Abstract
DNA (cytosine-5)-methyltransferases (DNMTs) catalyze the transfer of a methyl group from S-adenosyl-L-methionine (AdoMet) to the 5-position of cytosine residues and thereby silence transcription of regulated genes. DNMTs are important epigenetic targets. However, isolated DNMTs are weak catalysts and are difficult to assay. We report an ultrasensitive luciferase-linked continuous assay that converts the S-adenosyl-L-homocysteine product of DNA methylation to a quantifiable luminescent signal. Results with this assay are compared with the commonly used DNA labeling from [methyl-3H]AdoMet. A 5′-methylthioadenosine-adenosylhomocysteine nucleosidase is used to hydrolyze AdoHcy to adenine. Adenine phosphoribosyl transferase converts adenine to AMP and pyruvate orthophosphate dikinase converts AMP to ATP. Firefly luciferase gives a stable luminescent signal that results from continuous AMP recycling to ATP. This assay exhibits a broad dynamic range (0.1–1000 pmol of AdoHcy). The rapid response time permits continuous assays of DNA methylation detected by light output. The assay is suitable for high-throughput screening of chemical libraries with DNMT activity. The kinetic properties of human and bacterial CpG methyltransferases are characterized using this assay. Human catalytic domain DNMT3b activation by DNMT3L is shown to involve two distinct kinetic states that alter kcat but not Km for AdoMet. The assay is shown to be robust in the presence of high concentrations of the pyrimidine analogues 5-azacytidine and 5-azacytosine.
Keywords: DNA methyltransferase, DNMT, CpG islands, luciferase, epigenetics, S-adenosyl-L-homocysteine
AdoMet methyltransferases catalyze the transfer of a methyl group from S-adenosyl-L-methionine (AdoMet) to a methyl acceptor. Biological methyl acceptors include proteins, lipids, carbohydrates, oligonucleotides, and various small molecule metabolites. DNA (cytosine 5)-methyltransferases (DNMTs, EC 2.1.1.37) catalyze the transfer of a methyl group from AdoMet to a DNA substrate. The methyl group is transferred to the 5-position of a cytosine base, usually when the cytosine precedes a guanosine residue at so-called “CpG islands”. Methylation effectively inhibits transcription of the associated gene (1). Although most DNMTs prefer to methylate CpG sites, there is increasing evidence that they can also methylate cytosines at CpA, CpT and CpC sites (2, 3). Thus, DNMTs are an important part of the mechanisms by which gene transcription is controlled in development, differentiation and cancer.
Human cancer cell lines possess hypermethylated CpG islands and it has been postulated that over-methylation decreases the transcription of tumor suppressor genes, promoting cancer proliferation (2). Inhibitors of DNMTs may provide a chemotherapeutic option by reducing CpG hypermethylation, allowing transcription of the tumor suppressor genes and reducing tumor proliferation (3). Development of DNMT biology and inhibitor design would be facilitated by a sensitive assay that can be used to quantitate DNMT activity and be used for inhibitor screens. The in vitro rates of DNA methylation by DNMTs are slow; therefore, the assay must be sensitive and also give stable signals.
Current DNMT assays are discontinuous, using [methyl-3H or methyl-14C]AdoMet followed by separation of the methylated DNA product from unreacted AdoMet. Radioactive AdoMet assays provide high sensitivity but require safety precautions and involve multiple sample handling procedures. Separation of DNA and AdoMet involves selective elution of unreacted AdoMet from a DNA-binding medium (4, 5). High background signals arise from nonspecific binding of AdoMet or AdoMet breakdown products to the medium. Phenol/chloroform extraction followed by DNA precipitation reduces the contaminating proteins and lipids but introduces recovery errors (6, 7). Digestion of DNA with nuclease P1 and alkaline phosphatase followed by HPLC separation of the nucleosides allows specific analysis of labeled 5-methylcytosine but is slow and error-prone (8). Biotinylated oligonucleotide substrates facilitate DNA isolation in avidin-coated 96-well plates (1, 9). However, the use of biotinylated substrates is not always desirable. A scintillation proximity assay avoids separation of AdoMet and DNA since DNA binds to yttrium silicate scintillant beads, but this approach also suffers from nonspecific AdoMet binding (10). Fluorescence-based assays use an anti-AdoHcy antibody in conjunction with AdoHcy bearing a fluorescent tag (11), or use S-adenosyl-L-homocysteine hydrolase (SAHH) to generate homocysteine that reacts with thiol-sensitive fluorophores (12, 13). These assays do not have a large range and require the use of thiol-free assay mixtures or the use of anaerobic conditions (12). Spectrophotometric assays have been developed for other AdoMet-dependent methyltransferases, one using SAHH and Ellman’s reagent under discontinuous anaerobic conditions (14), and another using S-adenosyl-L-homocysteine nucleosidase to generate adenine from AdoHcy hydrolysis which is then converted to hypoxanthine using adenine deaminase (15). These cannot accommodate low AdoMet or DNA concentrations due to low sensitivity and also encounter interfering absorption from DNA substrates. Another method involves LCMS/MS and has been applied to the activity of DNMT1(16).
The luciferase-based luminescent assay described here for AdoMet-dependent methyltransferases can be used under continuous or discontinuous conditions in high-throughput plate formats. This assay evolved from our adenine detection assay developed for ribosome-inactivating proteins, including ricin A-chain (17) and subsequently adapted to protein methyltrasferases (18). The common feature of these luciferase-linked assays is the conversion of adenine to adenosine 5′-monophosphate (AMP) by adenine phosphoribosyl transferase (APRTase, EC 2.4.2.7) using 5-phospho-α-D-ribosyl-1-pyrophosphate (PRPP). AMP is converted to adenosine 5′-triphosphate (ATP) in a single step by pyruvate orthophosphate dikinase (PPDK, EC 2.7.9.1) using phosphoenolpyruvate (PEP) and inorganic pyrophosphate; the resulting ATP reacts with firefly luciferase to generate light (17, 18). This assay has a wide operating range and is sensitive to low adenine concentrations. For DNMT reactions, the product S-adenosyl-L-homocysteine is converted to adenine by recombinant 5′-methylthioadenosine/S-adenosyl-L-homocysteine nucleosidase (MTAN, EC 3.2.2.9). Light produced in this assay is directly proportional to AdoHcy concentration. As AdoHcy is an inhibitor of AdoMet-dependent methyltransferases (19), its removal by MTAN also prevents product inhibition. The continuous nature of this assay eliminates product separation and sample preparation steps. Recombinant catalytic domains of human DNMT3a (CD-DNMT3a) and DNMT3b (CD-DNMT3b) are compared to the bacterial M.SssI CpG methyltransferase and to recombinant human DNMT1. The full-length human DNMT1 is thought to be responsible for maintaining the methylation pattern of the genome during cell division and genome replication (1, 20). DNMT3a and DNMT3b are thought to play roles in de novo methylation of the genome during embryonic development and have also been implicated in cancers (1, 20). We have characterized activation of human DNMT3b by DNMT3L, a catalytically inactive accessory protein that interacts with DNMT3a and DNMT3b to enhance their catalytic activities. Assays in the presence of high concentrations of the pyrimidine analogues 5-azacytidine and 5-azacytosine demonstrate robust responses of this assay for screening chemical libraries.
Results and Discussion
Detection and Quantification of AdoHcy
Recombinant MTAN from S. enterica was used to hydrolyze AdoHcy to give adenine, the purine base converted to ATP in continuous assay buffer. The product ATP is linked to luciferase to generate a light signal and regenerate AMP (Figure 1). AdoHcy (25 μL) solutions were mixed in 96-well microplates with continuous assay buffer (25 μL) containing MTAN concentrations varying from 6 μM to 125 nM and the luminescence measured in the continuous kinetic acquisition mode. In this concentration range, MTAN hydrolyzed AdoHcy solutions to completion in less than 2 min; thus, 125 nM MTAN was used for subsequent assays. MTAN does not hydrolyze AdoMet, thus optimal and/or variable AdoMet concentrations are compatible with this assay.
Figure 1.
Schematic overview of the AdoHcy detection assay. AdoHcy is formed from AdoMet-dependent DNA methylation via DNMT and is converted to adenine using MTAN. The adenine is converted to AMP in an APRTase-catalyzed reaction with PRPP, then to ATP using PPDK with phosphoenolpyruvate. ATP reacts with luciferin and O2 in a firefly luciferase-catalyzed process to generate a stable light signal as the resulting AMP is continuously cycled to ATP.
A response curve was constructed by mixing 25 μL of the 2x continuous assay buffer with 25 μL AdoHcy standard solutions in a 96-well plate (Figure 2A). Luminescence measured in the continuous acquisition mode gave maximum luminescence from the AdoHcy signal within 2 min. Light output as a function of AdoHcy (pmol) gave a linear correlation (Figure 2B). Alternatively, all solutions could be mixed in several wells and the luminescence measured after several minutes of incubation in the discontinuous acquisition mode of the luminometer to construct the same curve (Figure 2B). The AdoHcy standard curve matched that obtained from adenine standard solutions, indicating that the coupling between AdoHcy and adenine is complete in terms of ATP yield (17). The range of this assay is broad as solutions containing 0.1–1000 pmol of AdoHcy in volumes of 50 μL fall within the detection range of the standard curve.
Figure 2.
AdoHcy assay and standard curve: (A) 36 pmol AdoHcy in 25 μL DNMT assay buffer and 25 μL continuous assay buffer measured in the kinetic acquisition mode of the luminometer at room temperature. The signal reached a maximum value within 90 s. (B) AdoHcy standard curve obtained by mixing 25 μL AdoHcy solutions (0.24 to 800 pmol) in DNMT assay buffer with 25 μL continuous assay buffer. After equilibration at room temperature for 3 to 5 min, the continuous luminescence output was measured. The slope of the line is 0.909 ± 0.023, corresponding to a slope 2.5% error in these measurements.
Expression of Human DNMTs
Full-length human DNMT3a and DNMT3b are composed of 912 and 853 amino acids, respectively. The 286 amino acid C-terminal catalytic domains were expressed for kinetic analysis. These domains were modified to contain an N-terminal His6 tag and a thrombin cleavage site. The catalytic domain was identified through sequence alignments of DNMT3a and DMNT3b from humans and mice which gave excellent homology in the C-terminal domains. All sequences contained a cysteine residue at the position for the proposed catalytic nucleophile (eg. 706 in human DNMT3a). The C-terminal regions of human DNMT3a/3b also aligned well with bacterial (cytosine 5)-methyltransferases. The bacterial DNMTs are smaller than mammalian DNMTs and consist primarily of the catalytic domain. The catalytic domains of mouse DNMT3a and DNMT3b have been previously expressed in E. coli (21, 22).
Synthetic gene constructs for CD-DNMT3a and CD-DNMT3b were transformed into strains of E. coli competent cells and it was found that BL21 star (DE3) cells afforded the highest yield of CD-DNMT3a while BL21 (DE3) pLysS cells were best for CD-DNMT3b expression. Expression was best at low temperatures (18 °C, 18 h) as much of the protein was expressed into inclusion bodies at 37 °C and could not be refolded after purification under denaturing conditions. Purification with a room-temperature Ni-NTA agarose column eluted CD-DNMT3a with 500 mM imidazole and CD-DNMT3b with 100 mM EDTA after washing away non-His6-tagged proteins with 100 mM imidazole. Pure fractions were dialyzed into a buffer containing 0.1% DTT as significant protein precipitation occurred during dialysis in the absence of DTT.
DNMT3L was also expressed since this protein has been reported to interact with DNMT3a and DNMT3b in both full-length and catalytic domain constructs and to enhance their catalytic activities (23). Expression of DNMT3L from a synthetic gene construct in E. coli BL21 (DE3) cells gave protein expression after 3 h induction at 37 °C and 2.5-fold increased expression after overnight expression at 18 °C. DNMT3L was purified by elution from a Ni-NTA agarose column with an imidazole gradient.
Human DNMT1 was also expressed. It is known to be significantly more active than DNMT3a, DNMT3b or their mixtures with DNMT3L. As the catalytic domain of DNMT1 has been reported to be inactive (24), the full-length 1620 amino acid construct was expressed in SF9 cells using a baculovirus-based expression system. Purification of DNMT1 was also achieved by elution from a Ni-NTA agarose column with an imidazole gradient.
DNMT Activity in Continuous Assays
The use of HPLC-purified AdoMet improved the accuracy and reproducibility of the AdoHcy production assays. AdoMet is unstable at neutral and alkaline pH and degrades via an intramolecular reaction giving 5′-methylthioadenosine (MTA) and homoserine lactone and a hydrolytic reaction giving adenine and S-pentosylmethionine (25). MTA is a substrate for MTAN and is hydrolyzed to give adenine. Thus, both breakdown pathways produce intermediates that contribute to the background in the luciferase-linked DNMT assay. Commercial AdoMet sources contain up to 10 % MTA and/or adenine. HPLC purification reduced these to below 0.5 % and can be efficiently performed on large scales using standard HPLC equipment. The purified AdoMet can be stored at −80 °C for > 6 months without loss.
AdoMet background decomposition rate for initial rate measurements is accomplished by incubating the complete reaction mixture without DNMTs for 3 min while measuring the background luminescence. This approach gives a stable linear background signal to be subtracted from the rate following DNMT addition (see ordinate intercepts in Figure 3A). The DNMT reaction is initiated by the addition of the enzyme followed by a 3 min incubation and 3 min measurement of luminescence. The order of DNA and DNMT addition can be reversed to allow background measurement of the AdoMet and DNMT solutions. The 3 min incubation after reaction initiation eliminates a lag phase prior to formation of a linear signal. The lag phase is attributed to enzyme-DNA binding and conformational changes since pre-incubating DNMTs with the DNA substrate shortens or eliminates the lag phase. Increasing the amounts of coupling enzymes and/or the amount of luciferase coupling agents (ATPlite) does not affect the lag phase. The reproducibility of continuous assay rates (multiple repeats of the same assay conditions) was ± 10% of the mean.
Figure 3.
Continuous assay measurements for initial rate reactions and for substrate saturation of M.SssI CpG methyltransferase. (A) Initial rates of the M.SssI CpG methyltransferase reaction with increasing AdoMet concentrations. (B) Initial reaction rates fit to theMichaelis-Menten equation for M.SssI CpG methyltransferase (45 nM) and 1 μg poly(dIdC) at room temperature. The standard errors of 9.1% for Km and 1.6% for kcat in this traditional steady-state kinetics assay are in agreement with published values for direct spectroscopic recording assays.
Bacterial M.SssI CpG methyltransferase is a well-characterized DNA methyltransferase and was also used to benchmark the assay. The commonly used synthetic oligonucleotide poly(2′-deoxyinosinic-2′-deoxycytidylic acid) (poly(dIdC)) was selected as the DNA substrate. It is reported to have high activity with DNMTs (26). Initial reaction rates were measured with a constant amount of poly(dIdC) while AdoMet concentration was varied in order to measure its Km (Figure 3B). Conversion of AdoHcy to a luminescent signal has the added benefit of removing the reaction product as AdoHcy is known to inhibit DNMTs. Reaction rates with 1 μg poly(dIdC) and 4 U M.SssI CpG methyltransferase (45 nM, in 50 μL reaction mixtures) were plotted against AdoMet concentration and fit to the Michaelis-Menten equation (Figure 3B). This gave a Km for AdoMet of 3.5 ± 0.32 μM and a kcat of 179 ± 2.8 h−1, consistent with reported values (Figure 3B, Table 1) (6). Thus, the error in rate measurement (kcat) in a practical kinetic assay of the method is ± 1.6%.
Table 1.
Kinetic parameters for DNMTs in the luciferase-coupled continuous assay.
| Enzyme | KmAdoMet (nM)a | KmDNA (nM)b | Vmax (pmol/h) | kcat (h−1) |
|---|---|---|---|---|
| M.SssI | 3500 ± 320 | N/Dc | 404 ± 6 | 179 ± 3 |
| DNMT1 | 2660 ± 750 | 837 ± 404 | 726 ± 75 | 28.1 ± 2.9 |
| CD-DNMT3a-DNMT3L | < 50 | 19 ± 36d | 9.5 ± 0.6 | 0.39 ± 0.02 |
| CD-DNMT3b-DNMT3L | 69 ± 40 | 329 ± 58 | 24.6 ± 1.6 | 1.00 ± 0.07 |
Near saturating poly(dIdC) concentration. dIdC base pair concentration = 16.4 μM. All assays were conducted at room temperature of approximately 22 °C.
Near saturating AdoMet concentration used: 35 μM for DNMT1, 10 μM for CD-DNMT3a and CD-DNMT3b in complexes with DNMT3L.
Not done.
Two factors contribute to the error; the lowest practical poly(dIdC) concentration (82 nM) is high relative to the Km value, and the high enzyme/substrates ratio requires fitting to the Morrison equation (see text) where the propagation of errors from equation complexity give rise to the large data fitting error.
In our hands, CD-DNMT3a and CD-DNMT3b alone gave a catalytic activity of < 0.50 pmol/hr, near the lower sensitivity of this assay and similar to most literature reports(27). The activities of the CD-DNMT3a and CD-DNMT3b constructs increased in the presence of 1:1 molar ratios of DNMT3L when the complexes were pre-incubated for 40 min at room temperature. Convenient enzyme concentrations were explored by measuring reaction rates at 10 μM AdoMet and 0.5 μg poly(dIdC) while varying the concentration of CD-DNMT3b + DNMT3L in 50 μL assay mixtures. Enzyme concentration over the range of 0.2–4.0 μM gave initial rates proportional to enzyme concentration. Michaelis-Menten curves with varying AdoMet and poly(dIdC) concentrations were analyzed with 500 nM CD-DNMT3 and DNMT3L (Figures 4A and 4B, respectively). The dIdC concentration of poly(dIdC) was calculated based on an average polymer length of 2100 bp (1200–3000 bp range) giving an average double-stranded molecular weight of 2.57 × 106 g/mol and a dIdC base pair concentration of 0.82 nmol dIdC/μg poly(dIdC) (7.8 nM DNA polymer in the assay). The Michaelis-Menten equation cannot be used to analyze the kinetic constants since enzyme (0.2 to 4.0 μM) greatly exceeds substrate concentration (7.8 nM (poly(dIdC)). When the amount of enzyme used is significant relative to the concentration of substrate, corrections can be made by fitting to the Morrison equation which includes terms for substrate depletion (28):
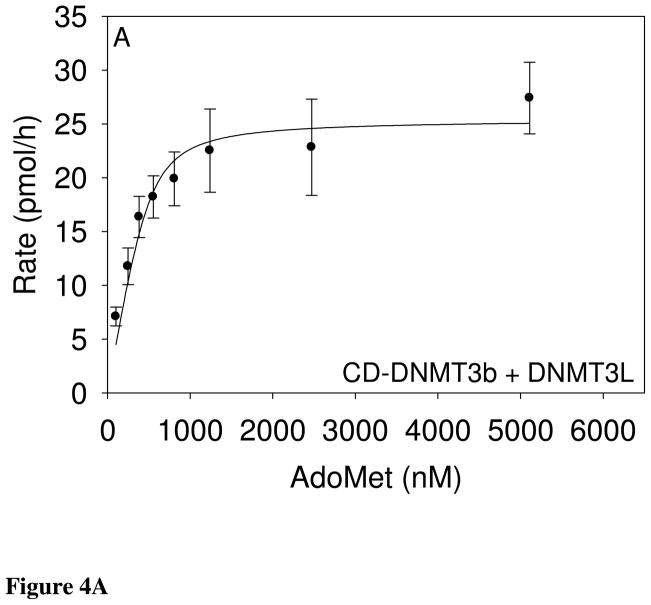
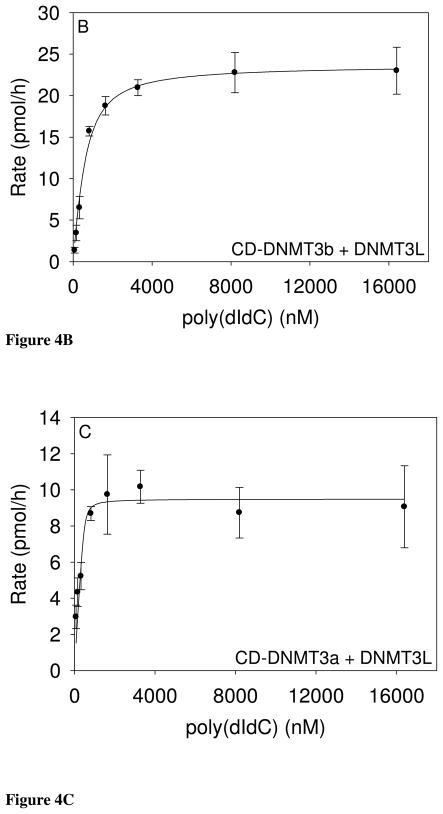
Figure 4.
Continuous assay measurements on human DNMTs at room temperature. Initial rates fit to the Morrison equation for significant depletion of substrate by enzyme: (A) 1 μg poly(dIdC), 492 nM CD-DNMT3b-DNMT3L, varied AdoMet concentration. (B) 10 μM AdoMet, 492 nM CD-DNMT3b-DNMT3L, varied poly(dIdC) concentration. (C) 10 μM AdoMet, 490 nM CD-DNMT3a-DNMT3L, varied poly(dIdC) concentration. The error bars represent the standard deviation of 2 or 3 individual rate measurements for each rate measurement.
The equation accounts for substrate bound to the enzyme, allowing valid analysis of kinetic constants. These fits gave an AdoMet Km of 69 ± 40 nM, a poly(dIdC) Km of 330 ± 58 nM, and kcat of 1.0 ± 0.07 h−1 (Table 1). For CD-DNMT3a with DNMT3L (490 nM) and 1 μg poly(dIdC), the reaction is saturated with AdoMet concentrations above 100 nM. Initial rates with decreasing AdoMet concentrations, indicated a Km value of less than 50 nM. These data fit well to a two-term Morrison equation containing two Km and two Vmax terms, suggesting that AdoMet concentration may also alter the CD-DNMT3a-DNMT3L interaction in addition to catalytic site saturation.
Data obtained with 490 nM CD-DNMT3a-DNMT3L, 10 μM AdoMet, and varying poly(dIdC) concentrations were fit to the Morrison equation to give a poly(dIdC) Km of 19 ± 36 nM and a kcat of 0.39 ± 0.02 h−1 (Figure 4C; Table 1). The relatively large error in the Km value results from propagation of errors from the repeated terms in the Morrison equation. [When the same data is inappropriately fitted to the Michaelis Menten equation, an apparent (but incorrect) Km of 201 ± 50 nM is obtained.] The enzyme complexes, CD-DNMT3a-DNMT3L and CD-DNMT3b-DNMT3L, have substantially lower Km values than the bacterial M.SssI CpG methyltransferase. The AdoMet Km for CD-DNMT3a-DNMT3L is more than 70-fold lower than that for M.SssI CpG methyltransferase. Both human CD-DNMT3-DNMT3L mixtures have kcat values two to three orders of magnitude lower than that for the M.SssI CpG methyltransferase.
Full-length human DNMT1 prefers hemimethylated DNA as a substrate in which one of the two strands contains methylated cytosine, however, it is more active on unmethylated poly(dIdC) DNA than CD-DNMT3a or CD-DNMT3b in complex with DNMT3L (26). DNMT1 (516 nM) initial rate data were fit to the Morrison equation to give a AdoMet Km of 2.7 ± 0.7 μM, a poly(dIdC) Km of 840 ± 400 nM, and a kcat of 28 ± 3 h−1 (Table 1). These values are similar to those for the bacterial M.SssI CpG methyltransferase, although its kcat value is 6.5 times greater than that for human DNMT1, consistent with reported values (6, 29).
The results with several DMNTs demonstrate the effectiveness of this luciferase-linked assay for characterizing DNA methyltransferase reactions. Measurement of highly- and less-active DNA methyltransferases is facilitated by the broad dynamic range of the assay. Finally, product inhibition by AdoHcy is prevented by its continual removal.
DNMT1 Activity Using Discontinuous Assays
Discontinuous assays have applications in endpoint assays and inhibitor screens. They are also suited to the measurement of kinetic data under experimental conditions not suited to luminometer reaction conditions.
Initial reaction rates for human DNMT1 were measured at room temperature and at 37 °C. Assays used 300 μL reaction mixtures containing 682 nM AdoMet, 6 μg poly(dIdC), and 258 nM DNMT1. Aliquots were removed every two min (2–10 min) and were quenched with 100 mM HCl. Aliquots from a no-enzyme control were treated in the same manner. The aliquots were neutralized with 100 mM KOH, mixed with one volume of 2x AdoHcy to ATP conversion buffer and incubated at room temperature for ~3 min. Aliquots of these samples were mixed with four volumes of ATPlite in a 96-well plate format to convert the ATP into a luminescent signal. After incubation at room temperature for ~3 min, the luminescence of each sample was measured and an initial rate plot was constructed (Figure 5A). Initial rates measured in RLU/s were converted to units of pmol/h with an AdoHcy standard curve constructed under the same assay conditions (Table 2). The rate at 37 °C was greater than that at room temperature, 7.5 vs 6.3 pmol/h, respectively. These rates are lower than the 30 pmol/h obtained from an identical reaction performed using the continuous luciferase-linked assay, thus the components of the continuous assay favor the methyltransfer reaction. The discontinuous rates were readily measured even at low AdoMet concentrations, and the assay is suitable for automated screening. Initial rate plots (eg. Figure 5A) could be constructed under various substrate concentrations using the luciferase-linked assay under discontinuous conditions in order to obtain kinetic data as in Figure 4.
Figure 5.
Initial rates for 258 nM DNMT1, 6 μg poly(dIdC), 300 μL total volume at room temperature and at 37 °C measured by discontinuous assay: (A) 682 nM AdoMet, luciferase-linked AdoHcy assay. (B) 672 nM [methyl-3H]-AdoMet, radiometric filter paper assay.
Table 2.
Initial rates of DNMT1 methylation of poly(dIdC) in luciferase-coupled (using AdoMet) or radiometric (using [methyl-3H]-AdoMet) assays.
| Assaya | Rate (pmol/h) |
|---|---|
| Discontinuous Luciferase 22 °C | 6.3 ± 1.2 |
| Discontinuous Luciferase 37 °C | 7.5 ± 0.8 |
| Radiometric 22 °C | 11.6 ± 0.7 |
| Radiometric 37 °C | 13.0 ± 0.4 |
DNMT1 was present at 258 nM in the presence of 0.2 μg/μL poly(dIdC). AdoMet was present at 682 nM for luciferase-linked assays and at 672 nM for radiometric assays.
Parallel reactions were performed with 672 nM [methyl-3H]AdoMet to compare the initial rates from the luciferase assay with those obtained from the radiometric filter paper assay (4, 5). The luciferase-linked assays were performed at the same AdoMet concentration. Aliquots were sampled at 2-min intervals (2–8 min) and spotted on 2.3 cm diameter Whatman DE81 filter paper circles. After drying (30 min), the circles were washed with 0.2 M NH4HCO3, water, and ethanol. The dried circles were analyzed for radioactivity in a scintillation counter. Background-correction for a no-enzyme control gave the reaction rates (Figure 5B). Initial rates in were converted to pmol/h using [methyl-3H]AdoMet standards (Table 2). Initial rates of 11.6 pmol/h at 22 °C and 13.0 pmol/h at 37 °C are similar to those obtained in the luciferase assays.
Effects of DNMT3L on CD-DNMT3b activity
The kinetic pattern for DNMT3L activation of DNMTs has not been resolved. One report using a truncated version of DNMT3a showed that maximum stimulation of DNMT activity occurred when equimolar amounts of DNMT3a and DNMT3L were preincubated for 5 min or 2 hr (5). Another report showed increased stimulation with DNMT3L concentrations to 70-fold excess with mouse CD-DNMT3a and to 20-fold excess with mouse CD-DNMT3b by examining initial rates under a single set of non-saturating substrate concentrations (23). A third report showed full stimulation of full-length mouse DNMT3a and DNMT3b with a 10-fold excess of DNMT3L (3). Most recently, the processivity of full-length human DNMT3a was shown to be increased in a 1:1 ratio with DNMT3L (30). Here we provide a titration of CD-DNMT3b with DNMT3L to demonstrate the utility of the luciferase assay system in kinetic activation experiments.
A 1:1 stoichiometric mixture of DNMT3L and CD-DNMT3a or CD-DNMT3b was used in the work reported above. Kinetic plots for AdoMet substrate saturation with near-saturating DNA concentrations and varied ratios of DNMT3L:CD-DNMT3b are useful to assist in resolution of mechanism of activation questions.
With ratios of 0.25 to 20, DNMT3L was incubated with 500 nM CD-DNMT3b protein and the results used to construct kinetic plots for activation and AdoMet saturation (Fig. 6). Data were fit to the Morrison equation, giving Km values that did not change but Vmax values that increased with increasing DNMT3L ratios (Figure 7). This change in Vmax was biphasic, increasing in a near-linear response at low DNMT3L concentrations and with continued activation at higher ratios, but with a lower response rate. The co-crystal structure has shown that the CD-DNMT3a/DNMT3L complex exists as a heterotetramer in which two CD-DNMT3a-DNMT3L complexes interact at the CD-DNMT3a/CD-DNMT3a interface (22). Although this kinetic patternof Figure 7 is possible from a number of different protein association models, the data fit well to the equation for two kinetically significant complexes with Km1 = 160 ± 5 nM and Vmax1 = 30 ± 5 pmol/h for the first complex. The second complex was not sufficiently saturated to give quantitative kinetic constants in this experiment but visual inspection suggests a Km2 above 9 μM and a Vmax2 above 100 pmol/h.
Figure 6.
Michaelis-Menten curves for AdoMet binding at 1 μg poly(dIdC), 500 nM CD-DNMT3b with varying DNMT3L concentrations: (A) 124 nM DNMT3L. (B) 252 nM DNMT3L. (C) 492 nM DNMT3L. (D) 996 nM DNMT3L. (E) 4940 nM DNMT3L. (F) 9800 nM DNMT3L. Data are fit to the Morrison equation to account for significant enzyme concentrations. Maximum catalytic rates are shown in Figure 7.
Figure 7.
Increasing DNMT3L concentration increases activity of CD-DNMT3b in a biphasic manner. Vmax values calculated from Michaelis-Menten curves for AdoMet binding with 1 μg poly(dIdC). The data was fitted to the equation for two independent saturable sites for DNMT3L.
5-Azacytidine and 5-azacytosine effects
The luciferase-based DNMT assays reported here have been developed in 96-well plates and are well-suited to larger high-throughput formats. We explored the sensitivity of this assay to interference by inhibitor fragments using two pyrimidine base analogues. Inhibition of DNA methylation in vivo is caused by 5-azacytidine and 5-azacytosine. The mechanism is known to involve metabolism to the deoxynucleoside triphosphates followed by incorporation into DNA and covalent inactivation of the DNMT enzymes 31. However, it is also possible that direct inhibition of DNMT activity could occur in the presence of the inhibitors.
The 5-azapyrimidines were tested for inhibition of the CD-DNMT3b-DNMT3L complex with DNMT3L in 1-fold excess to CD-DNMT3b. Under assay conditions with up to 1.28 mM of the 5-azapyrimidines (2.6 × 106-fold molar excess), neither showed inhibition. Free 5-azapyrimidines are unlikely to be involved in the mechanism of action of these agents. The robust action of the luciferase assay even in the presence of millimolar pyrimidine analogues supports the use of this assay for the purpose of screening chemical libraries.
Conclusions
A light-based continuous detection assay has been developed for the detection of AdoHcy released from DNA methyltransferase reactions. This assay uses an MTAN that hydrolyzes AdoHcy to give adenine and S-ribosylhomocysteine but does not interact with AdoMet. Adenine is converted to ATP using APRTase and PPDK. ATP is quantitated by a luciferase-catalyzed reaction with luciferin. This reaction produces a stable luminescent signal since the AMP product is cycled to ATP to sustain the signal. This assay has the added benefit of removing AdoHcy from reaction mixtures to prevent product inhibition. The assay is capable of detecting sub-picomole quantities of AdoHcy and has a broad dynamic range from 0.1 to 1000 pmol of AdoHcy. This continuous assay is a unique development for AdoMet-dependent DNA-methyltransferases because of the sensitivity, dynamic range and link to light production with recycling of AMP for stable light signal. The range of applications is broadened by use under both continuous and discontinuous conditions, as needed for chemical library screening and hit validation.
The utility of the assay is demonstrated by the ability to observe distinct kinetic parameters for the activation of CD-DNMT3b at low and high concentrations of DNMT3L. Examples of the assay in inhibitor screening are provided by appropriate catalytic responses in the presence of 5-azacytidine and 5-azacytosine at concentrations in excess of 1 millimolar.
Materials and Methods
M.SssI CpG methyltransferase and AdoMet were purchased from New England Biolabs (Ipswich, Massachusetts). AdoMet was further purified by reverse-phase HPLC using a Waters C18 column (4.6 × 250 mm), 20% acetonitrile at 0.5 mL/min. Poly(2′-deoxyinosinic-2′-deoxycytidylic acid) was purchased from Sigma (Ashland, Massachusetts; Cat. No. P4929). Firefly luciferase ATP assay kit (ATPlite) and Betaplate Scint scintillation fluid were purchased from Perkin-Elmer, as was S-adenosyl-L-[methyl-3H]methionine with a specific activity of 70.8 Ci/mmol (Waltham, Massachusetts). EDTA-free Complete protease inhibitor cocktail tablets and PhosSTOP phosphatase inhibitor tablets were purchased from Roche Applied Science (Indianapolis, Indiana). Recombinant S. cerevisiae APRTase bearing an N-terminal His6 tag and recombinant Clostridium symbiosum PPDK were prepared as previously described (16). Synthetic DNA constructs coding for DNMTs and MTAN were purchased from DNA 2.0 (Menlo Park, California). The Gateway LR Clonase II enzyme mix was purchased from Invitrogen (Carlsbad, California). Plasmid DNA was isolated and purified using the Qiagen HiSpeed plasmid midi kit (Valencia, California). Ni-NTA agarose was purchased from Qiagen. Corning 96-well non-binding surface microplates were purchased through Fisher Scientific (Pittsburgh, Pennsylvania). Protein concentrations were measured by Bradford assay in comparison with bovine serum albumin (BSA) standards obtained from Bio-Rad (Hercules, California). All other reagents used were from Fisher Scientific or Sigma-Aldrich and were the highest quality available. Luminescence was measured using a Glomax 96-well luminometer from Promega (Madison, Wisconsin). Scintillation counting was performed using a TriCarb 2910TR Liquid Scintillation Counter from Perkin Elmer.
Expression and Purification of Human DNMT1
Human DNMT1 expression constructs containing an N-terminal His6 tag and an rTEV protease cleavage site were subcloned from the pFastBac HTa plasmid (a generous gift of Prof. Keith D. Robertson, University of Florida). Polymerase chain reaction of human DNMT1 cDNA was used to incorporate an SpeI restriction endonuclease site at the 3′ end (sense primer: 5′-CCAACTCGGTCCGAAACCATGTCGTACTACC -3′ and antisense primer: 5′-GGCTCACTAGTTGTACTTCTCGACAAGCTTGGTACCGC -3′. The restriction sites are underlined). This PCR product was digested with RsrII and SpeI and ligated downstream of the polyhedrin promoter of a pFastDual vector that contains green fluorescent protein cDNA downstream of the p10 promoter (the generous gift of Dr. Kartik Chandran, Department of Microbiology and Immunology, Albert Einstein College of Medicine, (32)). The ligation of human DNMT1 was verified by DNA sequencing and contains an I311V mutation in human DNMT1 isoform b (GenBank Identifier: 4503351). A complete description of the expression and purification of DNMT1 is provided in the Supplementary Information.
Expression and Purification of Human DNMT3a Catalytic Domain (CD-DNMT3a)
The catalytic domain of human DNMT3a was identified by sequence alignment with bacterial cytosine methyltransferases that consist mainly of catalytic domains as well as through previous reports of the mouse DNMT catalytic domains (21). A construct coding for a 286 amino acid C-terminal domain of human DNMT3a (residues 627–912 of human DNMT3a, NCBI accession number Q9Y6K1) containing an N-terminal His6 tag with a thrombin cleavage site was purchased from DNA 2.0 in a pDONR221 Gateway donor vector (Invitrogen). A complete description of the expression and purification of CD-DNMT3a is provided in the Supplementary Information.
Expression and Purification of Human DNMT3b Catalytic Domain (CD-DNMT3b)
The catalytic domain for human DNMT3b was identified as described for CD-DNMT3a. A construct coding for a 286 amino acid C-terminal domain of human DNMT3b (residues 568–853 of human DNMT3b, NCBI accession number Q9UBC3) containing an N-terminal His6 tag with a thrombin cleavage site was purchased from DNA 2.0 in a pDONR221 Gateway donor vector (Invitrogen). A complete description of the expression and purification of CD-DNMT3b is provided in the Supplementary Information.
Expression and Purification of Human DNMT3L
A construct coding for the full-length human DNMT3L (NCBI accession number NP-037501) containing an N-terminal His6 tag with a thrombin cleavage site was purchased from DNA 2.0 in a pDONR221 Gateway donor vector (Invitrogen). A complete description of the expression and purification of DNMT3L is provided in the Supplementary Information.
Expression and Purification of S. enterica MTAN
The gene encoding MTAN in Salmonella enterica was synthesized and cloned into pDONR221 vector by DNA 2.0 (Menlo Park, CA), incorporating an N-terminal His6 tag sequence and terminal attB sequences. The gene was swapped into pDEST14 vector (ampR) using Invitrogen Gateway Cloning Technology (Carlsbad, CA), and successful clones were transformed into BL21 (DE3) competent cells. Several colonies were screened for protein expression under IPTG induction, and glycerol stocks were prepared from the best expression clones. For protein expression, LB medium was seeded with an overnight culture of BL21 (DE3) grown in the presence of ampicillin (100 μg/mL), and incubated at 37 °C with shaking at 250 rpm. Induction was initiated at A600 = 0.6 with 0.5 mM IPTG for 4 h. Cells were harvested by centrifugation at 4000 rpm for 20 min and resuspended in lysis buffer (40 mM TEA-HCl pH 7.8, 300 mM NaCl, 0.2 mg/mL lysozyme, 0.1 mg/mL DNaseI, protease inhibitors). Cells were lysed with three rounds of sonication for 15 s each round with 1 min cooling time between bursts. Cell debris was removed by centrifugation at 16,000 rpm for 30 min, and the cleared supernatant was loaded on a 5 mL Ni-NTA column equilibrated in lysis buffer. The column was washed with 5 column volumes of 10 mM imidazole in running buffer (20 mM TEA-HCl pH 7.8, 300 mM NaCl). Elution was done using a step gradient of 25 mM imidazole in running buffer with 250 mM imidazole in the final fraction. Fractions were analyzed by SDS-PAGE and pertinent fractions were pooled together and dialyzed overnight in 20 mM TEA-HCl pH 7.8, 50 mM NaCl. Glycerol was added to 10% of the volume prior to storage at −80 °C.
AdoHcy to ATP Conversion Buffer
The 2x AdoHcy to ATP conversion buffer was prepared as previously described with minor modifications (16). In brief, 100 mL of buffer contained 100 mM Tris-acetate pH 7.7, 2 mM PEP, 2 mM sodium pyrophosphate, 2 mM PRPP, 15 mM (NH4)2SO4, 15 mM (NH4)2MoO4, and phosphatase inhibitors (Roche PhosSTOP, 10 tablets). A 4:1 cellulose:charcoal mixture was suspended in water and packed into a disposable mini-column to a bed volume of 1.5 mL, through which a 10 mL aliquot of the previously prepared buffer was passed at 2 mL/min to remove traces of adenine, AMP, and ATP. Treated buffer was filtered (0.2 μm) and stored in 1 mL aliquots at −80 °C. Immediately prior to use, the 2x conversion buffer was completed by additions to give final concentrations of 10 mM MgSO4, 6 U APRTase, 18 U PPDK, and 250 nM MTAN and an increase in volume of 14 μL.
DNMT Kinetics using Continuous Luciferase Assay
Continuous assay buffer (2x) was prepared by adding 200 μL luciferin/luciferase (ATPlite) to 1 mL of the AdoHcy to ATP conversion buffer and warmed to room temperature. To 25 μL of this mixture was added AdoMet and the DNA substrate in DNMT assay buffer (25 mM HEPES pH 7.6, 50 mM KCl, 5 % glycerol, 0.1 % DTT) in a 96-well non-binding surface microplate. The mixture was incubated at room temperature for 3 min to permit any contaminating adenine, AMP, ATP or 5′-methylthioadenosine (MTA) to be converted to a stable background light signal as measured in the kinetic acquisition mode of the luminometer for 3 min. DNMT was added to give a final volume of 50 μL. After 3 min, the luminescent signal was measured for the following 3 min. Reactions with the less active DNMT3L-CD-DNMT3a and DNMT3L-CD-DNMT3b complexes were performed in triplicate. Initial rates were calculated by subtracting the background rate from that obtained after DNMT addition. Initial rates were fitted to the Morrison equation for tight binding substrates with the exception of data obtained while using M.SssI CpG methyltransferase which were fit to the Michaelis-Menten equation. The Morrison equation was used as high enzyme concentrations were necessary and it could no longer be assumed that free substrate concentration was equal to total substrate concentration. The kinetic parameters Km and Vmax were obtained from these fits after converting the luminescent rate (RLU/s) to an enzymatic rate (pmol AdoHcy/h) using the AdoHcy standard curve. The AdoHcy standard curve was constructed by mixing 25 μL of DNMT assay buffer containing varied concentrations of AdoHcy with 25 μL of continuous assay buffer, incubating for several minutes, and then measuring the luminescence of each sample in the discontinuous acquisition mode of the luminometer. Values were background corrected using a mixture containing no AdoHcy.
DNMT1 Rate Measurement using Discontinuous Luciferase Assay
DNMT1 was added to a solution of 682 nM AdoMet and 1 μg poly(dIdC) in DNMT assay buffer to a concentration of 258 nM in 300 μL total volume which had been equilibrated to 22 °C or 37 °C. A control reaction containing no DNMT1 was also performed at 37 °C. 15 μL aliquots of the reaction were quenched with 5 μL of 100 mM HCl at 2 min intervals from 2 to 10 min in triplicate. These were neutralized with 5 μL of 100 mM KOH and were mixed with 25 μL of 2x AdoHcy to ATP conversion buffer and incubated at room temperature for several minutes. 20 μL of each mixture were then mixed with 80 μL of ATPlite in a 96-well non-binding surface microplate. These mixtures were incubated at room temperature for several minutes and their luminescence was measured in the single measurement mode of the luminometer. The luminescent signal (RLU) was converted to pmol AdoHcy by constructing an AdoHcy standard curve in which 15 μL aliquots of AdoHcy solutions with varied concentrations in DNMT assay buffer were treated as above. Values were background corrected with a mixture containing no AdoHcy.
DNMT1 Rate Measurement using Radiometric Assay
The same two reactions described above for the discontinuous luciferase assay were also conducted with the radiolabel assay. In that case, 672 nM S-adenosyl-L-[methyl-3H]methionine was used in place of non-radioactive AdoMet. Aliquots (15 μL) were removed from each reaction in triplicate at 2 min intervals from 2 to 8 min and spotted on 2.3 cm diameter Whatman DE81 filter paper circles which were allowed to dry for 30 min. The circles were washed with 0.2 M NH4HCO3 (4 × 2 mL), water (4 × 2 mL), and absolute ethanol (1 × 2 mL); each wash lasted 5 min before decanting the liquid. Circles were then dried for 1 h, then placed in scintillation vials and covered with 10 mL Betaplate Scint scintillation fluid. Radioactivity was measured on a Tricarb scintillation counter and background was corrected with a no-enzyme control. A conversion factor of 12 CPM/fmol was used.
Supplementary Material
Acknowledgments
This work was supported by NIH Research Grant CA135405 and an NSERC Post-Doctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada to I.H. (PDF-434344-2007).
References
- 1.Gowher H, Jeltsch A. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 2.Grandjean V, Yaman R, Cuzin F, Rassoulzadegan M. PLoS ONE. 2007;2:e1136. doi: 10.1371/journal.pone.0001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suetaki I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 4.Datta J, Ghoshal K, Denny WA, Gamage SA, Brooke DG, Phiasivongsa P, Redkar S, Jacob ST. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. J Biol Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 6.Lee WJ, Zhu BT. Carcinogenesis. 2006;27:269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 7.Morey SR, Smiraglia DJ, James SR, Yu J, Moser MT, Foster BA, Karpf AR. Cancer Res. 2006;66:11659–11667. doi: 10.1158/0008-5472.CAN-06-1937. [DOI] [PubMed] [Google Scholar]
- 8.Hogarth LA, Redfern CP, Teodoridis JM, Hall AG, Anderson H, Case MC, Coulthard SA. Biochem Pharmacol. 2008;76:1024–1035. doi: 10.1016/j.bcp.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Roth M, Jeltsch A. Biol Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]
- 10.Baker MR, Zarubica T, Wright HT, Rife JP. Nucleic Acids Res. 2009;37:e32. doi: 10.1093/nar/gkn1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves TL, Zhang Y, Scott JE. Anal Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Leffler S, Thompson DH, Hárycyna CA. Biochem Biophys Res Commun. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 13.Collazo E, Couture J-F, Bulfer S, Trievel RC. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. Anal Biochem. 2004;326:100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Wang Y. Biochemistry. 2009;48:2290–2299. doi: 10.1021/bi801467z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturm MB, Schramm VL. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibáñex G, McBean JL, Astudillo YM, Luo M. Anal Bioichem. 2010;401:203–210. doi: 10.1016/j.ab.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Bacolla A, Pradhan S, Roberts RJ, Wells RD. J Biol Chem. 1999;274:33011–33019. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 20.Jurkowska RZ, Jurkowski TP, Jeltsch A. Chembiochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 21.Gowher H, Jeltsch A. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 22.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 24.Fatemi M, Hermann A, Pradhan S, Jeltsch A. J Mol Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman JL. Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- 26.Svedruzic ZM, Reich NO. Biochemistry. 2005;44:9472–9485. doi: 10.1021/bi050295z. [DOI] [PubMed] [Google Scholar]
- 27.Jurkowska RZ, Siddique AN, Jurkowski TP, Jeltsch A. ChemBioChem. 2011 doi: 10.1002/cbic.20100673. in press. [DOI] [PubMed] [Google Scholar]
- 28.Morrison JF, Walsh CT. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 29.Flynn J, Glickman JF, Reich NO. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 30.Holz-Schietinger C, Reich NO. J Biol Chem. 2010;285:29091–29100. doi: 10.1074/jbc.M110.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champion C, Guianvarc’h D, Sénamaud-Beaufort C, Jurkowska RZ, Jeltsch A, Ponger L, Arimondo PB, Guieysse-Peugeot AL. PLoS ONE. 2010;5:e12388. doi: 10.1371/journal.pone.0012388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Rahmeh A, Morelli M, Whelan SPJ. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.