SUMMARY
Definitive identification of Treponema pallidum (Tp) rare outer membrane proteins (OMPs) has long eluded researchers. TP0326, the sole protein in Tp with sequence homology to a Gram-negative OMP, belongs to the BamA family of proteins essential for OM biogenesis. Structural modeling predicted that five polypeptide transport-associated (POTRA) domains comprise the N-terminus of TP0326, while the C-terminus forms an 18-stranded amphipathic β-barrel. Circular dichroism, heat-modifiability by SDS-PAGE, Triton X-114 phase partitioning and liposome incorporation supported these topological predictions and confirmed that the β-barrel is responsible for the native protein's amphiphilicity. Expression analyses revealed that native TP0326 is expressed at low abundance, while a protease-surface accessibility assay confirmed surface exposure. Size-exclusion chromatography and blue native polyacrylamide gel electrophoresis revealed a modular Bam complex in Tp considerably larger than that of E. coli. Non-orthologous ancillary factors and self-association of TP0326 via its β-barrel may both contribute to the Bam complex. Tp-infected rabbits mount a vigorous antibody response to both POTRA and β-barrel portions of TP0326, whereas humans with secondary syphilis respond predominantly to POTRA. The syphilis spirochete appears to have devised a stratagem for harnessing the Bam pathway while satisfying its need to limit surface antigenicity.
Keywords: Treponema pallidum, Syphilis, BamA, Outer Membrane Protein, POTRA, β Barrel
INTRODUCTION
The cell envelope of Gram-negative bacteria is composed of outer and inner membranes (OM and IM, respectively) which demarcate the intervening periplasmic space and peptidoglycan (PG) layer (Silhavy et al., 2010). While the IM is a phospholipid bilayer composed entirely of glycerophospholipids, the prototypical OM is an asymmetrical bilayer with lipopolysaccharide (LPS) in the outer leaflet and glycerophospholipids in the inner. The general topologies of IM and OM integral membrane proteins differ markedly. Integral membrane proteins span the IM via one or more hydrophobic α-helices, whereas OM-spanning proteins generally are β-barrels consisting of anti-parallel amphipathic β-strands (Schulz, 2002; Wimley, 2003). The OM functions as a permeability barrier, protecting the bacterium against noxious environmental compounds, such as antibiotics and bile salts (Nikaido, 2003; Delcour, 2009), its numerous β-barrel outer membrane proteins (OMPs) forming aqueous channels for the passive or selective uptake of nutrients (Pages et al., 2008; Silhavy et al., 2010). Little was understood about the process by which Gram-negative bacteria assemble β-barrel OMPs into the OM until the discoveries of Omp85 of Neisseria meningitidis and YaeT of Escherichia coli (Voulhoux et al., 2003; Wu et al., 2005). These OMPs, since renamed BamA (β-barrel assembly machinery protein A), represent a family of highly conserved proteins essential for OM biogenesis (Knowles et al., 2009; Silhavy et al., 2010; Tommassen, 2010). Members of the BamA family also are required for insertion of β-barrel precursors into the OMs of mitochondria and chloroplasts, eukaryotic organelles derived from ancestral proto-bacterial and cyanobacterial endosymbionts, respectively (Walther et al., 2009; Tommassen, 2010). Bacterial BamA orthologs have a bipartite structure comprised of a periplasmic N-terminal region with five polypeptide-transport-associated (POTRA) domains and a C-terminal OM spanning β-barrel region (Knowles et al., 2009; Silhavy et al., 2010; Tommassen, 2010). In E. coli, BamA is the central component of a multi-protein complex consisting of four ancillary factors (BamB-E) which function collectively in OM biogenesis (Silhavy et al., 2010; Tommassen, 2010). Comparative studies of the Bam ancillary factors have demonstrated varying degrees of conservation across bacterial species (Gatsos et al., 2008; Tommassen, 2010; Silhavy et al., 2010; Volokhina et al., 2009; Anwari et al., 2010). Current thinking holds that, as nascent OMPs exit from the Sec translocon, the principal periplasmic chaperone SurA ferries them to the Bam complex, which promotes proper folding into the OM (Sklar et al., 2007; Silhavy et al., 2010; Tommassen, 2010). This is believed to occur via a two-step process in which individual β-strands form alongside the β-sheets of POTRA segments (β-augmentation) while bending of the POTRA arm assists in the creation of β-hairpins that can then insert into the bilayer (Tamm et al., 2004; Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008).
Syphilis is a multistage, sexually transmitted disease caused by the spirochetal pathogen Treponema pallidum. Although T. pallidum is often analogized to Gram-negative bacteria, the structure, composition, and physical properties of its cell envelope diverge markedly from those of Gram-negative bacteria. For example, the OM of T. pallidum is a fluid and fragile lipid bilayer devoid of LPS (Belisle et al., 1994; Fraser et al., 1998). Whereas in Gram-negative bacteria the peptidoglycan (PG) layer is affixed to the underside of the OM, enhancing its rigidity, in T. pallidum the PG resides approximately midway within the periplasmic space and lacks biochemical linkage with the OM; above the treponemal PG layer are soluble proteins and flagella, while below are lipoproteins and integral IM proteins (Izard et al., 2009; Liu et al., 2010). In contrast to the canonical protein-rich Gram-negative OM, the T. pallidum OM contains a paucity of membrane-spanning proteins; freeze-fracture electron microscopy studies estimate that the OMPs of T. pallidum total <1% of those found in E. coli (Walker et al., 1989; Radolf et al., 1989). The dearth of surface antigens is the ultrastructural basis for the syphilis spirochete's impressive capacity to evade the robust immune responses it elicits within its obligate human host and its well earned reputation as a stealth pathogen (Radolf, 1995; Lafond & Lukehart, 2006; Radolf & Lukehart, 2006). Definitive identification of these so-called rare OMPs has eluded investigators for nearly three decades due to a variety of factors, most notably their extremely low abundance, their lack of sequence relatedness to known OMPs of Gram-negative bacteria, and the fragility of the spirochete's OM, all set against the backdrop of the bacterium's recalcitrance to in vitro cultivation (Radolf, 1995; Fraser et al., 1998; Lafond & Lukehart, 2006; Cameron, 2006).
In 2000, Cameron et al. (2000) discovered the BamA ortholog TP0326 (Tp92) using a differential screening strategy to identify E. coli clones expressing T. pallidum opsonic targets. At that time, virtually nothing was known about the Bam complex so the potential contribution of TP0326 to maintenance of the syphilis spirochete's OM was not appreciated for several years (Gentle et al., 2005). In a recent report (Cox et al., 2010), we employed a battery of cellular localization and topological prediction tools to generate ranked clusters of candidate rare OMPs, collectively referred to as the putative T. pallidum OMPeome. TP0326, the only protein encoded by the T. pallidum genome with sequence homology to a known Gram-negative β-barrel OMP (Fraser et al., 1998), emerged from this analysis as one of our two top-ranked candidates; attention also was called to its sequence and predicted structural relatedness to BamA. The function of TP0326 as the central component of an OMP assembly platform also finds support in the report by Lenhart and Akins (2010) who showed that the related protein BB0795 is essential for the assembly of OMPs in Borrelia burgdorferi, the Lyme disease spirochete. The work reported herein demonstrates not only that TP0326 possesses the hallmark bipartite topology of a BamA, but that it also possesses the key properties required to establish it as a bona fide rare OMP (Radolf, 1995; Nikaido, 2003; Silhavy et al., 2010): low abundance, amphiphilicity, surface-exposure, and β-barrel structure. Our studies, therefore, represent a major step in the longstanding quest for T. pallidum OMPs in addition to advancing our primitive understanding of OM biogenesis in the syphilis spirochete (Radolf, 1995). Interestingly, in contrast to T. pallidum-infected rabbits, which produce antibodies against both the POTRA and β-barrel regions of TP0326, humans with syphilis appear to direct their humoral responses only against the antibody-inaccessible portion of the molecule (i.e., POTRA domains). Thus, the syphilis spirochete appears to have devised a stratagem for harnessing the Bam pathway while satisfying its need to limit surface antigenicity, a prerequisite for stealth pathogenicity.
RESULTS
Genetic organization of the bamA locus in T. pallidum
More than ten years ago, it was reported that TP0326 (Tp92) is orthologous to several Gram-negative OMPs of then unknown function (Cameron et al., 2000), two of which are the now well characterized BamAs of N. meningitidis and E. coli. Analysis using the most recent version of the BLAST algorithm (Altschul et al., 1990) confirmed these relationships with high probability values as well as the relatedness of TP0326 to the BamA orthologs of T. denticola (Jun et al., 2008), L. interrogans (Haake & Matsunaga, 2010), and B. burgdorferi (Lenhart & Akins, 2010). Of note, recent experimental evidence has confirmed that BB0795 is required for biogenesis of the B. burgdorferi OM (Lenhart & Akins, 2010). In many Gram-negative bacteria, bamA is flanked upstream by rseP, which encodes an IM zinc-metalloprotease that cleaves the anti-σE factor RseA in response to misfolded OMPs in the periplasm (Li et al., 2009), and downstream by skp, which encodes a periplasmic OMP chaperone (Fig. S1). In T. pallidum, skp (tp0327) is immediately downstream of tp0326, while rseP (tp0600) is remotely downstream (Fig. S1). As in T. pallidum, the skp orthologs of T. denticola (Seshadri et al., 2004) and B. burgdorferi (Fraser et al., 1997) are immediately downstream of bamA while L. interrogans (Nascimento et al., 2004) lacks this gene. In contrast to T. pallidum, in the other spirochetes the rseP genes are upstream and quite distant from bamA.
BamAs have a characteristic bipartite architecture consisting of a C-terminal β-barrel and one or more POTRA domains at the N-terminus (Tommassen, 2010; Silhavy et al., 2010). The SMART (Schultz et al., 1998) and Pfam (Finn et al., 2010) databases predict the N-terminal half of TP0326 contains five POTRA domains, while the C-terminal half consists of a “bacterial surface antigen” domain belonging to the OMP β-barrel superfamily (Fig. 1A). There are no solved structures containing all 5 POTRA domains, but there are, however, two structures representing flexed and extended POTRA1-4 (Kim et al., 2007; Gatzeva-Topalova et al., 2008) and a recently solved structure of POTRA4-5 (Gatzeva-Topalova et al., 2010) of the E. coli BamA. By aligning all three structures centered on POTRA4, Gatzeva-Topalova et al. (2010) generated spliced models depicting the 5 POTRA domains in both extended and flexed conformations. To create a structural representation of all 5 POTRA domains of TP0326, we modeled POTRA1-5 using the SWISS-MODEL server (Arnold et al., 2006) and the combined E. coli structures. The root mean square deviation (RMSD) values of the predicted structures of the T. pallidum proteins (flexed and extended POTRA1-4 and POTRA45) are all < 0.1 Å with respect to the corresponding E. coli structures. In addition, the RMSD values of the T. pallidum spliced flexed and extended models containing POTRA1-5, both aligned to POTRA4 of the POTRA45 model, are 0.896 and 1.135 Å, respectively, which are in-line with those reported for the E. coli spliced models (Gatzeva-Topalova et al., 2010). As shown in Fig. 1B, all five domains are predicted to contain the hallmark POTRA fold consisting of a three-stranded β-sheet overlaid by a pair of anti-parallel α-helices (Kim et al., 2007; Gatzeva-Topalova et al., 2008). Furthermore, the edge of each POTRA domain is predicted to form a groove between α2 and β2 lined with hydrophobic residues (data not shown). In E. coli, these grooves are thought to bind and fold individual β-strands by β-augmentation (Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008). Also noteworthy is the β-bulge unique to POTRA3, which consists of V225 and E226 (data not shown) (Kim et al., 2007). In tandem, the five domains can assume either flexed or extended conformations with the hinge located at the interface between POTRA2 and 3. Depending on its overall orientation in relation to the plane of the OM (Gatzeva-Topalova et al., 2010), the extended POTRA arm could extend as much as 10 nm into the periplasm.
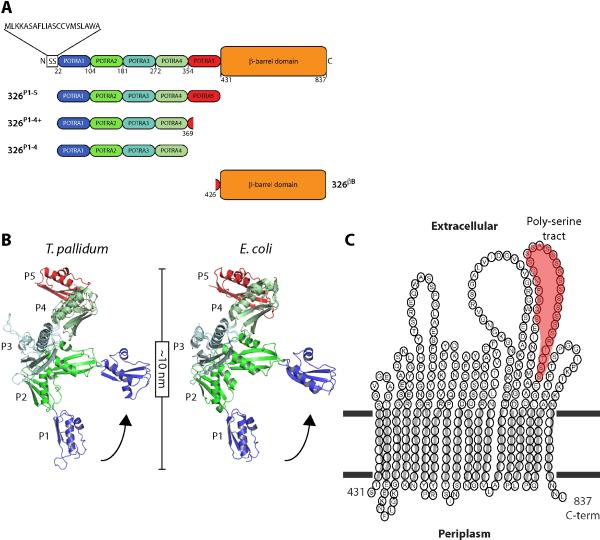
Figure 1. Domain architecture and in silico structural analysis of TP0326.
(A) Schematic representation of TP0326 and the POTRA and β-barrel constructs utilized in this study. A putative signal sequence (SS) spans amino acids (aa) 1-21 (Cameron et al., 2000; Cox et al., 2010) and is followed by 5 POTRA domains (POTRA1 aa 22-104, POTRA2 aa 105-181, POTRA3 aa 182-272, POTRA4 aa 273-354, and POTRA5 aa 355-430) and a β-barrel (aa 431-837). (B) Spliced model of TP0326 POTRA1-5, represented as P1-5, shown in flexed and extended conformations compared to those of E. coli POTRA1-5. (C) Predicted topology of the TP0326 β-barrel. Residues of the 8 short periplasmic turns and 9 long extracellular loops are indicated, while the poly-serine tract is highlighted in red.
Currently, there are no structural templates that can be utilized to generate a three dimensional representation of the putative TP0326 β-barrel. The TbbPred (Natt et al., 2004) and PRED-TMBB (Bagos et al., 2004) servers predicted, however, that the β-barrel contains an even number (18) of OM-spanning, anti-parallel segments with both N- and C-termini within the periplasm, short periplasmic turns, and long extracellular loops (Fig. 1C), all attributes shared by Gram-negative OMPs (Schulz, 2002; Wimley, 2003). Moreover, the ClustalW alignment of the β-barrel domains from geographically diverse strains of T. pallidum indicates a high degree of conservation, including the predicted surface-exposed loops (Fig. S2). A serine-rich tract extending from residues 763 to 786, a unique feature of TP0326, is predicted to reside on 1 of the 9 extracellular loops (Fig. 1C).
The C-terminal portion of TP0326 forms a β-barrel
Previous studies revealed that full-length recombinant TP0326 lacking a signal sequence is partially degraded when expressed in E. coli (Cameron et al., 2000; Tomson et al., 2007). In our hands as well, full-length recombinant TP0326 both expressed poorly and appeared to be truncated in E. coli lysates (data not shown). To circumvent this problem, we generated separate POTRA and β-barrel constructs (Fig. 1A and Table 1). Both 326P1-5 and 326P1-4 expressed well but exhibited some degradation (data not shown). During the course of our studies, Kim et al. (2007) reported that addition of several residues from the 5th POTRA domain greatly enhanced the stability of the E. coli BamA POTRA1-4. Similarly, a POTRA1-4 construct containing 14 residues from the 5th POTRA domain (326P1-4+, Table 1) was stable throughout purification (Fig. S3A). A β-barrel construct with a C-terminal His-tag (326βb) expressed in E. coli and was stable during purification (Fig. S3B).
TABLE 1.
List of Primers
| Cloning | Sequence (5’ – 3’) | Coordinates |
|---|---|---|
| 5’ tp0326* | GCCCATATGCAGGCAAACGACAATTGGTACG | 344387-344408 |
| 3’ tp0326* | GATACTCGAGCTACAAATTATTTACCGTGAACGAC | 346834-346813 |
| 3’ tp0326 326P1-4 | GGCGAATTCTCACTATTCCACCACAGTGTACGTAAGC | 345139-345118 |
| 3’ tp0326 326P1-4+ | GGCGAATTCTCACTAATTCTTCGTTCCCTTAATGATAAT | 345430-345407 |
| 3’ tp0326 326P1-5 | GGCGAATTCTTACTAGTTTGCCGTCGACTGCTCCTC | 345625-345605 |
| 5’ tp0326 326βB | GATAGCTAGCATGGATGTGCGGCCCGGCTCTGAG | 345599-345619 |
| qRT-PCR primers | Sequence (5’ – 3’) | Coordinates |
| tp0249 (flaA) fwd | GCGGTTGCACAGTGGGAG | 261664-261681 |
| tp0249 (flaA) rev | CAGCATGGGCGACAGGAT | 261724-261707 |
| tp0117/0131 (tprC/D) fwd | CAAGAGAGAGCTATCCTCAAAG | 135833-135812 |
| 152033-152012^ | ||
| Tp0117/0131 (tprC/D) rev | GTTTAGCAGTGACAACTCTTGG | 135545-135566 |
| 151745-151766^ | ||
| tp0453 (p30.5) fwd | TCTACGAAAACGGAACACGGAATC | 482639-482620 |
| tp0453 (p30.5) rev | CAAAGCGAGGCGGTAACTGC | 482639-482620 |
| tp0574 (tp47) fwd | GCAGGAGACCGAAGACAGCAG | 623271-623251 |
| tp0574 (tp47) rev | GCACGCACCATCTTAGTAACGC | 623194-623215 |
| tp0326 fwd | ACGAGGTTATCCTGCGTGAAATG | 345438-345460 |
| tp0326 rev | CTTCCCACTGACAAAAGAGCGAA | 345701-345679 |
Underlined sequence indicates a restriction site:
Coordinates for tprD.
Upon folding, the fluorescence emission maximum of tryptophan-containing integral membrane proteins “blue-shifts” as the tryptophan residues move from an aqueous to a hydrophobic environment (Heuck & Johnson, 2002). We, therefore, used tryptophan fluorescence to monitor folding of 326βb and a control β-barrel, E. coli OmpG, which have ten and 12 tryptophans, respectively. Fig. 2A shows that unfolded 326βB and OmpG had emission maxima at 346 nm as compared to the maxima of 337 nm for the folded proteins in detergent buffers; the folded proteins also displayed increased emission intensity. We next employed circular dichroism (CD) spectroscopy to assess the β-sheet content of the two folded proteins. Both 326βB and OmpG displayed broad minima centering on 218 nm (Fig. 2B), indicating a predominance of β-structure. Deconvolution of the spectra (Whitmore & Wallace, 2004) calculated β-structure contents of 48 % for both proteins. The spectrum of 326P1-4+, in contrast, indicated a mixed α/β fold with a distinct minimum at 208 nm followed by a broad shoulder ranging from 215-222 nm (Fig. S3C). Deconvolution calculated α-helical (24%) and β-sheet (28%) contents similar to those reported for E. coli POTRA (Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008).
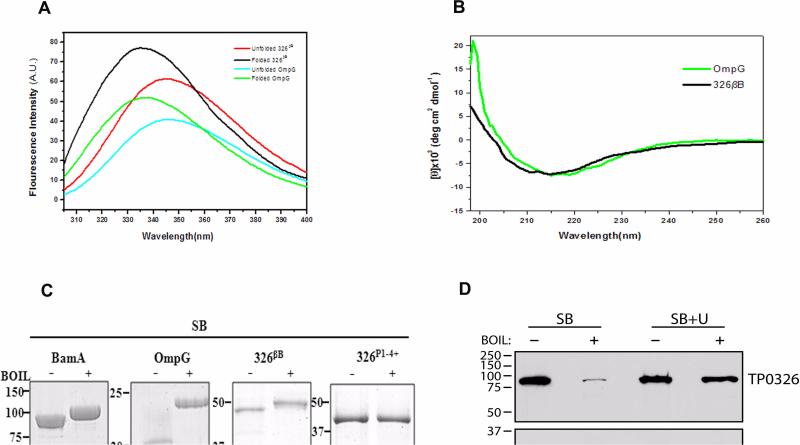
Figure 2. The C-terminal portion of TP0326 forms a β-barrel.
(A) Tryptophan fluorescence emission spectra of unfolded 326βB and OmpG and folded forms in 0.5% DDM and 1% OG micelles, respectively. (B) CD spectra of folded 326βB (5 μM) and OmpG (2.6 μM) in DDM and OG buffer, respectively. (C) Heat-modifiability of folded E. coli BamA, OmpG, 326βB, and 326P1-4+. Proteins were stained with GelCode® Blue following SDS-PAGE with (+) or without (-) boiling in sample buffer (SB). (D) T. pallidum samples were dissolved in SB without (SB) or with (SB + U) 8M urea prior to SDS-PAGE with (+) or without (-) boiling and immunoblotted against POTRA or TroA (loading control) antiserum. MW standards (kDA) for SDS-PAGE and immunoblot analyses are on the left of each panel.
β-barrel forming proteins characteristically retain a high degree of β-sheet content when solubilized in SDS at room temperature and, consequently, run with lower apparent molecular masses by SDS-PAGE than when denatured by boiling; this property, termed heat-modifiability, has often been used to distinguish folded from unfolded populations of OMPs and assess their structural stability (Conlan & Bayley, 2003; Burgess et al., 2008). As expected, boiling did not affect the electrophoretic mobility of 326P1-4+ (Fig. 2C). Folded 326βB, in contrast, displayed heat-modifiability comparable to that of the E. coli BamA and OmpG controls (Fig. 2C). Surprisingly, given the results for 326βB and recombinant E. coli BamA, immunoblot analysis of T. pallidum lysates failed to reveal heat modifiability of native TP0326 (Fig. 2D). The immunoblot studies (included the stacking gels) also revealed that a substantial amount of the native protein is lost (presumably degraded) during heating even in the presence of protease inhibitors; this odd but highly reproducible phenomenon could be prevented by addition of 8M urea to the sample buffer (Fig. 2D). Lastly, we confirmed the native protein's lack of heat modifiability by immunoblotting equal amounts of boiled and unboiled TP0326 in lysates that had been electrophoresed side by side (data not shown). Of note, all subsequent immunoblot analyses requiring denaturing conditions were performed using sample buffer containing 8M urea.
Membrane insertion of TP0326 occurs via the β-barrel
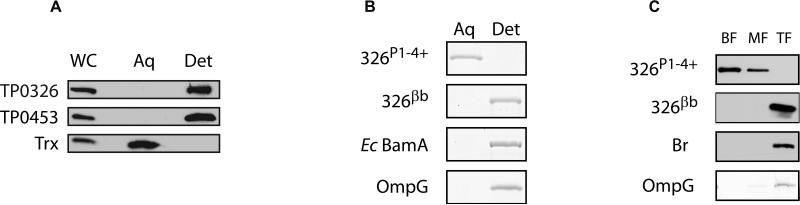
The bioinformatics and structural analyses described earlier predicted that TP0326 consists of an OM-spanning β-barrel and tandem POTRA domains forming a distensible arm within the periplasm. Experiments were conducted to evaluate this topological model. We first determined by Triton X-114 phase-partitioning (Brusca & Radolf, 1994) that native TP0326 possesses the amphiphilic character expected of a polypeptide containing an OM-spanning β-barrel domain (Fig. 3A). As shown in Fig. 3B, 326P1-4+ and 326βB partitioned exclusively into the aqueous and detergent-enriched phases, respectively. These results imply that the β-barrel domain is solely responsible for the amphiphilicity of the native protein. To extend these findings, we compared the abilities of 326P1-4+ and 326βB to incorporate into liposomes that simulate the phospholipid composition of the T. pallidum OM (Belisle et al., 1994; Rigaud & Lévy, 2003; Hazlett et al., 2005); following incorporation, proteoliposomes and unincoporated proteins were separated on discontinuous sucrose gradients. Fig. 3C shows that 326βB, as well as the OmpG and bacteriorhodopsin controls, representing both an OMP and an IMP, respectively, were recovered only from the top (liposome containing, TF) fractions, whereas 326P1-4+ was detected in the heavier bottom and middle fractions (BF and TF, respectively) containing unincorporated material.
Figure 3. The β-barrel is responsible for the amphiphilicity of TP0326.
(A) Freshly harvested T. pallidum, solubilized in 2% Triton X-114, was phase-partitioned and subjected to immunoblot analysis with monospecific antisera directed against 326P1-4+, TP0453, and thioredoxin (Trx). Lanes: whole cells (WC), aqueous phase (Aq), and detergent-enriched phase (Det). (B) Purified recombinant 326P1-4+, 326βB, and E. coli BamA and E. coli OmpG were phase-partitioned in Triton X-114 and stained with GelCode® Blue following SDS-PAGE. (C) Liposomes were reconstituted with 326P1-4+, 326βB, Br, and OmpG followed by sucrose density gradient ultracentrifugation. Fractions either were subjected to immunoblotting with monospecific antisera directed against 326P1-4+, the poly-histidine tag (326βB), or Br or stained with GelCode® Blue (OmpG) following SDS-PAGE. Lanes: bottom, middle, and top fractions (BF, MF, and TF, respectively).
Native TP0326 is expressed at extremely low levels and is surface-exposed
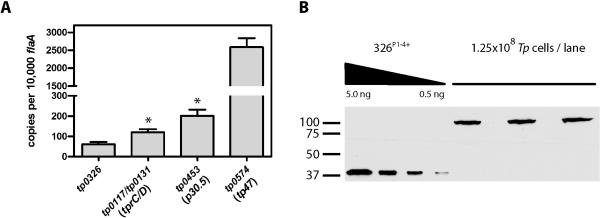
To further the characterization of TP0326 as a potential rare OMP, we examined its expression in T. pallidum at both the transcriptional (Fig. 4A) and translational (Fig. 4B) levels. The number of tp0326 transcripts was miniscule (approx. 0.05%) compared to that of flaA and was significantly lower than those encoding either TP0117/TP0131 (TprC/D), a prime candidate OMP (Cox et al., 2010), or the OM-associated lipoprotein TP0453 (Hazlett et al., 2005). The copy numbers for all three transcripts were markedly lower than that determined for tp0574, which encodes the abundant lipoprotein carboxypeptidase Tp47 (Deka et al., 2002). Expression of native TP0326 on a per cell basis was determined by quantitative immunoblot analysis. As shown in Fig. 4B, the anti-POTRA antiserum proved to be exquisitely sensitive, capable of detecting sub-nanogram amounts of 326P1-4+. The mean copy number (141 ± 52) calculated from two independent experiments agrees well with the above qRT-PCR data and our previous determination that treponemes express approximately 15,000 copies of FlaA protein per cell (Parsonage et al., 2010).
Figure 4. TP0326 is expressed at extremely low levels.
(A) Copy numbers of tp0326, tp0117/tp0131, tp0453, and tp0574 transcripts, normalized to flaA. Bars represent the means ± SEM. The asterisks indicate levels of significance between tp0326 and both tp0117/tp0131 (P = 0.0049) and tp0453 (P = 0.0194) transcript copy numbers. (B) Quantitative immunoblot analysis of native TP0326. Immunoblots of the 326P1-4+ and treponemal lysates were subjected to densitometric analysis. Standard curves were generated from the densitometric values obtained for recombinant 326P1-4+. The amount of TP0326 per T. pallidum cell was determined by dividing the corresponding densitometrically-derived value in the lysates by 1.25 × 108 T. pallidum, the amount loaded per lane. Copy numbers were determined by the molecular mass of TP0326 (MWTP0326= 92,039 Da). MW standards (kDA) are indicated on the left.
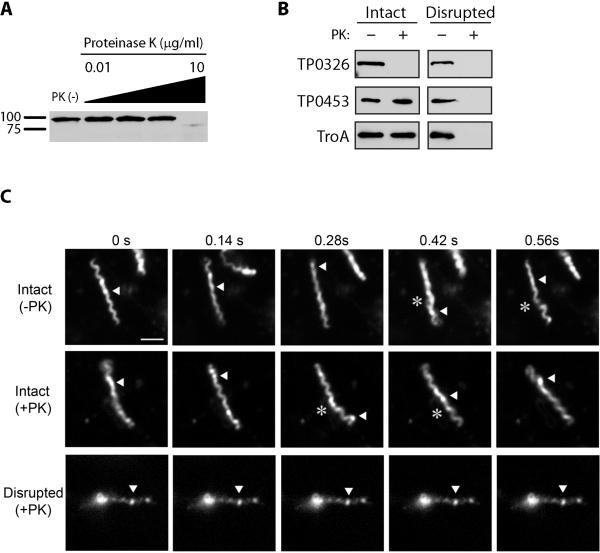
We examined the surface accessibility of TP0326 by proteinase K (PK) treatment in freshly harvested treponemes, an approach which could be combined with immunoblot analysis and the use of our ultra-sensitive POTRA antiserum. In light of the well recognized fragility of the T. pallidum OM, we monitored the motility of organisms by darkfield microscopy and live-imaging as a means of confirming that their OMs remained intact. β-barrels OMPs are known to be resistant to proteolysis when properly folded (Hoenger et al., 1993; Werner et al., 2003); as a further precaution to minimize inadvertent damage to the bacteria, we first determined the lowest concentration of PK required for surface proteolysis. As shown in Fig. 5A, with intact organisms, TP0326 was resistant to concentrations of PK up to 1 μg/ml but was degraded at 10 μg/ml. Figure 5B shows that PK treatment (10 μg/ml) of intact organisms degraded TP0326 but not the periplasmic controls TP0453 and TP0163 (TroA). TP0453 and TP0163 were degraded in treponemes treated with Triton X-100 and lysozyme, confirming that these two periplasmic proteins are, in fact, intracellular and not intrinsically protease-resistant. The lack of degradation of TP0453 in intact organisms is particularly noteworthy because this polypeptide is lipid-anchored to the inner leaflet of the OM and is susceptible to proteolysis following minor perturbations of this bilayer (Hazlett et al., 2005). Treponemes become non-motile upon minor disruption of their OMs. Fig. 5C (also see Supplemental Videos 1-3) shows that PK treatment alone had no discernible effect on the motility of freshly harvested organisms, whereas detergent-lysozyme treated organisms exposed to the same concentration of PK were non-motile and noticeably thinner.
Figure 5. TP0326 is surface-exposed.
(A) Immunoblot analysis of TP0326, detected by the POTRA1-5 antiserum, in intact T. pallidum (1.0 × 108 Tp/lane) treated with graded concentrations (0.1 - 10μg/ml) of proteinase K (PK). MW standards (kDA) are shown on the left. (B) PK accessibility of TP0326 and two periplasmic proteins in intact and detergent-lysozyme treated organisms. Intact spirochetes (5.0 × 108/ml) in a 1ml volume were treated with 10 μg/ml of PK, while detergent-lysozyme treated spirochetes (5.0 × 108 resuspended in 200μl of PK disruption buffer) were treated with a total of 10 μg of PK. Each lane represents 1.0 × 108 T. pallidum immunblotted with antisera to POTRA1-5 of TP0326, TP0453, and TroA. (C) Live imaging of intact (−PK and +PK) and detergent-lysozyme treated (+PK) organisms. For intact spirochetes (−PK and +PK), asterisks and arrowheads, respectively, indicate flexing and waveform propagation, while for detergent-lysozyme treated spirochetes (+PK) identically positioned arrowheads indicate lack of flexing and waveform propagation. Time scale is 0-0.56s and the scale bar = 5.0μM.
Native TP0326 forms part of a high molecular mass complex
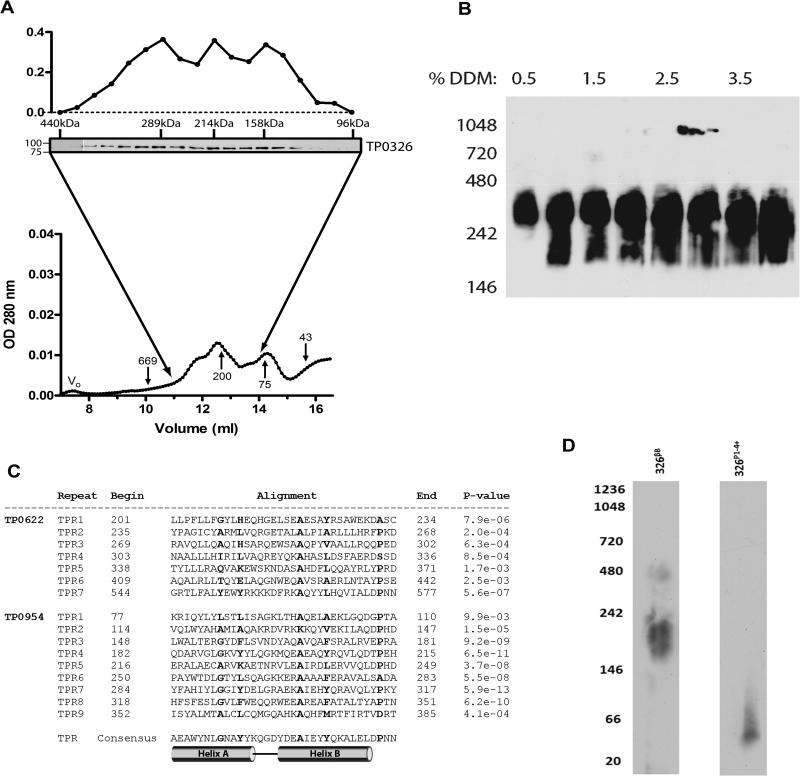
In Gram-negative bacteria, BamA functions as the central component of a complex with ancillary factors which collectively promote OM biogenesis (Wu et al., 2005; Gatsos et al., 2008; Tommassen, 2010; Silhavy et al., 2010). To determine whether TP0326 forms complexes, we used size-exclusion chromatography (SEC) to examine the oligomerization state of the native protein in T. pallidum lysates solubilized with 2% DDM. As shown in Fig. 6A, immunoblot analysis revealed that TP0326 eluted with a broad profile spanning ~100-440 kDa by SEC. Densitometry indicated that the immunoreactive material containing TP0326 appears to consist of sub-complexes with MWs of ~289 kDa and ~214 kDa and a presumptive monomer centered about 150 kDa (the MW of a DDM micelle ~50 kDa). Recent studies in Caulobacter crescentus showing that high concentrations of DDM caused dissociation of the Bam complex (Anwari et al., 2010), raised the possibility that a similar phenomenon might have occurred with TP0326. We, therefore, utilized blue native polyacrylamide gel electrophoresis (BN-PAGE), shown in Fig. 6B, to further characterize the TP0326 complex and evaluate its stability in graded concentrations of DDM under non-denaturing conditions. In 0.5% DDM, native TP0326 formed a discrete complex of ~300-400 kDa. Partial dissociation occurred at higher detergent concentrations, although a substantial amount of the high MW complex remained even in 4% DDM. Mass spectrometric analysis was performed on the un-dissociated complex resolved by BN-PAGE (0.5% DDM) in an attempt to identify the components of the native complex; the results were inconclusive because the samples did not contain enough material to detect TP0326 (data not shown).
Figure 6. Native TP0326 forms a complex.
(A) A size-exclusion chromatography of freshly harvested T. pallidum (~2.5×109) solubilized with 25mM Tris (pH 7.0) and 2% DDM. VO indicates the void volume, whereas arrows with numbers demarcate the elution volumes of the MW (kDa) standards. Above the chromatogram are immunoblots detecting TP0326 with the POTRA antiserum in fractions spanning a MW range of 96 to 400 kDa. The left two lanes in the immunoblot image come from a separate film. MW standards for the immunoblot analysis are shown to the left. TP0326 was not detected above or below this range. (B) Immunoblot analysis of T. pallidum lysates solubilized with 25mM Tris (pH 7.0) and graded concentrations of DDM following separation by BN-PAGE (left panel). A total of 5.0 × 108 T. pallidum was loaded per lane. (C) Sequence alignments of predicted tetratricopeptide repeat of the potential BamD candidates of T. pallidum, TP0622 and TP0954, compared to a TPR consensus sequence (D'Andrea & Regan, 2003). Probability-values (far right column) of being a TPR were calculated by the TPRpred server (http://toolkit.tuebingen.mpg.de/tprpred/). (D) BN-PAGE of 326P1-4+ and 326βB. MW standards (kDa) are shown to the left.
Bioinformatic analysis revealed that T. pallidum does not encode recognizable orthologs for the Bam ancillary factors of E. coli, N. meningitidis, or C. crescentus (Table 2). By searching the T. pallidum genome for the β-propeller (Jawad & Paoli, 2002) and tetratricopeptide repeat (TPR) (D'Andrea & Regan, 2003) protein-protein interaction motifs characteristic of BamB and BamD (Gatsos et al., 2008), respectively, we discovered three possible ancillary factors (Table 2). TP0629 contains at least 1 β-propeller repeat but, because it lacks an N-terminal signal sequence, seems an unlikely BamB candidate. TP0622 (MW = 66.7 kDa) and TP0954 (MW = 54.6 kDa) are both predicted to be lipoproteins with 7 and 9 TPRs, respectively (Fig. 6C). Of note, BamD is the only ancillary factor essential for cell viability and OMP assembly in E. coli (Onufryk et al., 2005; Malinverni et al., 2006). Strengthening the argument for TP0622 as the authentic BamD is the finding that tp0622 lies immediately upstream of tp0620 and tp0621, which encode candidate OMPs TprI and TprJ, respectively (Cox et al., 2010; Cameron, 2006). Not surprisingly, these possible T. pallidum ancillary factors have orthologs in T. denticola. TDE2565 is predicted to have at least 1 β-propeller motif and, unlike TP0629, an N-terminal signal sequence. TDE1053 and TDE0993, homologs of TP0622 and TP0954, respectively, are both putative lipoproteins and predicted to have 9 and 10 TPRs, respectively (data not shown). Neither B. burgdorferi nor L. interrogans encode recognizable orthologs to the Bam ancillary factors, although non-orthologous proteins with β-propeller and TPR motifs were indentified (Table 2).
TABLE 2.
Possible candidate BAM ancillary factors in T. pallidum and other spirochetes
| BamB | BamC | BamD | BamE | Others | |
|---|---|---|---|---|---|
| E. coli | B2512 (YfgL) | B2477 (NlpB) | B2595 (YfiO) | B2617 (SmpA) | |
| N. meningitidis | -* | NMB0928 | NMB0703 (ComL) | NMB0204 | NMB0382 (RmpM) |
| C. crescentus | CC1653 | - | CC1984 | CC1365 | CC3229 (Pal) |
| T. pallidum | TP0629^ | - | TP0954^^ TP0622^^ |
- | - |
| T. denticola | TDE2565^ | - | TDE0993^^ TDE1053^^ |
- | - |
| B. burgdorferi | BB0743^ | - | BB0324^^ | - | - |
| L. interrogans | LIC12208^ | - | -# | - | - |
A dash (-) indicates that no orthologs were identified.
Identified by the presence of β-propeller motifs.
Identified by the presence of tetratricopeptide repeats (TPR).
>10 proteins were identified.
Studies of BamA superfamily members have noted varying degrees of homo-oligomerization of the recombinant proteins mediated by POTRA and/or β-barrel (Surana et al., 2004; Robert et al., 2006; Bredemeier et al., 2007; Nesper et al., 2008; Meng et al., 2009). We, therefore, used the recombinant constructs to examine whether self-association of native TP0326 could contribute to complex formation. As reported by Kim et al. (Kim et al., 2007) for the comparable E. coli construct, SEC analysis of 326P1-4+ (Fig. S3A) shows a single peak representing a calculated MW of a monomer (~40 kDa). BN-PAGE also showed that 326P1-4+ is monomeric (Fig. 6D). In contrast, folded 326βB migrated in BN-PAGE predominantly with a calculated molecular mass in the vicinity of 200-kDA (Fig. 6D), most consistent with a trimer after correcting for the approximate 50-kDa size of the DDM micelles.
Identification of potential C-terminal recognition sequences for TP0326
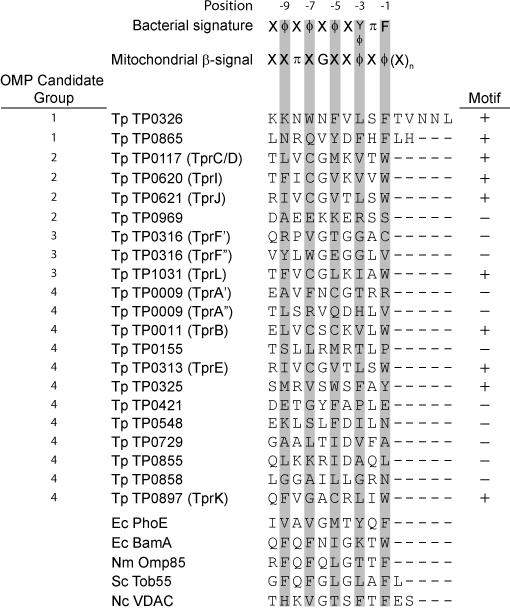
Recognition of the C-terminus of a precursor β-barrel polypeptide by BamA, recently shown to occur via POTRA1 in E. coli (Bennion et al., 2010), is an important determinant of the efficiency of folding and OM insertion (Robert et al., 2006; Walther et al., 2009; Tommassen, 2010). For many Gram-negative OMPs, the C-terminal signature sequence ends with an aromatic residue preceded by hydrophobic residues at alternating positions (-3, -5, -7, and -9) (Struyve et al., 1991; Robert et al., 2006) (Fig. 7). For mitochondrial β-barrel OMPs (e.g., Tob55 and VDAC), the recently identified C-terminal signature sequence, referred to as the β-signal, is highly similar to that of Gram-negative bacteria but need not be at the extreme C-terminus (i.e., it can be followed by up to 28 residues) (Kutik et al., 2008; Tommassen, 2010). When we aligned the C-termini of the in silico-derived T. pallidum OMPeome (Cox et al., 2010) to identify potential signature sequences, quite interestingly, we found evidence that T. pallidum may utilize both types of recognition motifs (Fig. 7). With the exception of TP0325, the candidates with bacterial-type signature sequences are members of the Tpr family without frameshifts, while TP0326 and TP0865, our other top-ranked candidate, contain mitochondrial-type signatures. Also noteworthy is that a number of the proteins without matches for either motif are lower-ranked candidates. Bennion et al. (Bennion et al., 2010) showed that self-recognition of E. coli BamA is SurA-independent and occurs via POTRA1. The result for TP0326 implies that it too self-recognizes via a signature sequence.
Figure 7. Identification of potential C-terminal signature sequences for POTRA recognition in candidate T. pallidum OMPs.
Above the alignment are the consensus bacterial signature sequence (Struyve et al., 1991; Robert et al., 2006) and mitochondrial β-signal (Kutik et al., 2008; Tommassen, 2010) for POTRA recognition by OMP precursors. The alignment shows the C-termini of TP0326, the 16 other proteins in the predicted T. pallidum (Tp) OMPeome (Cox et al., 2010), and representative β-barrel OMPs: E. coli (Ec) PhoE and BamA; N. meningitidis (Nm) Omp85; Saccharomyces cerevisiae (Sc) Tob55; and Neurospora crassa (Nc) VDAC. Accession numbers of Sc Tob55 and Nc VDAC are DAA10518 and EAA31714, respectively. All other sequences were obtained from the Comprehensive Microbial Resource database (http://cmr.jcvi.org/). “OMP candidate group numbers” designate the ranked clusters containing candidate OMPs determined recently by consensus computational framework (Cox et al., 2010). “+” and “-“ designate the presence or absence, respectively, of a C-terminal sequence that meets either the bacterial or mitochondrial consensus. Single aa codes are used. X, any aa; ϕ, hydrophobic aa; π, polar aa; n, 1 to 28 aa. . The sequences were aligned manually.
T. pallidum-infected rabbits, but not humans, mount a vigorous antibody response against the TP0326 β-barrel
TP0326 induces a surprisingly robust antibody response in humans and rabbits with syphilitic infection (Van Voorhis et al., 2003; McKevitt et al., 2005; Brinkman et al., 2006) despite its extremely low level of expression. To gain insights into the potential relationship of TP0326-specific antibodies to both immune evasion and protective immunity during syphilitic infection, we used 326P1-4+ and 326βB to examine antibody responses of T. pallidum-infected rabbits and humans to the molecule's periplasmic and OM-associated domains. The results are presented in Fig. 8. Whereas sera from three different animals reacted strongly with both 326P1-4+ and 326βB, pooled secondary human syphilitic sera (HSS) reacted only with 326P1-4+. We expanded these analyses by separately examining the reactivities of the six sera used to form the pool. Of the six, three individuals did not respond to either construct. Three individuals responded strongly to POTRA while one (#40) mounted a barely detectable response to the β-barrel. Particularly noteworthy, identical results were obtained when the patient sera were immunoblotted against folded 326βB, arguing against the possibility that humans respond exclusively to conformational epitopes not detected by conventional immunoblotting. Heterogeneity of surface-exposed epitopes could explain the lack of reactivity of human syphilitic sera with the β-barrel construct derived from the Nichols-Farmington strain. This is unlikely, however, given the high degree of sequence conservation among the predicted extracellular loops of TP0326 proteins in geographically diverse T. pallidum strains (Fig. S2)
Figure 8. T. pallidum-infected rabbits, but not humans, mount a vigorous antibody response against the TP0326 β-barrel.
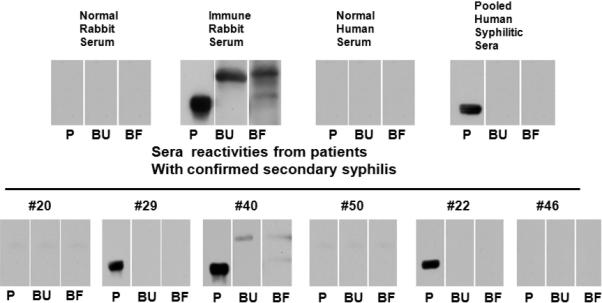
Antibody reactivities of normal rabbit serum, immune rabbit serum (IRS, representative of 3 different animals), normal human serum, pooled and individual sera from six patients with secondary syphilis (Cruz et al., 2010) against 326P1-4+ (P) and unfolded and folded 326βB (BU and BF, respectively). Each immunoblot was performed using 100 ng of purified recombinant protein.
DISCUSSION
More than five decades have elapsed since syphilis researchers first noted the poor surface immunoreactivity of live T. pallidum (Nelson & Mayer, 1949; Hardy & Nell, 1957; Metzger et al., 1961). Beginning in the late 1980s, there emerged a steady stream of evidence revealing that the spirochete's OM contains an extraordinarily low density of integral membrane proteins which present few surface antigenic targets to its obligate human host (Radolf, 1995; Cameron, 2006; Lafond & Lukehart, 2006; Izard et al., 2009; Cox et al., 2010; Liu et al., 2010). Understandably, past efforts to characterize the T. pallidum OM have underscored the structure's many physical, compositional, and ultrastructural differences from its Gram-negative counterparts (Fraser et al., 1998; Radolf, 1995; Hazlett et al., 2005; Cameron, 2006). The discovery of the BamA family provides an avenue for delineating the distinctive features and biogenesis of the T. pallidum OM, as well as their relationship to syphilis pathogenesis (Cruz et al., 2010), within a mechanistic framework more closely aligned with the experimental microbiology of prototypical diderm organisms.
Full-length TP0326 expressed in E. coli is unstable, as shown here and elsewhere (Cameron et al., 2000; Tomson et al., 2007). The strategy adopted to circumvent this problem, separate expression of N- and C-terminal segments, enabled us to demonstrate unequivocally, in combination with structural modeling, that native TP0326 possesses the dual domain architecture characteristic of BamA proteins (Knowles et al., 2009; Silhavy et al., 2010; Tommassen, 2010). The N-terminal segment not only contains five predicted POTRA motifs with a mixture of α-helical and β-structure, it completely lacks amphiphilic character based on both Triton X-114 phase partitioning and liposome incorporation studies. Consistent with the “construction rules” enumerated for β-barrels (Schulz, 2000), the C-terminal segment is predicted to contain (i) an even number (18) of anti-parallel, amphipathic β-strands, (ii) N- and C-termini within the periplasm, (iii) short periplasmic turns, and (iv) large, extracellular loops, one of which contains a polyserine tract not present in other members of the BamA family (Cameron et al., 2000). Experimental analysis fulfilled these predictions to the extent that the folded C-terminal fragment of TP0326 was shown to be rich in β-sheet content, heat-modifiable, and capable of inserting into a lipid bilayer. Native TP0326 is amphiphilic and surface-exposed based on surface proteolysis, as shown herein, and opsonophagocytosis (Cameron et al., 2000). The only membrane topology compatible with our experimental and in silico results is one in which the N-terminal POTRA domains are periplasmic and the C-terminal β-barrel resides within the OM. The extraordinarily low abundance of TP0326 based on both qRT-PCR and quantitative immunoblotting analysis is consistent with the very low particle densities in T. pallidum OMs revealed by freeze-fracture EM (Radolf et al., 1989; Walker et al., 1989) and justifies the conclusion that it is a true rare OMP.
In E. coli, the POTRA arm of BamA provides docking sites for four lipoproteins, designated BamB-E, forming the macromolecular machine that catalyzes the folding and membrane insertion of newly exported OMP precursors (Wu et al., 2005; Gatsos et al., 2008; Tommassen, 2010; Silhavy et al., 2010). Other than BamA, the Bam complex is not well conserved in Gram-negative bacteria (Gatsos et al., 2008; Tommassen, 2010; Silhavy et al., 2010; Volokhina et al., 2009; Anwari et al., 2010), suggesting that bacterial species have tailored the OM assembly process to meet the specialized requirements of their OMs and OMP repertoires. The syphilis spirochete appears to have taken this phylogenetic diversity to an extreme by dispensing with readily identifiable orthologs of the known ancillary factors. Nevertheless, fractionation of detergent-solubilized T. pallidum by SEC and BN-PAGE yielded unequivocal evidence that native TP0326 exists as part of higher order complexes. Moreover, both methods revealed that the complexes contain labile components that dissociate from a putative TP0326-based ‘core’ with increasing detergent concentration, a result reminiscent of the ‘modular’ Bam complex described for Caulobacter crescentus (Anwari et al., 2010). Using bioinformatics, we were able to identify two candidate BamD lipoproteins based on the presence of hallmark TPR motifs. As noted earlier, we prefer TP0622 for the sole reason that its encoding gene resides upstream of genes for two OMP candidates (Cox et al., 2010). Binding of BamB to BamA is theorized to involve an induced fit mechanism of β-augmentation between the eight-bladed β-propeller fold of BamB and β-strands on one or more of BamA's five POTRA domains (Gatsos et al., 2008; Kim & Paetzel, 2011). The prediction that POTRA3 contains a ‘β-bulge’ (Kim et al., 2007) suggests the existence of a functional BamB ortholog. Investigators have reported varying degrees of homo-oligomerization of BamA proteins, either by the POTRA domain or β-barrel (Surana et al., 2004; Robert et al., 2006; Bredemeier et al., 2007; Nesper et al., 2008; Meng et al., 2009). BN-PAGE of the recombinant constructs raised the possibility that self-association of TP0326 via the β-barrel also contributes to complex formation.
Two explanations, both likely consequences of the spirochete's unorthodox cell envelope ultrastructure (Cameron, 2006; Izard et al., 2009; Liu et al., 2010), can be envisioned to explain the divergence of the T. pallidum Bam complex. First, ancillary factors containing OmpA-like PG binding motifs are thought to play a generalized role in anchoring the Bam complex to the closely apposed PG layer in Gram-negative bacteria, perhaps helping to guide strands of nascent OMPs through the murein meshwork as they insert into the OM (Walther et al., 2009; Anwari et al., 2010). T. pallidum presumably would have no need for such functions because its PG layer has no covalent linkage with the OM (Cameron, 2006; Izard et al., 2009; Liu et al., 2010). The other could reflect the spirochete's distinctive mechanism for chaperoning nascent OMPs across the periplasm. E. coli utilizes two pathways, SurA and Skp/DegP, for chaperoning OMP precursors within the periplasm (Mogensen & Otzen, 2005), with the former considered predominant and known to interact with POTRA domains (Sklar et al., 2007; Bennion et al., 2010). That T. pallidum appears to lack SurA, relying instead upon Skp and DegP to ferry nascent OMPs to the assembly machinery. If so, one would anticipate that the components and interactivity of its OM assembly platform would differ fundamentally from those of E. coli and closely related Gram-negatives.
For T. pallidum, an extracellular bacterium, stealth pathogenicity hinges on maintaining a sufficient density of OM proteins to meet physiological and virulence-related needs, while minimizing the number of surface antigenic targets that render the bacterium vulnerable to antibody-dependent clearance mechanisms (Lukehart, 2008). Indications are emerging as to how the syphilis spirochete maintains this delicate balance. The Sec system of Gram-negative bacteria includes a soluble cytoplasmic chaperone, SecB, that captures polypeptides destined for export and directs them to the Sec translocon (Bechtluft et al., 2010). T. pallidum's lack of SecB could create a natural bottleneck to export of OM precursors across the IM, while exclusive reliance on the Skp/DegP pathway could limit the efficiency with which substrates are delivered to the POTRA arm of TP0326. The nature of the C-terminal recognition sequence is an important determinant of substrate recognition by the POTRA arm that also affects the conformational state of the β-barrel, as measured by pore formation, and presumably its catalytic function (Robert et al., 2006). Diversity of C-terminal sequences, or even lack thereof, represents an additional mechanism for fine tuning the kinetics of folding and assembly of individual OMPs (Walther et al., 2009; Tommassen, 2010).
A number of years ago, we predicted that rare OMPs would be poorly immunogenic because they are nonlipidated and expressed at extremely low levels (Radolf, 1995). Serologic studies using full-length TP0326 disproved this idea (Van Voorhis et al., 2003; McKevitt et al., 2005; Brinkman et al., 2006), suggesting that the spirochete employs an alternative stratagem to harness the Bam pathway while avoiding the deleterious consequences of the antibody responses TP0326 elicits. Immunoblot analysis with the POTRA and β-barrel constructs revealed what appears to be the spirochete's solution to this dilemma and an ostensibly striking dichotomy in the antibody responses to TP0326 elicited in humans and rabbits. Anti-TP0326 antibodies generated by humans, the pathogen's natural host, appear to be directed primarily against the periplasmic portion of the molecule, while T. pallidum-infected rabbits generate antibodies against both antibody-inaccessible and accessible regions of the protein. These results support our longstanding contention of a direct linkage between lack of antibody binding by the pathogen and immune evasion during the secondary stage of human syphilis (Radolf, 1995), a notion receiving clinical support from PCR-based assessments of spirochetal burdens in the blood of patients with secondary syphilis (Gayet-Ageron et al., 2009; Martin et al., 2009; Cruz et al., 2010). They also suggest that the antibodies in human secondary syphilitic sera that surface label or promote opsonophagocytosis of subpopulations of treponemes (Cox et al., 2010; Cruz et al., 2008; Moore et al., 2007) are directed against other, as yet unidentified, rare OMPs. Finally, our findings raise the exciting and testable possibility that antibodies against surface-exposed determinants of TP0326 contribute to bacterial clearance and protective immunity in the experimental rabbit model (Radolf & Lukehart, 2006).
EXPERIMENTAL PROCEDURES
Propagation and harvesting of Treponema pallidum
Animal protocols described in this work strictly follow the recommendations of the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the University of Connecticut Health Center Animal Care Committee under the auspices of Animal Welfare Assurance A347-01. The Nichols-Farmington strains of T. pallidum subspecies pallidum was propagated by intratesticular inoculation of adult New Zealand White rabbits with 1 × 108 treponemes per testis and harvested approximately 10 days later (Hazlett et al., 2001). To extract the treponemes, the testes were surgically removed, minced, and subsequently incubated on a rotator for 45-60 min in a 50-ml conical tube at room temperature in 5 ml of CMRL medium (Invitrogen, Carlsbad, CA) supplemented with 100 μl of protease inhibitor cocktail (PIC) (Sigma-Aldrich, St. Louis, MO). For the OMP surface accessibility and motility assays (below), PIC was not added to the extraction media. To remove rabbit testicular debris, the treponemal suspension was transferred to a sterile tube and centrifuged for 10 min at 500 × g. T. pallidum was enumerated using a Petroff-Hausser counting chamber (Hausser Scientific Company, Horsham, PA). The treponemes were harvested by centrifugation for 20 min at 10,000 × g at 4°C and the pellet was washed with ice-cold PBS prior to processing for experiments described below.
Immunologic reagents
Rat polyclonal antiserum directed against the POTRA domains of TP0326 (Cox et al.), TroA (TP0163) (Akins et al., 1997), p30.5 (TP0453) (Hazlett et al., 2005), thioredoxin (Trx, TP0919) (Parsonage et al., 2010), and Helicobacterium salinarium bacteriorhodopsin (Br) (Hazlett et al., 2005) were described previously. The mouse monoclonal antibody (hybridoma clone HIS-1) specific for polyhistidine tags was purchased from Sigma-Aldrich. Immune rabbit serum (IRS) was described previously (Hazlett et al., 2001; Cox et al., 2010); normal rabbit serum (NRS) was obtained from healthy, uninfected animals. Normal human serum (NHS) was obtained from a healthy volunteer without a history of syphilis and confirmed to be nonreactive by rapid plasma reagent testing. Sera from HIV-seronegative persons with secondary syphilis were obtained from individuals enrolled at a study site located in Cali, Colombia (Cruz et al., 2010). Serum specimens were obtained following informed consent in protocols approved by the human subjects boards at the Connecticut Children's Medical Center, the University of Connecticut Health Center (UCHC), and Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM).
Bioinformatics and in silico structural analysis
Sequence analysis was performed using BLASTp (Altschul et al., 1990), which compared TP0326 to other bacterial proteins, and the SMART (Schultz et al., 1998) and Pfam (Finn et al., 2010) databases, which identified the POTRA and β-barrel domains of TP0326. ClustalW alignments of the β-barrel of TP0326 from various strains of T. pallidum were performed in MacVector (Cary, NC, v 11.1.0) using sequences in the NCBI database. To predict 3D models of the POTRA domains, the amino acid sequence of the 5 domains was submitted to the SWISS-MODEL server (Arnold et al., 2006). Three structures of the E. coli BamA POTRA domains solved by X-ray crystallography served as a templates (PDB IDs: 2QCZ, 3EFC, and 3OG5) (Kim et al., 2007; Gatzeva-Topalova et al., 2008; Gatzeva-Topalova et al., 2010). The program PyMOL (Schrödinger, LLC, New York, NY, v. 1.3) was used to create a model of all 5 POTRA domains aligning all three models centered on POTRA4 as recently described (Gatzeva-Topalova et al., 2010), calculate RMSD values, and generate the structural representations shown in Fig. 1.. The TbbPred (Natt et al., 2004) and PRED-TMBB (Bagos et al., 2004) servers were used to generate a topological model of the β-barrel domain of TP0326, while the TPRpred server (http://toolkit.tuebingen.mpg.de/tprpred/) identified tetratricopeptide repeats (TPR) in TP0622 and TP0954.
Cloning
The TP0326 POTRA (326P1-5, 326P1-4, and 326P1-4+) and β-barrel (326βB) constructs were PCR-amplified from T. pallidum DNA using the primers listed in Table 1. POTRA constructs, all with N-terminal histidine tags cleavable with thrombin, were cloned into the NdeI (5’-end) and EcoRI (3’-end) restriction sites of expression vector pET28a (Novagen, San Diego, CA). 326βB, which contains a C-terminal His-tag, was cloned into the NheI (5’-end) and XhoI (3’-end) restriction sites of pET23b (Novagen, San Diego, CA). Nucleotide sequencing was performed to confirm that the sequences of the protein constructs were correct.
Expression, purification and folding of the TP0326 β-barrel, E. coli BamA, and E. coli OmpG
POTRA constructs were expressed and purified as previously described (Cox et al., 2010). 326βB was expressed in the BL21(DE3) Rosetta-gami strain (Agilent Technologies, Inc., Santa Clara, CA). For each batch purification, 1 l of LB was inoculated with 50 ml of overnight culture grown at 37°C; IPTG (final concentration 0.1 mM) was added when the culture reached an optical density (600 nm) between 0.2 and 0.3. Cells were grown for an additional 3 h and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The pellets were resuspended with 20 ml of 50 mM Tris (pH 7.5), 100 μg of lysozyme, and 100 μl of PIC and stored at -20°C. After thawing, the bacterial suspension was lysed by sonication for three 30 s pulses interspersed with 30 s of rest on ice. The pellet was recovered by centrifugation at 20,000 × g for 30 min at 4°C and then incubated in solubilization buffer [100mM NaH2PO4; pH 8.0, 10mM Tris, 8M urea] for 30min at 4°C; the remaining insoluble material was removed by centrifugation at 20,000g for 30 min at 4 °C. The supernatant was added to Ni-NTA agarose matrix (Qiagen) that had been equilibrated in Solubilization Buffer and incubated with shaking at room temperature for 30 min. The matrix was washed with Wash Buffer (100mM NaH2PO4; pH 6.3, 10mM Tris, 8 M urea) and subsequently eluted with elution buffer (100mM NaH2PO4; pH 4.5, 10mM Tris, 8 M urea). SDS-PAGE and immunoblot analysis using the poly-histidine tag antibody were employed to identify the protein during purification and assess its purity (Fig. S1B). The purified protein was incubated in folding buffer (2% DDM, 100mM NaCl, 50mM Tris) for 24 h at 4°C to ensure complete folding of the protein. The samples were centrifuged at 20,000 × g for 30 min at 4°C to remove misfolded aggregates.
The expression vectors containing the E. coli ompGm2 (pET29A:OmpGm2) and E. coli bamA (pET15b::Ec-yaeT) genes were generous gifts from Jörg Kleinschmidt (Universität Konstanz). Both proteins were expressed, purified and folded as previously described (Qu et al., 2007; Cox et al., 2010). The Halobacterium salinarium membrane protein bacteriorhodopsin (Br) (Sigma-Aldrich) was resuspended in 50 mM HEPES (pH 7.5), 50 mM NaCl, and 2% OG (Anatrace).
Protein concentration was determined by measuring A280 in 20 mM sodium phosphate (pH 6.5) and 6 M guanidine hydrochloride (Edelhoch, 1967). The ProtParam tool provided by the ExPASy proteomics server (Gasteiger et al., 2003) was used to calculate the molar extinction coefficients (M-1 cm-1) of 326P1-4+, 326βB, E. coli BamA and OmpG, respectively.
Tryptophan fluorescence to monitor folding of 326βB and OmpG
Spectra were obtained using a Hitachi F-2500 fluorescence spectrophotometer with samples placed in a 5-mm path length quartz cell at 25 °C. The excitation wavelength was 295nm, and the bandwidth of the excitation monochromator was 2.5 nm. The folding buffers for 326βB and OmpG were 50mM Tris (pH7.5), 50mM NaCl, 0.5% DDM (DDM buffer) and 50mM Tris (pH7.5), 50mM NaCl, 2% OG, (OG buffer) respectively. Tryptophan emission spectra were recorded between 300 and 400 nm. Background spectra without 326βB and OmpG were subtracted to obtain the final emission curves.
Circular dichroism spectroscopy
CD analyses were performed using a Jasco J-715 spectropolarimeter (Jasco, Easton, MD). Far-UV CD spectra were acquired at 20°C in a 1 mm path-length cuvette, with a 1 nm bandwidth, 8 s response time, and a scan rate of 20 nm/min. Each protein spectra, representing the average of nine scans, were baseline corrected by subtracting the spectral attributes of the buffer. The DICHROWEB server was utilized to assess the secondary structure contents of the proteins from their spectra (Whitmore & Wallace, 2004).
Heat-modifiability
Recombinant proteins solubilized in SB were subjected to SDS-PAGE with or without boiling (10 min) followed by staining with GelCode® Blue. To generate native or denatured lysates from freshly harvested T. pallidum, respectively, samples were either incubated overnight at 4°C with 50 mM Tris (pH 7.0), 0.5% DDM, and 5% PIC (native lysis buffer) or incubated for 30 min at 25°C with SB+8M. Insoluble material was removed from the sample lysed with native lysis buffer by centrifugation at 20,000 g for 20 min at 4 °C. To examine the heat-modifiability of native TP0326, lysates solubilized in SB (native) or SB+8M (denatured) were split in half and one aliquot was boiled for 10 min. Subsequently, the lysates were resolved by SDS-PAGE and the gel including the stack was transferred to nitrocellulose membranes (0.45 μM pore size, GE Healthcare) at 25 V for 25 min using a semi-dry apparatus (Bio-Rad). Membranes were blocked for 1 h with PBS, 5% non-fat dry milk, 5% fetal bovine serum, and 0.1% Tween-20 and probed overnight at 4°C with primary antibodies directed against the POTRA domains or TroA at a dilution of 1:1,000 or 1:3,000, respectively. After washing with PBS and 0.05% Tween-20 (PBST), the membranes were incubated for 1 h at 4°C with a HRP-conjugated goat anti-rat antibody (Southern Biotech, Birmingham, AL) at dilutions of 1:30,000. Following washes with PBST, the immunoblots were developed using the SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific).
Triton X-114 phase-partitioning
Phase-partitioning of T. pallidum proteins with Triton X-114 has been described previously (Cox et al., 2010). Recombinant E. coli BamA (10 μg), OmpG (10 μg), 326P1-4+ (10 μg), and 326βB (1 μg) were added to 2% Triton X-114 in PBS supplemented with 0.5% PIC and incubated overnight at 4°C. The partitioned materials were phase-separated, and the detergent-enriched and aqueous phases were washed five times. All samples were precipitated with 10 volumes of acetone overnight at -80°C for subsequent SDS-PAGE analysis and staining with GelCode® Blue.
Incorporation into liposomes
All phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL) dissolved in chloroform. To simulate the OM lipid composition of T. pallidum, the lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phosphor-L-serine] (PS), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phosphor-rac-(1-glycerol)] (PG), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE) were mixed at a ratio of 69.3:17:13:0.7, respectively, dried at room temperature under argon, and placed in a vacuum for ~6 h. The lipids were rehydrated with 350 μl of 50mM HEPES (pH 7.5) and 50 mM NaCl (HEPES buffer), incubated at 37°C for 30 min, and resuspended by vortexing. To generate large unilamellar vesicles (LUV), the rehydrated lipid suspension was frozen in liquid N2 and then thawed at 37°C. After 5 freeze-thaw cycles, the sample was passed 21 times through a mini-extruder (Avanti Polar Lipids) equipped with a polycarbonate filter having a pore size of 0.1 μM. The resulting LUVs were stored at 4°C.
To incorporate OmpG and Br into liposomes, the proteins (1 μg each) were diluted in half to a 200 μl volume, mixed with 2 mg of LUVs, rapidly diluted to 4 ml with HEPES buffer, and subsequently dialyzed overnight at 4°C against 2 liters of HEPES buffer and 5 g of BIO-BEADS SM-2 (Bio-Rad). For the incorporation of 326βB into liposomes, 50 μl of a purified sample in 100mM NaH2PO4, (pH 4.5), 50 Tris and 8 M urea was diluted into 150 μl of 25 mM MES (pH 5.5), 0.1% β-mercaptoethanol, 0.03% DDM. LUVs (2 mg) were added to the sample, which was incubated on ice for 6 h and then diluted to 4 ml with 25 mM MES (pH 5.5). Following overnight incubations, the samples were cleared of precipitated material by centrifugation at 20,000 × g for 25 min at 4°C. Proteoliposomes were harvested by centrifugation at 125,000 × g for 30 min at 4°C and resuspended to remove residual detergent with either 200 μl of 25 mM Tris (pH 7.0), for OmpG and Br, or 200 μl of 25 mM MES (pH 5.5), for 326βB. A total of 5 μg of 326P1-4+ in 200 μl was mixed and incubated with LUVs, as described for 326βB; the LUV-protein mixture was neither diluted nor harvested. Sucrose gradient ultracentrifugation was employed to separate the proteoliposomes from un-incorporated material. Sucrose was added to the proteoliposomes to yield a 50% concentration. On top of this layer, two successive layers of 40 and 6% sucrose, in the corresponding resuspension buffer (above), were added and subsequently centrifuged at 300,000 × g for 1 h at 4 C. For Br, 326βB, and 326P1-4+, the proteoliposome and non-liposome fractions were assessed for protein content by immunoblotting (above). For OmpG, the fractions were precipitated with 10 volumes of ice-cold acetone overnight at -80°C for subsequent SDS-PAGE analysis and staining with GelCode® Blue.
Quantitative real-time reverse transcription (qRT)-PCR
Small pieces (approximately 1-2 mm3) of testicular tissue, freshly isolated from infected rabbits, were placed in a 500 μl aliquot of TRIzol (Invitrogen) immediately following extraction and homogenized using silicon carbide beads (Beadbeater; Biocore, MD). The TRizol-tissue suspension was transferred to a new microfuge tube. The beads then were washed with an additional 500 μl aliquot of TRIzol that was combined with the Trizol-tissue suspension. RNA was isolated according to the manufacturer's instructions. Contaminating genomic DNA was removed from RNA samples by treatment with 10 U of TurboDNA-free (Ambion, Austin, TX), followed by phenol-chloroform extraction and ethanol precipitation. DNA-free RNAs were stored at -80°C.
Total RNA (2 μg) was converted to cDNA in the presence and absence of reverse transcriptase using the Superscript III First-strand synthesis for RT-PCR kit (Invitrogen) with random hexamer primers according to the manufacturer's instructions. The resulting cDNA was amplified in an iCycler thermal cycler (Bio-Rad) using the gene-specific primer pairs listed in Table 1. Amplification of cDNAs was carried out in quadruplicate using 1 × iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions with the annealing temperature and concentration of MgCl2 optimized for each primer pair. Amplicons corresponding to each gene target were cloned into pCR®2.1-TOPO® (Invitrogen) and purified recombinant plasmid DNAs were diluted (107 to 102 copies/μl) to generate standard curves. Transcript copy numbers were calculated using the iCycler post-run analysis software based on internal standard curves. Values were background-subtracted using “No RT” and “No template” control reactions prior to being normalized against copies of flaA (tp0249) present in the same cDNA. To determine the statistical significance of observed differences between tp0326 and tp0117/tp0131 and tp0326 and tp0453, data points were compared within GraphPad Prism v. 5.00 (GraphPad Software, San Diego,CA) using an unpaired t-test with two-tailed P-values and a 95% confidence interval.
Quantitative immunoblot analysis
Freshly harvested treponemes (see above) were lysed with SB+8M, at a concentration of 5 × 107 T. pallidum per μl, and incubated at room temperature for 30 min. To quantitate the level of TP0326 in T. pallidum, lysates from 1.25 × 108 treponemes and graded amounts of recombinant 326P1-4+ were electrophoresed on 10% SDS polyacrylamide gels and transferred to nitrocellulose membranes as described above. Standard curves of 326P1-4+ were generated by densitometric analysis of the scanned immunoblots using ImageJ (NIH, v. 1.44c). The standard curve was used to calculate the total amount of native TP0326 in the resolved treponemal lysates. The amount of TP0326 per T. pallidum cell was determined by dividing the corresponding densitometrically derived value in the lysates by 1.25 × 108 T. pallidum. Copy numbers were determined by the molecular mass (MWTP0326 = 92,039 Da).
Surface accessibility of TP0326 to proteinase K (PK) digestion
To determine the lowest concentration of PK (Invitrogen) required to achieve surface proteolysis of TP0326, freshly isolated organisms (5 × 108 / ml of CMRL), representing intact treponemes, were subjected to digestion with graded concentrations (0.1-10 μg) of PK for 1 h. All steps were performed at room temperature unless stated otherwise. For proteolysis of periplasmic controls, spirochetes were harvested by centrifugation at 10,000 × g for 20 min, resuspended with 200 μl of PK lysis buffer [50 mM Tris (pH 7.0), 0.5% Triton X-100, 0.1% β-mercaptoethanol, and 50 μg of lysozyme (Sigma-Aldrich)], incubated for 1 h, and then treated with 10 μg of PK for 1 h. The breakdown of the PG layer by lysozyme was found to be necessary for the complete disruption of the periplasmic compartment (Izard et al., 2009; Liu et al., 2010). The activity of the protease was stopped upon the addition of phenylmethylsulfonyl fluoride (PMSF) to 1 mg / ml. Intact treponemes were pelleted by centrifugation at 20,000 × g for 20 min and subsequently resuspended with SB+8M, while detergent-lysozyme treated organisms were resuspended with SB+8M. Immunoblotting was performed as described above.
To assess motility, aliquots (10 μl) of each sample immediately following PK treatment (1 h incubation) were transferred onto a glass microscope slide and gently overlaid with coverglass, and then viewed by darkfield on an Olympus BX41 microscope (Center Valley, PA) using a 100 × (1.4 NA) oil immersion objective. The motility of the organisms was observed visually and recorded using a Retiga Exi CCD camera (QImaging, Surrey, BC, Canada) and StreamPix (NorPix, Montreal, QC, Canada) software. ImageJ was used to adjust the brightness and contrast of the images. Images were converted into movies using QuickTime Pro (Apple Inc., Cupertino, CA, v. 7.0).
Size exclusion chromatography (SEC) of native TP0326
Approximately 2.5 × 109 of freshly harvested spirochetes were lysed with 50 mM Tris (pH 7.0), 2.0% DDM, and 5% PIC, cleared of insoluble material by centrifugation at 20,000 × g for 20 min, and subsequently loaded onto an Superdex™ 200 10/300 analytical grade size-exclusion column (GE Healthcare BioSciences) that had been equilibrated with 50 mM Tris (pH 7.0) and 2% DDM. Fractions of 100 μl were collected, resolved on a 10% SDS-polyacrylamide gel, and then transferred onto a nitrocellulose membrane for immunoblotting (above).
Blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis of native and recombinant BamA complexes
To determine the appropriate detergent concentration for lysing T. pallidum under native conditions, freshly harvested treponemes were solubilized overnight at 4°C with 50 mM Tris (pH 7.0), 0.5-4.0% DDM, and 5% PIC. Prior to electrophoresis, lysates were cleared of detergent insoluble material by centrifugation at 20,000 × g for 20 min at 4°C. Native lysates consisting of 5.0 × 108 to 1.0 × 109 spirochetes were resolved in a 4-12% Bis-Tris acrylamide gel (Bio-Rad) at 4°C using the BN-PAGE method (Schagger & von Jagow, 1991; Wittig et al., 2006). The cathode buffer [50 mM tricine (pH 7.0) and 15 mM bis-tris] contained 0.02% Coomassie brilliant blue G-250 (CBB-G250) for the first 1/3rd of the run, after which the gel was run with fresh cathode buffer without CBB-G250. For the duration of the run, the anode buffer consisted of 50 mM bis-tris (pH 7.0). Resolved lysates were transferred to a nitrocellulose membrane in 50 mM tricine (pH 7.0) followed by immunoblotting using POTRA antiserum. Samples containing 326βB were diluted to reduce the concentration of DDM to 0.5 % or 1% and incubated on ice for 30 min prior to BN-PAGE. The resolved samples were transferred as above to nitrocellulose followed by immunoblotting using poly-histidine tag antiserum.
Mass-spectrometry of TP0326 complexes
For mass-spectrometry analysis, native lysates in 0.5% DDM were subjected to BN-PAGE and half of the gel was used for immunoblotting (see above) which served as a guide to localize TP0326 within the gel. The region of the gel containing the un-dissociated complex was divided into 4 slices, and all were submitted to the University of Victoria Genome BC Proteomics Centre (Victoria, BC, Canada) for in-gel trypsin digestion and liquid chromatography-electrospray ionization tandem mass spectrometry (LC-MS/MS) analysis. Briefly, protein bands were de-stained in a solution of 1M ammonium bicarbonate (Sigma-Aldrich) and 20% acetonitrile (Thermo Fisher Scientific) followed by a solution of 50% methanol (Thermo Fisher Scientific) and 5% acetic acid (Anachemia, Montreal, QC, Canada). Proteins were reduced using 10mM DTT (dithiothreitol; Fluka Chemical Company, Ronkonkoma, NY, USA) and alkylated using 100mM iodoacetamide (Fluka). Proteins were digested overnight using a 20ng/μl solution (in 50 mM ammonium bicarbonate) of porcine modified trypsin (Promega, Madison, WI, USA), and peptides were extracted with successive washes of 50% acetonitrile (Thermo Fisher Scientific), 10% formic acid (Sigma-Aldrich), and 50mM ammonium bicarbonate (Sigma-Aldrich). The resulting peptides were analyzed by LC-MS/MS as previously described (Eshghi et al., 2009).
Supplementary Material
Figure S1. bamA loci of spirochetes. The bamA loci of T. pallidum, T. denticola, B. burgdorferi, and L. interrogans aligned to those of N. meningitidis and E. coli. Dark green arrows indicate bamA. The periplasmic OMP chaperone skp is represented as a light green arrow. Red arrows highlight rseP, a zinc IM-protease involved in the extracytoplasmic stress response mediated by the sigma factor rpoE.
Figure S2. Minimal variance of TP0326 β-barrel sequences. ClustalW alignment of TP0326 β-barrel sequences from geographically diverse T. pallidum strains (on the left). The dark and light shading indicates identical and similar residues, respectively. As shown in Fig. 2C, green and red bars demarcate the predicted intra- and extracellular loops, respectively, whereas blue arrows indicate predicted membrane-spanning β-strands. The sequence for the Nichols strain (Fraser et al., 1998) is identical to that of the Nichols-Farmington strain used in this study. Accession numbers: Nichols AAC65313; Chicago ADD72452; SS14 ACD70752; Baltimore-2 and -3 AAF61468 and AAF61469, respectively; Sea81-3 AAF61471; Haiti-b AAF61473; Dallas-1 ABW94728; Grady ABW94731, Chi-b ABW94727; South Africa ABW94745; Philadelphia-1 ABW94738; Mexica-a ABW94735; Madras ABW94734. Only partial sequences are available for strains Dallas-1 through Madras (Harper et al., 2008).
Figure S3. Purification of 326P1-4+ 326βb. (A) SEC elution profile and SDS-PAGE analysis (inset) of purified of 326P1-4+. The nominal MW of 326P1-4+ was calculated based on a calibration curve. (B) SDS-PAGE of purified 326βB. (C) CD spectrum of purified 326P1-4+.
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grants AI-26756 (J.D.R.), AI-051334 (C.E.C), and 5R03TW008023 (J.C.S.) from the National Institutes of Health, by an award from the Michael Smith Foundation for Health Research (CEC), and by the Connecticut Children's Medical Center (CCMC) Arrison and Burr Curtis Research Funds (J.C.S.). S. D.-E. is the recipient of a career development award from the Northeastern Research Center for Excellence (NIH grant U54 AI057159).
REFERENCES
- Akins DR, Robinson E, Shevchenko D, Elkins C, Cox DL, Radolf JD. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved signal sequence to the Treponema pallidum cytoplasmic membrane. J. Bacteriol. 1997;179:5076–5086. doi: 10.1128/jb.179.16.5076-5086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anwari K, Poggio S, Perry A, Gatsos X, Ramarathinam SH, Williamson NA, Noinaj N, Buchanan S, Gabriel K, Purcell AW, Jacobs-Wagner C, Lithgow T. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus. PLoS One. 2010;5:e8619. doi: 10.1371/journal.pone.0008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtluft P, Nouwen N, Tans SJ, Driessen AJ. SecB--a chaperone dedicated to protein translocation. Mol Biosyst. 2010;6:620–627. doi: 10.1039/b915435c. [DOI] [PubMed] [Google Scholar]
- Belisle JT, Brandt ME, Radolf JD, Norgard MV. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J. Bacteriol. 1994;176:2151–2157. doi: 10.1128/jb.176.8.2151-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennion D, Charlson ES, Coon E, Misra R. Dissection of B-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E. Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J. Biol. Chem. 2007;282:1882–1890. doi: 10.1074/jbc.M609598200. [DOI] [PubMed] [Google Scholar]
- Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 2006;44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusca JS, Radolf JD. Isolation of integral membrane proteins by phase partitioning with Triton X-114. Methods Enzymol. 1994;228:182–193. doi: 10.1016/0076-6879(94)28019-3. [DOI] [PubMed] [Google Scholar]
- Burgess NK, Dao TP, Stanley AM, Fleming KG. B-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE. The T. pallidum outer membrane and outer membrane proteins. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponema: Molecular and Cellular Biology. Caister Academic Press; Norwich,UK: 2006. pp. 237–266. [Google Scholar]
- Cameron CE, Lukehart SA, Castro C, Molini B, Godornes C, Van Voorhis WC. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J. Infect. Dis. 2000;181:1401–1413. doi: 10.1086/315399. [DOI] [PubMed] [Google Scholar]
- Conlan S, Bayley H. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry (Mosc) 2003;42:9453–9465. doi: 10.1021/bi0344228. [DOI] [PubMed] [Google Scholar]
- Cox DL, Luthra A, Dunham-Ems S, Desrosiers DC, Salazar JC, Caimano MJ, Radolf JD. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect. Immun. 2010;78:5178–5194. doi: 10.1128/IAI.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz AR, Moore MW, La Vake CJ, Eggers CH, Salazar JC, Radolf JD. Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect. Immun. 2008;76:56–70. doi: 10.1128/IAI.01039-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz AR, Pillay A, Zuluaga AV, Ramirez LG, Duque JE, Aristizabal GE, Fiel-Gan MD, Jaramillo R, Trujillo R, Valencia C, Jagodzinski L, Cox DL, Radolf JD, Salazar JC. Secondary syphilis in cali, Colombia: new concepts in disease pathogenesis. PLoS Negl Trop Dis. 2010;4:e690. doi: 10.1371/journal.pntd.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Deka RK, Machius M, Norgard MV, Tomchick DR. Crystal structure of the 47-kilodalton lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein. J. Biol. Chem. 2002;277:41857–41864. doi: 10.1074/jbc.M207402200. [DOI] [PubMed] [Google Scholar]
- Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 2009;1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry (Mosc) 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Eshghi A, Cullen PA, Cowen L, Zuerner RL, Cameron CE. Global proteome analysis of Leptospira interrogans. J Proteome Res. 2009;8:4564–4578. doi: 10.1021/pr9004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsos X, Perry AJ, Anwari K, Dolezal P, Wolynec PP, Likic VA, Purcell AW, Buchanan SK, Lithgow T. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol. Rev. 2008;32:995–1009. doi: 10.1111/j.1574-6976.2008.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayet-Ageron A, Ninet B, Toutous-Trellu L, Lautenschlager S, Furrer H, Piguet V, Schrenzel J, Hirschel B. Assessment of a real-time PCR test to diagnose syphilis from diverse biological samples. Sex. Transm. Infect. 2009;85:264–269. doi: 10.1136/sti.2008.034314. [DOI] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Haake DA, Matsunaga J. Leptospira: a spirochaete with a hybrid outer membrane. Mol. Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy PH, Jr., Nell EE. Study of the antigenic structure of Treponema pallidum by specific agglutination. Am J Hyg. 1957;66:160–172. doi: 10.1093/oxfordjournals.aje.a119893. [DOI] [PubMed] [Google Scholar]
- Harper KN, Ocampo PS, Steiner BM, George RW, Silverman MS, Bolotin S, Pillay A, Saunders NJ, Armelagos GJ. On the origin of the treponematoses: a phylogenetic approach. PLoS.Negl.Trop.Dis. 2008;2:e148. doi: 10.1371/journal.pntd.0000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Cox DL, Decaffmeyer M, Bennett MP, Desrosiers DC, La Vake CJ, La Vake ME, Bourell KW, Robinson EJ, Brasseur R, Radolf JD. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 2005;187:6499–6508. doi: 10.1128/JB.187.18.6499-6508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Sellati TJ, Nguyen TT, Cox DL, Clawson ML, Caimano MJ, Radolf JD. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 2001;193:1015–1026. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuck AP, Johnson AE. Pore-forming protein structure analysis in membranes using multiple independent fluorescence techniques. Cell Biochem. Biophys. 2002;36:89–101. doi: 10.1385/CBB:36:1:89. [DOI] [PubMed] [Google Scholar]
- Hoenger A, Pages JM, Fourel D, Engel A. The orientation of porin OmpF in the outer membrane of Escherichia coli. J. Mol. Biol. 1993;233:400–413. doi: 10.1006/jmbi.1993.1520. [DOI] [PubMed] [Google Scholar]
- Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 2009;191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad Z, Paoli M. Novel sequences propel familiar folds. Structure. 2002;10:447–454. doi: 10.1016/s0969-2126(02)00750-5. [DOI] [PubMed] [Google Scholar]
- Jun HK, Kang YM, Lee HR, Lee SH, Choi BK. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect. Immun. 2008;76:2428–2438. doi: 10.1128/IAI.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex. J. Mol. Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Kruger V, Prinz C, Meisinger C, Guiard B, Wagner R, Pfanner N, Wiedemann N. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Lafond RE, Lukehart SA. Biological basis for syphilis. Clin. Microbiol. Rev. 2006;19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol. Microbiol. 2010;75:692–709. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang B, Feng L, Kang H, Qi Y, Wang J, Shi Y. Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage. Proceedings of the National Academy of Sciences. 2009;106:14837–14842. doi: 10.1073/pnas.0903289106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J. Mol. Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukehart SA. Scientific monogamy: thirty years dancing with the same bug: 2007 Thomas Parran Award Lecture. Sex. Transm. Dis. 2008;35:2–7. doi: 10.1097/OLQ.0b013e318162c4f2. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- Martin IE, Tsang RS, Sutherland K, Tilley P, Read R, Anderson B, Roy C, Singh AE. Molecular characterization of syphilis in patients in Canada: azithromycin resistance and detection of Treponema pallidum DNA in whole-blood samples versus ulcerative swabs. J. Clin. Microbiol. 2009;47:1668–1673. doi: 10.1128/JCM.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKevitt M, Brinkman MB, McLoughlin M, Perez C, Howell JK, Weinstock GM, Norris SJ, Palzkill T. Genome scale identification of Treponema pallidum antigens. Infect. Immun. 2005;73:4445–4450. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Fronzes R, Chandran V, Remaut H, Waksman G. Protein oligomerization in the bacterial outer membrane (Review). Mol. Membr. Biol. 2009;26:136–145. doi: 10.1080/09687680802712422. [DOI] [PubMed] [Google Scholar]
- Metzger M, Hardy PH, Jr., Nell EE. Influence of lysozyme upon the treponeme immobilization reaction. Am J Hyg. 1961;73:236–244. doi: 10.1093/oxfordjournals.aje.a120182. [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- Moore MW, Cruz AR, LaVake CJ, Marzo AL, Eggers CH, Salazar JC, Radolf JD. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun. 2007;75:2046–2062. doi: 10.1128/IAI.01666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, Monteiro-Vitorello CB, Ho PL, Haake DA, Verjovski-Almeida S, Hartskeerl RA, Marques MV, Oliveira MC, Menck CF, Leite LC, Carrer H, Coutinho LL, Degrave WM, Dellagostin OA, El Dorry H, Ferro ES, Ferro MI, Furlan LR, Gamberini M, Giglioti EA, Goes-Neto A, Goldman GH, Goldman MH, Harakava R, Jeronimo SM, Junqueira-de-Azevedo IL, Kimura ET, Kuramae EE, Lemos EG, Lemos MV, Marino CL, Nunes LR, de Oliveira RC, Pereira GG, Reis MS, Schriefer A, Siqueira WJ, Sommer P, Tsai SM, Simpson AJ, Ferro JA, Camargo LE, Kitajima JP, Setubal JC, Van Sluys MA. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natt NK, Kaur H, Raghava GP. Prediction of transmembrane regions of beta-barrel proteins using ANN- and SVM-based methods. Proteins. 2004;56:11–18. doi: 10.1002/prot.20092. [DOI] [PubMed] [Google Scholar]
- Nelson RA, Jr., Mayer MM. Immobilization of Treponema pallidum in vitro by antibody produced in syphilitic infection. J. Exp. Med. 1949;89:369–393. doi: 10.1084/jem.89.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesper J, Brosig A, Ringler P, Patel GJ, Muller SA, Kleinschmidt JH, Boos W, Diederichs K, Welte W. Omp85(Tt) from Thermus thermophilus HB27: an ancestral type of the Omp85 protein family. J. Bacteriol. 2008;190:4568–4575. doi: 10.1128/JB.00369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- Parsonage D, Desrosiers DC, Hazlett KR, Sun Y, Nelson KJ, Cox DL, Radolf JD, Poole LB. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6240–6245. doi: 10.1073/pnas.0910057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J. Mol. Biol. 2007;374:91–105. doi: 10.1016/j.jmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Radolf JD. Treponema pallidum and the quest for outer membrane proteins. Mol. Microbiol. 1995;16:1067–1073. doi: 10.1111/j.1365-2958.1995.tb02332.x. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Lukehart SA. Immunology of Syphilis. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponemes: Cellular and Molecular Biology. Caister Academic Press; Norfolk, UK: 2006. pp. 285–322. [Google Scholar]
- Radolf JD, Norgard MV, Schulz WW. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2051–2055. doi: 10.1073/pnas.86.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud J-L, Lévy D. Reconstitution of membrane proteins into liposomes. In: Nejat D, editor. Methods Enzymol. Academic Press; 2003. pp. 65–86. [DOI] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE. B-Barrel membrane proteins. Curr. Opin. Struct. Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Schulz GE. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Smajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Surana NK, Grass S, Hardy GG, Li H, Thanassi DG, Geme JW., 3rd Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14497–14502. doi: 10.1073/pnas.0404679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm LK, Hong H, Liang B. Folding and assembly of B-barrel membrane proteins. Biochim. Biophys. Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Tommassen J. Assembly of outer-membrane proteins in bacteria and mitochondria. Microbiology. 2010;156:2587–2596. doi: 10.1099/mic.0.042689-0. [DOI] [PubMed] [Google Scholar]
- Tomson FL, Conley PG, Norgard MV, Hagman KE. Assessment of cell-surface exposure and vaccinogenic potentials of Treponema pallidum candidate outer membrane proteins. Microbes Infect. 2007;9:1267–1275. doi: 10.1016/j.micinf.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhis WC, Barrett LK, Lukehart SA, Schmidt B, Schriefer M, Cameron CE. Serodiagnosis of syphilis: antibodies to recombinant Tp0453, Tp92, and Gpd proteins are sensitive and specific indicators of infection by Treponema pallidum. J. Clin. Microbiol. 2003;41:3668–3674. doi: 10.1128/JCM.41.8.3668-3674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volokhina EB, Beckers F, Tommassen J, Bos MP. The B-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 2009;191:7074–7085. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Walker EM, Zampighi GA, Blanco DR, Miller JN, Lovett MA. Demonstration of rare protein in the outer membrane of Treponema pallidum subsp. pallidum by freeze-fracture analysis. J. Bacteriol. 1989;171:5005–5011. doi: 10.1128/jb.171.9.5005-5011.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D, Tommassen J. Biogenesis of beta-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell. Mol. Life Sci. 2009;66:2789–2804. doi: 10.1007/s00018-009-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J, Augustus AM, Misra R. Assembly of TolC, a structurally unique and multifunctional outer membrane protein of Escherichia coli K-12. J. Bacteriol. 2003;185:6540–6547. doi: 10.1128/JB.185.22.6540-6547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC. The versatile B-barrel membrane protein. Curr. Opin. Struct. Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. bamA loci of spirochetes. The bamA loci of T. pallidum, T. denticola, B. burgdorferi, and L. interrogans aligned to those of N. meningitidis and E. coli. Dark green arrows indicate bamA. The periplasmic OMP chaperone skp is represented as a light green arrow. Red arrows highlight rseP, a zinc IM-protease involved in the extracytoplasmic stress response mediated by the sigma factor rpoE.
Figure S2. Minimal variance of TP0326 β-barrel sequences. ClustalW alignment of TP0326 β-barrel sequences from geographically diverse T. pallidum strains (on the left). The dark and light shading indicates identical and similar residues, respectively. As shown in Fig. 2C, green and red bars demarcate the predicted intra- and extracellular loops, respectively, whereas blue arrows indicate predicted membrane-spanning β-strands. The sequence for the Nichols strain (Fraser et al., 1998) is identical to that of the Nichols-Farmington strain used in this study. Accession numbers: Nichols AAC65313; Chicago ADD72452; SS14 ACD70752; Baltimore-2 and -3 AAF61468 and AAF61469, respectively; Sea81-3 AAF61471; Haiti-b AAF61473; Dallas-1 ABW94728; Grady ABW94731, Chi-b ABW94727; South Africa ABW94745; Philadelphia-1 ABW94738; Mexica-a ABW94735; Madras ABW94734. Only partial sequences are available for strains Dallas-1 through Madras (Harper et al., 2008).
Figure S3. Purification of 326P1-4+ 326βb. (A) SEC elution profile and SDS-PAGE analysis (inset) of purified of 326P1-4+. The nominal MW of 326P1-4+ was calculated based on a calibration curve. (B) SDS-PAGE of purified 326βB. (C) CD spectrum of purified 326P1-4+.