cdc42-binding protein, CIP4, leading to N-WASP activation [73]. Finally, the Cdc42 specific GEF, FGD1, activates Cdc42-mediated invadopodia formation, and knockdown of FGD1 in human breast and prostate cancer cell lines results in a substantial decrease in matrix degradation [74]. Interestingly, FGD1 expression levels are elevated in human breast and prostate cancer samples compared to normal tissue, furthermore, FGD1 elevation is associated with aggressiveness of tumor grade [74]. Combined, these studies suggest that signaling downstream of Cdc42 is important for invadopodia formation.
Figure 4. Cdc42 activation contributes to tumor cell invasion and migration.
Cdc42 activation promotes cancer cell invasion through the regulation of invadopodia formation (A). Via its activity in mediating actin polymerization through N-WASP-Arp2/3 complex, Cdc42 promotes invadopodia structure formation. Additionally, through the IQGAP-mediated regulation of exocyst function, Cdc42 regulates the accumulation of matrix metalloproteinases at the tips of invadopodia, facilitating extracellular matrix degradation. Cdc42 activity is involved in both mesenchymal and amoeboid cell migration, while its ability to regulate MMP-mediated matrix degradation may be important for both mesenchymal and collective cell migration (B).
Aside from the ability of Cdc42 to regulate actin polymerization through N-WASP and Arp2/3, which is critical for invadopodia formation and persistence, there is also evidence suggesting a role for Cdc42 in the matrix-degrading function of the invadopodia structure through modulation of MMP activities. MT1-MMP is a transmembrane MMP capable of digesting a number of ECM components and cleaving pro-MMP2 leading to its activation [75]. While normal epithelial cells do not express MT1-MMP, MT1-MMP has been shown to accumulate at the invasive front of tumors and is concentrated at invadopodia [75, 76]. Studies in MDA-MB-231 breast cancer cells reveal trafficking of MT1-MMP to invadopodia following Cdc42 activation. Upon Cdc42 activaiton, one its effectors, IQGAP, complexes with components of the exocyst at invadopodia. RNAi-mediated silencing of either exocyst components or IQGAP resulted in diminished level of MT1-MMP at invadopodia, suggesting that Cdc42-induced IQGAP binding to the exocyst is crucial for the trafficking of MT1-MMP to invadopodia [76, 77]. Thus, these studies highlight a critical role for Cdc42 activity in the formation of invadopodia structures through the regulation of actin polymerization and in the modulation of invadopodia function through the regulation of MMP trafficking.
4.2. Cdc42 in cancer cell invasion and migration
Cancer metastasis is dependent on cancer cell migration, as well as on the ability of the tumor cell to invade the surrounding tissue, break through the basement membrane, and intravasate into circulation. Cancer cell invasion can occur via single or collective cell migration. Often, single cell migration is thought to involve a process termed epithelial-to-mesenchymal transition (EMT), in which tumor cells dissolve their cell-to-cell contacts and take on a more mesenchymal morphology to move through the extracellular matrix aided by the secretion of proteinases [66]. One of the hallmarks of EMT is the loss of the adhesion molecule, E-cadherin, and the acquisition of mesenchymal cadherins, such as N-cadherin, by cadherin switching [66]. One consequence of cadherin switching is the release of the E-cadherin binding protein, p120-catenin, from the cell membrane into the cytoplasm. Once in the cytoplasm, p120-catenin represses RhoA activity and activates Rac and Cdc42 through Vav2 to promote actin cytoskeleton remodeling [69, 78, 79]. Additionally, activation of Cdc42 during mesenchymal cell movement is required to generate the necessary actomyosin contractility for cell movement in the absence of Rho-ROCK signaling. In this context, the Cdc42 effector, MRCK, is necessary for phosphorylation of MLC2, actomyosin contractility, and subsequent cell invasion [80]. Consistent with the involvement of Cdc42 in EMT-mediated cell movement, overexpression of the Cdc42 GEF, ARHGEF9, is commonly observed in hepatocellular carcinoma samples and correlates with increased Cdc42 activation, reduced E-cadherin expression, and an EMT phenotype. Conversely, silencing of ARHGEF9 resulted in a reduction in Cdc42-GTP and restored an epithelial phenotype [68], correlating Cdc42 signaling with EMT in both mouse models and human cancer. These studies suggest an important role for Cdc42 in EMT-mediated, single tumor cell migration and invasion.
In addition to EMT-mediated cell movement, individual tumor cells can take on an amoeboid morphology while migrating through the extracellular matrix. This amoeboid migration is particularly evident in three-dimensional environments and, in contrast to mesenchymal movement, is characterized by a rounded, blebbing cell appearance and invasion through the extracellular matrix independent of proteolytic matrix remodeling [81]. Studies in melanoma cells suggest that in addition to mesenchymal cell movement, Cdc42 activity is also critical for amoeboid cell movement through a 3D collagen matrix. Activation of Cdc42 by the GEF, DOCK10, and subsequent activation of the effectors, PAK2 and N-WASP, is critical for amoeboid cell movement as RNAi-mediated targeting of DOCK10, PAK2, or N-WASP results in a transition of melanoma cells from an amoeboid to a mesenchymal morphology. However, Cdc42 targeting blocks both amoeboid and mesenchymal movement and overall tumor cell invasion, consistent with the idea that Cdc42 is important for both mesenchymal and amoeboid tumor cell movement [82]. These studies suggest that there may be significant plasticity associated with single tumor cell invasion. Therefore, given the critical role for Cdc42 in both amoeboid and mesenchymal invasion, targeting of Cdc42 would be ideal to inhibit broadly inhibit tumor cell invasion.
Tumor cells may also migrate collectively in clusters, a process which is commonly observed in many developmental morphogenesis processes [66, 83]. During collective cell migration, cells at the leading front demonstrate an invasive phenotype and secrete MMPs to make way for the migration of follower cells. In one model of collective cell invasion, expression of podoplanin, a protein which is upregulated at the invasive front of many human carcinomas, induces collective cell migration in the absence of EMT through a process in which Rho, Rac and Cdc42 activities are suppresed [84]. Interestingly, while podoplanin-induced cell migration was characterized by filopodia formation, a characteristic of cell morphology often attributed to Cdc42 activation, in this instance filopodia formation does not require induction of Cdc42 activity. In addition to cancer cell-autonomous mediation of collective cell migration, stromal cells such as fibroblasts may also function as leader cells to initiate collective cancer cell migration. In this context, leading fibroblasts deform the extracellular matrix as they pass through and leave tracks that cancer cells can follow. In contrast to podoplanin-induced collective cell migration, models of fibroblast-led invasion exhibit a requirement for Cdc42-mediated activation of MRCK to allow migration of carcinoma cells behind leading fibroblasts [85]. Therefore, while Cdc42 activity is not required for podoplanin-induced collective cell migration, it is required for collective migration of carcinoma cells behind leading fibroblasts. It remains unclear what accounts for the differential requirement of Cdc42 activity in these two models of collective cancer cell migration. Future studies are needed to elucidate the migratory role of Cdc42 in distinct cellular contexts.
5. Cdc42 as a putative “tumor suppressor”
5.1. Cdc42-mediated “tumor suppressor” activities
While there are a multitude of studies suggesting a role for activated Cdc42 in oncogenesis, several recent studies have sprung forth to suggest that in other pathologic contexts, Cdc42 may also function as a tumor suppressor. One such study examined the effect of a hepatocyte-specific deletion of Cdc42 in mice. Loss of Cdc42 in the liver resulted in chronic liver damage, hepatomegaly and development of hepatacellular carcinoma (HCC) by 8 months of age [86]. However, the mechanism by which Cdc42 loss results in hyperproliferation and subsequent HCC formation in the liver is not entirely clear - whether hyperproliferation upon Cdc42 loss is directly related to Cdc42 deficiency or is a byproduct of liver damage induced upon Cdc42 loss has not been determined. It is interesting to note that in a model of liver regeneration, elevated level of Cdc42 transcript correlated with hepatocyte proliferation following partial hepatectomy, suggesting that in this condition, Cdc42 may actually promote hepatocyte proliferation [87]. Furthermore, there are additional reports of Cdc42 upregulation in human HCC samples [88, 89], which would correlate with an oncogenic role for Cdc42 in promoting human liver tumors. Together, these studies point to a complex role for Cdc42 function in hepatocyte physiology and liver tumorigenesis.
A second example of a putative tumor suppressor role for Cdc42 was from a study examining hematopoietic cell specific deletion of Cdc42. Induced gene targeting of Cdc42 in murine bone marrow hematopoietic stem/progenitor cells resulted in a loss of hematopoietic stem cell quiescence and hyperproliferation of blood progenitors, causing lethality in mice approximately 20 days later. Examination of the Cdc42-deficient mice revealed a skewing of hematopoetic differentiation from erythroid toward a myeloid fate, resulting in the myeloproliferative disease (MPD) [90]. While MPD is not cancer, these studies suggest that Cdc42 loss in blood stem/progenitor cells could initiate a first step of transformation, which, when accompanied by another oncogenic event, could lead to the development of leukemia. This study also demonstrated that Cdc42 plays a critical role in normal hematopoesis by regulating transcriptional programs responsible for determining a myeloid or erythroid fate [90]. Therefore, the “tumor suppressor” function of Cdc42 in this context appears to stem from the ability of Cdc42 to maintain proper cell differentiation as well as regulate progenitor proliferation.
A third example of the putative tumor suppressor function of Cdc42 can be seen in models of neuroblastoma characterized by N-myc amplification. Neuroblastomas with N-myc amplification display deletions of the short arm of chromosome 1 in 90–95% of cases. This chromosomal region contains the cdc42 gene, and one copy of Cdc42 is consistently lost in these cancers [91]. Furthermore, N-myc overexpression in neuroblastoma cells results in reduced Cdc42 expression and ectopic expression of activated Cdc42 in these cells results in neuroblastoma cell differentiation. These data suggest that in certain models of neuroblastoma, Cdc42 may function as a tumor suppressor by promoting differentiation. It is interesting to note however, that while Cdc42 levels were reduced in neuroblastoma cells, they were never lost completely. Additionally, further depletion of Cdc42 in these cells through siRNA-mediated knockdown actually resulted in cell death, complicating the tumor suppressive role of Cdc42 in neuroblastoma. These results suggest that while reduced Cdc42 signaling may promote a less differentiated phenotype, complete loss of Cdc42 is not conducive to tumorigenesis [91].
Together, these studies support the notion that the effect of Cdc42 signaling in cancer is complex. While Cdc42 and its downstream effectors can be positive regulators in cancer development and progression, it may also function with a tumor suppressive activity in defined physiological conditions. It remains to be seen whether the ability of Cdc42 to prevent tumorigenesis can be generalized beyond the above reported cases.
5.2. Maintaining cell polarity: a putative mechanism for Cdc42-mediated tumor suppression?
While the underlying mechanism of Cdc42-mediated “suppression” of hyperproliferation and tumorigenesis remains largely unknown, tissue specific gene targeting studies in mice indicate that the role of Cdc42 in cell cycle progression, survival or proliferation varies between cell types [13]. There is evidence suggesting that maintenance of cell polarity could be one way in which Cdc42 contributes to tumor suppression. Most internal epithelial tissues contain a monolyer of polarized epithelial cells enclosing a central lumen. In 3D culture models of Madin-Darby canine kidney (MDCK) cells, Cdc42 is important for establishment of polarized epithelial cysts by regulating the vesicular trafficking of proteins to the apical surface of the cell. This Cdc42-mediated apical exocytosis occurred in a Par6-aPKC-dependent manner and is required for lumen formation [92]. In addition to its ability to regulate epithelial polarity via modulation of protein trafficking to distinct cellular surfaces, the Cdc42-Par6-aPKC signaling complex has been shown to control the formation of normal 3D polarized epithelial cyst structures by altering mitotic spindle orientation and the direction of cell division. In particular, loss of Cdc42, or Par6/aPKC, resulted in aberrant cysts with multiple lumens [93, 94]. The involvement of Cdc42 in the maintenance of planar cell polarity is further corroborated in mouse studies examining pancreatic development. Deletion of Cdc42 in the developing pancreas revealed that Cdc42 is required for apical polarization and is responsible for microlumen formation during the early stages of pancreatic development as well as for maintaining apical cell polarity [95]. However, while the role of Cdc42 in the establishment and maintenance of epithelial polarization is clearly important in development, whether Cdc42 exerts such a role in tumor initiation and/or progression has not been explored. One example of the close association of Cdc42-regulated epithelial cell polarity with deregulated proliferation comes from a mouse model of forebrain specific deletion of Cdc42, in which mislocalization of the polarity components Par6, aPKC, F-actin and Numb was found associated with a hyperproliferative phenotype of the neural progenitor cells [96]. Given the frequent commonality between developmental pathways and those deregulated in cancer, it may be extrapolated that the Cdc42-mediated maintenance of cell polarity is involved in cancer initiation and/or progression.
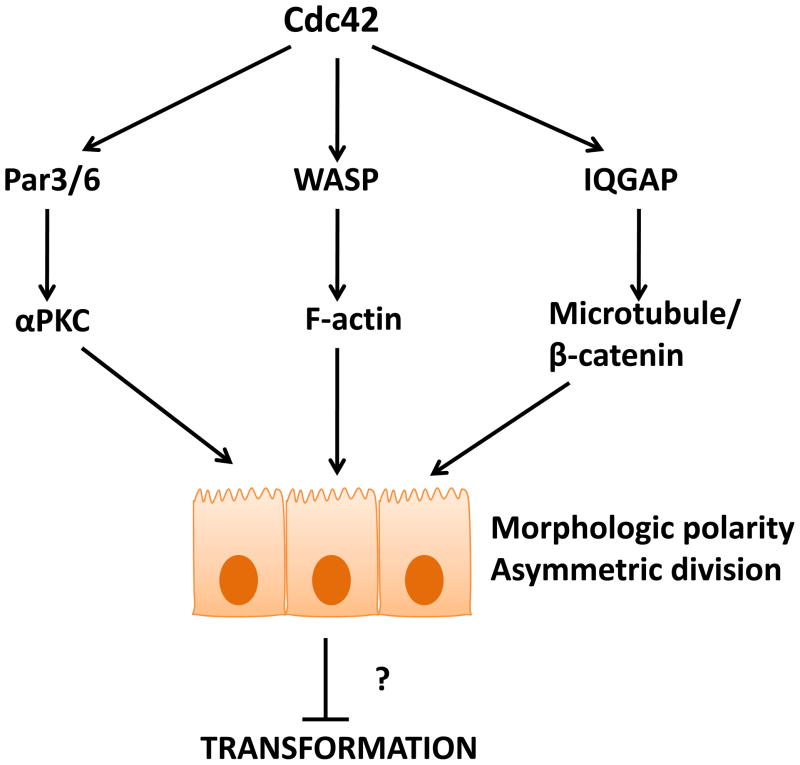
Cdc42 regulates the establishment of cell polarity through modulating intracellular vesicle trafficking to the apical cell surface and orientation of cell division spindle, and help maintain cell polarity by the strengthening of cell-cell junctions. E-cadherin-containing adherens junctions (AJs) are specialized cell-cell adhesion complexes which help maintain proper barrier function and apical-basolateral polarity in epithelial cells; furthermore, adherens junction disruption has been associated with tumor progression [97]. Activation of Cdc42 is required for the sustained E-cadherin-mediated adhesion at the adherens junction, and strong E-cadherin adhesion requires the linking of adherens junctions to the actin cytoskeleton [98, 99]. E-cadherin is linked to the actin cytoskeleton through interactions with β-catenin which further binds to α-catenin to anchor the cadherin/catenins complex to the actin cytoskeleton. Cdc42 can be linked to the adherens junction via its effector, IQGAP. In the absence of Cdc42 activity, IQGAP binds to β-catenin, displacing α-catenin from the cadherin/catenins complex at the AJ, resulting in the loss of α-catenin-linked actin filaments from the adherens junction and reduced cell-cell adhesion. In contrast, active Cdc42 binds to IQGAP, preventing its binding to β-catenin and subsequent AJ disruption [99, 100]. Therefore, loss of Cdc42 could result in AJ disruption and a loss of epithelial cell polarity that is associated with tumorigenicity. However, it is interesting to note that in the model of HCC induction upon Cdc42 deletion described above, loss of Cdc42 did not cause any significant changes in the localization of the cadherin/catenins complex [86], whereas aged mice deficient for Cdc42 in the skin exhibited a loss of cell-cell contacts [101]. This suggests that disruption of adherens junctions upon Cdc42 depletion may be a tissue specific event.
Aside from the role in regulating morphologic cell polarity, Cdc42 may also be involved in the regulation of asymmetric cell division by stem and progenitor cells to affect cell fate determination. Recent studies have suggested that upon loss of polarity, stem and/or progenitor cells lose the ability to undergo asymmetric cell division, and this further contributes to tumor initiation [102, 103]. What role Cdc42 may play in this phenomenon requires further elucidation, particularly in the MPD-phenotype manifesting in the hematopoietic knockout mouse model [90]. However, the idea that Cdc42-associated polarity pathway is important for cell fate determination has been demonstrated by a Cdc42 knockout study in the skin in which epidermal-specific deletion of Cdc42 resulted in altered cell fate decisions by epidermal progenitor cells [93]. This is correlated to the ability of Cdc42 to regulate levels of nuclear β-catenin downstream of Par6/aPKC-GSK3β. Additionally, while loss of Cdc42 in the skin did not result in tumor formation, epidermal hyperplasia was reported [101]. These studies in skin are consistent with Cdc42 deletion in blood and myc-driven neuroblastomas in which a loss or reduction in Cdc42 seem to promote hyperplasia by preventing normal cell differentiation [90, 91].
One recent study presented another intriguing explanation for Cdc42 mediated tumor suppression. Warner et al. reported that in Drosophila, as a component of the Par6/aPKC polarity complex, Cdc42 functions as a negative regulator of apoptosis-induced compensatory proliferation in response to tissue damage [96]. Therefore, Cdc42 loss in pre-cancerous or cancerous tissues in which the apoptotic process is compromised could allow proliferation in the absence of cell death, thus promoting tissue/tumor hyperproliferation [104]. These studies illustrate yet another possible tumor suppressive role for Cdc42 by maintaining cell polarity and keeping the compensatory proliferation in check.
6. Summary
Current evidence indicate that Cdc42 is capable of functioning as either a mitogenic or tumor suppressive signal transducer, a concept that requires further examinations in specific physiologic and pathologic contexts. For instance, it will be important to determine the activity and expression pattern of Cdc42 and its signaling effects during various stages of tumor initiation, tumor maintenance, or tumor metastasis. Whether the pro-oncogenic versus tumor suppressive role of Cdc42 can be accounted for by tissue-type or microenvironment differences is another area for future investigation. Mechanistically, given the seemingly universal role of Cdc42 in establishing and maintaining tissue/cell polarity [13], it is important to bear in mind that suppression of mitogenic signals that flow through the Cdc42 signaling axis may come at the price of disrupted cell polarity machinery, at least transiently. The combined considerations of these effects may help conclude the feasibility, risk/benefit, and potential efficacy related to the therapeutic targeting of Cdc42 in specific cancer types/stages. Alternatively, it will be interesting to explore the possibility that targeting Cdc42 could synergize with existing chemotherapy for improved outcomes. For instance, targeting leukemia stem cells with chemotherapy may be more effective if the cancer stem cells can be mobilized from their protective bone marrow niche by transiently suppressing Cdc42-mediated cancer stem cell adhesion and polarity. Like the recently appreciated diverse physiological functions of Cdc42 in mouse model studies [13], future studies of the role of Cdc42 in various tumor types and tumor progression stages promise to reveal much of the complexity and unknowns of the signaling involved.
Figure 3. Cdc42 mediates cellular transformation by oncogenic mutations of EGFR or Ras.
Loss of Cdc42 prevents transformation by oncogenic Ras, possibly mediated by its ability to enhance the activity PI3K and Raf, two well-characterized Ras effector pathways. In addition, Cdc42 is required for EGFR-mediated cellular transformation. Normal level of Cdc42 activity, through the regulation of the ubiquitin ligase c-Cbl, is necessary for the endocytosis, degradation, and/or recycling of EGFR receptor. In the absence of Cdc42, EGFR is rapidly degraded, suppressing EGFR-mediated transformation.
Figure 5. Cdc42 can be tumor suppressive by maintaining proper tissue/cell polarity.
Cdc42 modulates the activity of a variety of effector pathways to control cell polarity. The ability of Cdc42 to maintain epithelial cell polarity and associated cell adhesion through the maintenance of adherens junctions, and its regulatory role in asymmetric cell division of stem and progenitor cells, are possible mechanisms for Cdc42-mediated tumor suppression.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sinha S, Yang W. Cell Signal. 2008;20:1927–1934. doi: 10.1016/j.cellsig.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Dovas A, Cox D. Commun Integr Biol. 2010;3:101–105. doi: 10.4161/cib.3.2.10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Mol Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- 4.Tcherkezian J, Lamarche-Vane N. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 5.Olofsson B. Cell Signal. 1999;11:545–554. doi: 10.1016/s0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 6.Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DI, Pringle JR. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson DI. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerione RA. Trends Cell Biol. 2004;14:127–132. doi: 10.1016/j.tcb.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Aznar S, Lacal JC. Cancer Lett. 2001;165:1–10. doi: 10.1016/s0304-3835(01)00412-8. [DOI] [PubMed] [Google Scholar]
- 11.Sahai E, Marshall CJ. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 12.Vega FM, Ridley AJ. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Melendez J, Grogg M, Zheng Y. J Biol Chem. 2011;286:2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, Fischer DJ, Santos MF, Tigyi G, Pasteris NG, Gorski JL, Xu Y. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Shang X, Guo F, Murphy K, Kirby M, Kelly P, Reeves L, Smith FO, Williams DA, Zheng Y, Pang Q. Blood. 2008;112:1683–1686. doi: 10.1182/blood-2008-03-147090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fidyk N, Wang JB, Cerione RA. Biochemistry. 2006;45:7750–7762. doi: 10.1021/bi060365h. [DOI] [PubMed] [Google Scholar]
- 17.Lin R, Bagrodia S, Cerione R, Manor D. Curr Biol. 1997;7:794–797. doi: 10.1016/s0960-9822(06)00338-1. [DOI] [PubMed] [Google Scholar]
- 18.Vanni C, Ottaviano C, Guo F, Puppo M, Varesio L, Zheng Y, Eva A. Cell Cycle. 2005;4:1675–1682. doi: 10.4161/cc.4.11.2170. [DOI] [PubMed] [Google Scholar]
- 19.Rihet S, Vielh P, Camonis J, Goud B, Chevillard S, de Gunzburg J. J Cancer Res Clin Oncol. 2001;127:733–738. doi: 10.1007/s004320100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Wang Y, Zhang Y, Miao Y, Zhao Y, Zhang PX, Jiang GY, Zhang JY, Han Y, Lin XY, Yang LH, Li QC, Zhao C, Wang EH. Lung Cancer. 2009;63:375–382. doi: 10.1016/j.lungcan.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Gomez Del Pulgar T, Valdes-Mora F, Bandres E, Perez-Palacios R, Espina C, Cejas P, Garcia-Cabezas MA, Nistal M, Casado E, Gonzalez-Baron M, Garcia-Foncillas J, Lacal JC. Int J Oncol. 2008;33:185–193. [PubMed] [Google Scholar]
- 22.Tucci MG, Lucarini G, Brancorsini D, Zizzi A, Pugnaloni A, Giacchetti A, Ricotti G, Biagini G. Br J Dermatol. 2007;157:1212–1216. doi: 10.1111/j.1365-2133.2007.08246.x. [DOI] [PubMed] [Google Scholar]
- 23.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritz G, Just I, Kaina B. Int J Cancer. 1999;81:682–687. doi: 10.1002/(sici)1097-0215(19990531)81:5<682::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 25.Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y, Yoshida K. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 26.Gjoerup O, Lukas J, Bartek J, Willumsen BM. J Biol Chem. 1998;273:18812–18818. doi: 10.1074/jbc.273.30.18812. [DOI] [PubMed] [Google Scholar]
- 27.Olson MF, Ashworth A, Hall A. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda S, Taniguchi H, Oceguera-Yanez F, Ando Y, Watanabe S, Monypenny J, Narumiya S. FEBS Lett. 2006;580:3375–3380. doi: 10.1016/j.febslet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Hill CS, Wynne J, Treisman R. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 30.Debidda M, Wang L, Zang H, Poli V, Zheng Y. J Biol Chem. 2005;280:17275–17285. doi: 10.1074/jbc.M413187200. [DOI] [PubMed] [Google Scholar]
- 31.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 32.Narumiya S, Yasuda S. Curr Opin Cell Biol. 2006;18:199–205. doi: 10.1016/j.ceb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ, Cheng T. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eva A, Aaronson SA. Nature. 1985;316:273–275. doi: 10.1038/316273a0. [DOI] [PubMed] [Google Scholar]
- 37.Zheng Y. Trends Biochem Sci. 2001;26:724–732. doi: 10.1016/s0968-0004(01)01973-9. [DOI] [PubMed] [Google Scholar]
- 38.Ron D, Tronick SR, Aaronson SA, Eva A. EMBO J. 1988;7:2465–2473. doi: 10.1002/j.1460-2075.1988.tb03093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin R, Cerione RA, Manor D. J Biol Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- 40.Hamann MJ, Lubking CM, Luchini DN, Billadeau DD. Mol Cell Biol. 2007;27:1380–1393. doi: 10.1128/MCB.01608-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki Y, Sagara M, Shibata Y, Shirouzu M, Yokoyama S, Akiyama T. Oncogene. 2007;26:7620–7267. doi: 10.1038/sj.onc.1210574. [DOI] [PubMed] [Google Scholar]
- 42.Kawasaki Y, Tsuji S, Muroya K, Furukawa S, Shibata Y, Okuno M, Ohwada S, Akiyama T. EMBO Rep. 2009;10:1355–1362. doi: 10.1038/embor.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, Popescu NC. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Thorgeirsson SS, Popescu NC. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Chang Z, Chiao LJ, Kang Y, Xia Q, Zhu C, Fleming JB, Evans DB, Chiao PJ. Cancer Res. 2009;69:7851–7859. doi: 10.1158/0008-5472.CAN-08-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moissoglu K, McRoberts KS, Meier JA, Theodorescu D, Schwartz MA. Cancer Res. 2009;69:2838–2844. doi: 10.1158/0008-5472.CAN-08-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theodorescu D, Sapinoso LM, Conaway MR, Oxford G, Hampton GM, Frierson HF., Jr Clin Cancer Res. 2004;10:3800–3806. doi: 10.1158/1078-0432.CCR-03-0653. [DOI] [PubMed] [Google Scholar]
- 48.Harding MA, Theodorescu D. Eur J Cancer. 2010;46:1252–1259. doi: 10.1016/j.ejca.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C, Yao J, Chen T, Wan D, Wang H, Gu J, Yao M, Li J, Tu H, He X. Nat Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 50.Koide N, Yamada T, Shibata R, Mori T, Fukuma M, Yamazaki K, Aiura K, Shimazu M, Hirohashi S, Nimura Y, Sakamoto M. Clin Cancer Res. 2006;12:2419–2426. doi: 10.1158/1078-0432.CCR-05-1852. [DOI] [PubMed] [Google Scholar]
- 51.Schubbert S, Shannon K, Bollag G. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 52.Qiu RG, Abo A, McCormick F, Symons M. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y, Bagrodia S, Cerione RA. J Biol Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
- 54.Beeser A, Jaffer ZM, Hofmann C, Chernoff J. J Biol Chem. 2005;280:36609–36615. doi: 10.1074/jbc.M502306200. [DOI] [PubMed] [Google Scholar]
- 55.Appledorn DM, Dao KH, O’Reilly S, Maher VM, McCormick JJ. BMC Cancer. 2010;10:13. doi: 10.1186/1471-2407-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng CM, Li H, Gasman S, Huang J, Schiff R, Chang EC. Mol Cell Biol. 2011;31:983–997. doi: 10.1128/MCB.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu WJ, Tu S, Cerione RA. Cell. 2003;114:715–725. doi: 10.1016/s0092-8674(03)00688-3. [DOI] [PubMed] [Google Scholar]
- 58.Hirsch DS, Shen Y, Wu WJ. Cancer Res. 2006;66:3523–3530. doi: 10.1158/0008-5472.CAN-05-1547. [DOI] [PubMed] [Google Scholar]
- 59.Wu WJ, Erickson JW, Lin R, Cerione RA. Nature. 2000;405:800–804. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- 60.Guo F, Zheng Y. Mol Cell Biol. 2004;24:1426–1438. doi: 10.1128/MCB.24.3.1426-1438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gadea G, Lapasset L, Gauthier-Rouviere C, Roux P. EMBO J. 2002;21:2373–2382. doi: 10.1093/emboj/21.10.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas A, Giesler T, White E. Oncogene. 2000;19:5259–5269. doi: 10.1038/sj.onc.1203895. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Yang L, Debidda M, Witte D, Zheng Y. Proc Natl Acad Sci U S A. 2007;104:1248–1253. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, Wu D. Nat Cell Biol. 2005;7:399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 65.Tang Y, Marwaha S, Rutkowski JL, Tennekoon GI, Phillips PC, Field J. Proc Natl Acad Sci U S A. 1998;95:5139–5144. doi: 10.1073/pnas.95.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yilmaz M, Christofori G. Mol Cancer Res. 2010;8:629–642. doi: 10.1158/1541-7786.MCR-10-0139. [DOI] [PubMed] [Google Scholar]
- 67.Bouzahzah B, Albanese C, Ahmed F, Pixley F, Lisanti MP, Segall JD, Condeelis J, Joyce D, Minden A, Der CJ, Chan A, Symons M, Pestell RG. Mol Med. 2001;7:816–830. [PMC free article] [PubMed] [Google Scholar]
- 68.Chen L, Chan TH, Yuan YF, Hu L, Huang J, Ma S, Wang J, Dong SS, Tang KH, Xie D, Li Y, Guan XY. J Clin Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson E, Seachrist DD, DeLeon-Rodriguez CM, Lozada KL, Miedler J, Abdul-Karim FW, Keri RA. J Biol Chem. 2010;285:29491–29501. doi: 10.1074/jbc.M110.136770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buccione R, Caldieri G, Ayala I. Cancer Metastasis Rev. 2009;28:137–149. doi: 10.1007/s10555-008-9176-1. [DOI] [PubMed] [Google Scholar]
- 71.Fisher KE, Sacharidou A, Stratman AN, Mayo AM, Fisher SB, Mahan RD, Davis MJ, Davis GE. J Cell Sci. 2009;122:4558–4569. doi: 10.1242/jcs.050724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pichot CS, Arvanitis C, Hartig SM, Jensen SA, Bechill J, Marzouk S, Yu J, Frost JA, Corey SJ. Cancer Res. 2010;70:8347–8356. doi: 10.1158/0008-5472.CAN-09-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ayala I, Giacchetti G, Caldieri G, Attanasio F, Mariggio S, Tete S, Polishchuk R, Castronovo V, Buccione R. Cancer Res. 2009;69:747–752. doi: 10.1158/0008-5472.CAN-08-1980. [DOI] [PubMed] [Google Scholar]
- 75.Seiki M. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 76.Poincloux R, Lizarraga F, Chavrier P. J Cell Sci. 2009;122:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- 77.Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D’Souza-Schorey C, Chavrier P. J Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheung LW, Leung PC, Wong AS. Oncogene. 2010;29:2427–2440. doi: 10.1038/onc.2009.523. [DOI] [PubMed] [Google Scholar]
- 79.Noren NK, Liu BP, Burridge K, Kreft B. J Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilkinson S, Paterson HF, Marshall CJ. Nat Cell Biol. 2005;7:255–261. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- 81.Sahai E. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 83.Friedl P, Gilmour D. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 84.Wicki A, Lehembre F, Wick N, Hantusch B, Kerjaschki D, Christofori G. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 85.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, Sahai E. Nat Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 86.van Hengel J, D’Hooge P, Hooghe B, Wu X, Libbrecht L, De Vos R, Quondamatteo F, Klempt M, Brakebusch C, van Roy F. Gastroenterology. 2008;134:781–792. doi: 10.1053/j.gastro.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 87.Cimica V, Batusic D, Chen Y, Hollemann T, Pieler T, Ramadori G. Genomics. 2005;86:352–364. doi: 10.1016/j.ygeno.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Chang CS, Huang SM, Lin HH, Wu CC, Wang CJ. Hepatogastroenterology. 2007;54:2061–2068. [PubMed] [Google Scholar]
- 89.Grise F, Bidaud A, Moreau V. Biochim Biophys Acta. 2009;1795:137–151. doi: 10.1016/j.bbcan.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 90.Yang L, Wang L, Kalfa TA, Cancelas JA, Shang X, Pushkaran S, Mo J, Williams DA, Zheng Y. Blood. 2007;110:3853–3861. doi: 10.1182/blood-2007-03-079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valentijn LJ, Koppen A, van Asperen R, Root HA, Haneveld F, Versteeg R. Cancer Res. 2005;65:3136–3145. doi: 10.1158/0008-5472.CAN-04-2469. [DOI] [PubMed] [Google Scholar]
- 92.Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaffe AB, Kaji N, Durgan J, Hall A. J Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Durgan J, Kaji N, Jin D, Hall A. J Biol Chem. 2011 doi: 10.1074/jbc.M110.174235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kesavan G, Sand FW, Greiner TU, Johansson JK, Kobberup S, Wu X, Brakebusch C, Semb H. Cell. 2009;139:791–801. doi: 10.1016/j.cell.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 96.Chen L, Liao G, Yang L, Campbell K, Nakafuku M, Kuan CY, Zheng Y. Proc Natl Acad Sci U S A. 2006;103:16520–16525. doi: 10.1073/pnas.0603533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Malliri A, Collard JG. Curr Opin Cell Biol. 2003;15:583–589. doi: 10.1016/s0955-0674(03)00098-x. [DOI] [PubMed] [Google Scholar]
- 98.Chu YS, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, Dufour S. J Cell Biol. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuroda S, Fukata M, Nakagawa M, Kaibuchi K. Biochem Biophys Res Commun. 1999;262:1–6. doi: 10.1006/bbrc.1999.1122. [DOI] [PubMed] [Google Scholar]
- 100.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, Shoji I, Matsuura Y, Yonehara S, Kaibuchi K. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 101.Wu X, Quondamatteo F, Lefever T, Czuchra A, Meyer H, Chrostek A, Paus R, Langbein L, Brakebusch C. Genes Dev. 2006;20:571–585. doi: 10.1101/gad.361406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Florian MC, Geiger H. Stem Cells. 2010;28:1623–1629. doi: 10.1002/stem.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 104.Warner SJ, Yashiro H, Longmore GD. Curr Biol. 2010 doi: 10.1016/j.cub.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo L, Liao YJ, Jan LY, Jan YN. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 106.Eaton S, Wepf R, Simons K. J Cell Biol. 1996;135:1277–1289. doi: 10.1083/jcb.135.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kerber RA, O’Brien E, Cawthon RM. Aging Cell. 2009;8:239–250. doi: 10.1111/j.1474-9726.2009.00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen F, Ma L, Parrini MC, Mao X, Lopez M, Wu C, Marks PW, Davidson L, Kwiatkowski DJ, Kirchhausen T, Orkin SH, Rosen FS, Mayer BJ, Kirschner MW, Alt FW. Curr Biol. 2000;10:758–765. doi: 10.1016/s0960-9822(00)00571-6. [DOI] [PubMed] [Google Scholar]