Abstract
Histone deacetylase inhibitors (HDACi) have been proposed as therapies for certain cancers and as an anti-reservoir therapy for HIV+ individuals with HAART, yet, their roles in glial inflammatory and innate antiviral gene expression have not been defined. In this study, we examined the effects of two non-selective HDACi, trichostatin A and valproic acid, on antiviral and cytokine gene expression in primary human microglia and astrocytes stimulated with TLR3 or TLR4 ligand. HDACi potently suppressed the expression of innate antiviral molecules such as IFNβ, interferon-simulated genes, and proteins involved in TLR3/TLR4 signaling. HDACi also suppressed microglial and astrocytic cytokine and chemokine gene expression, but with different effects on different groups of cytokines. These results have important implications for the clinical use of HDACi.
INTRODUCTION
In the central nervous system (CNS), astrocytes and microglia constitute important modulators of immune and inflammatory reactions, participating in both innate and acquired immune responses. Astrocytes and microglia express a number of toll-like-receptor (TLR) family member proteins, such as TLR3 and TLR4 (Carpentier et al., 2008; Rivieccio et al., 2006; Suh et al., 2007; Suh et al., 2009b). Ligand binding to TLR receptors activates specific sets of transcription factors that lead to the production of secretory inflammatory mediators and antimicrobial factors (Akira et al., 2006; Hiscott et al., 2006; Suh et al., 2009a). For example, TLR3 and TLR4 signal through an adaptor called toll/interleukin-1 receptor domain-containing adaptor protein inducing IFNβ (TRIF), which then activates the transcription factor IFN regulatory factor 3 (IRF3). IRF3 in turn activates the IFNβ gene, which then sets off the secondary wave of interferon-stimulated gene (ISG) production through induction of additional transcription factors such as IRF7. Astrocytes or microglia stimulated with synthetic dsRNA polyinosinic-polycytidylic acid (PIC: TLR3 ligand) or microglia activated with lipopolysaccharide (LPS: TLR4 ligand) show induction of many ISGs, as well as cytokine and chemokine genes. TLR3/4 activation in glial cells also leads to induction of antiviral activity against intracellular pathogens such as HIV and HCMV in an IRF3- and IFNβ-dependent manner.
Acetylation of various histones has emerged as an important posttranslational modification involved in regulation of gene expression or silencing (Dokmanovic and Marks, 2005; Minucci and Pelicci, 2006; Monneret, 2005). Histone acetylation is regulated by histone acetyl transferases (HATs) and histone deacetylases (HDACs). In general, HATs induce gene transcription whereas HDACs suppress transcription. The human class I HDACs (nuclear localized proteins of 22–55 kDa) include HDAC 1, 2, 3 and 8, and class II HDACs (nuclear and cytosolic localized proteins of 120–135 kDa) include HDAC 4, 5, 6, and 9. The class III HDACs are SIRT 1–7, the human homologues of mouse sirt 2, and do not have histone deacetylase activity. Most non-selective HDAC inhibitors (HDACi) such as trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA) inhibit all class I and class II HDACs but not class III HDACs. Owing to the general activities of HDACi on tumor cells (inhibition of growth and differentiation, and promotion of apoptosis), many are being developed as anti-cancer therapies (Dokmanovic and Marks, 2005; Marks and Xu, 2009; Minucci and Pelicci, 2006; Monneret, 2005). For example, SAHA is now approved for treatment of cutaneous lymphomas (Marks, 2007). Additional areas in which HDACi are being investigated include “HIV anti-reservoir” therapy. For example, valproic acid (VA) has been promoted as an adjunct therapy for viral eradication in HIV-infected individuals receiving highly active anti-retroviral therapy (HAART) (Archin et al., 2009; Lehrman et al., 2005). Since HIV can hide in the host cell in a latent form (reservoirs) in individuals with HAART, HDACi can induce transcription of the virus rendering them once again susceptible to antiviral therapy.
Although initially defined as enzymes facilitating deacetylation of histones, more recent studies have indicated that HDACs also deacetylate non-histone proteins, and that they act to both enhance and inhibit gene transcription. For example, histone deacetylase activity is required for STAT1 and STAT5 signaling (Klampfer et al., 2004; Rascle et al., 2003) and TSA, SAHA and VA suppress type I IFN-stimulated anti-viral gene expression in several different systems (Chang et al., 2004; Genin et al., 2003; Nusinzon and Horvath, 2003; Sakamoto et al., 2004; Vlasakova et al., 2007). In contrast to their consistent inhibitory activities on the innate antiviral immune response, the effects of HDACi on inflammation and cell activation are more complex. While some studies have shown cytokine-enhancing effects of HDACi (Chen et al., 2001; Kiernan et al., 2003; Mahlknecht et al., 2004; Suuronen et al., 2003), others have reported suppression of cytokine and nitric oxide production, including in rodent models of trauma, experimental autoimmune encephalomyelitis, ischemic stroke, and Parkinson’s disease (Camelo et al., 2005; Kim et al., 2007; Leoni et al., 2005; Peng et al., 2005; Zhang et al., 2008). Studies of microglia are rare and one such study in murine microglia reported that HDACi increased cytokine and nitric oxide production (Suuronen et al., 2003).
Despite the current and proposed use of HDACi in clinical settings, their role on human glial cytokine production and innate antiviral protein expression have not been defined. Using a well-established model of primary human glial cell cultures, we investigated the effect of TSA and VA on PIC- and LPS-induced gene expression. We hypothesized that HDACi will inhibit transcription of the genes involved in innate immune responses that are dependent on IRF3, and we indeed confirm these findings in both microglia and astrocytes. In addition, HDACi also suppressed microglial and astrocytic cytokine and chemokine gene expression, but with different effects on different groups of cytokines. These results have important implications for the clinical use of HDACi.
MATERIALS AND METHODS
Human fetal brain cell culture
Human CNS cell cultures were prepared from human fetal abortuses as described (Lee et al., 1992; Suh et al., 2005). All tissue collection was approved by the Albert Einstein College of Medicine Institutional Review Board. Primary mixed CNS cultures were prepared by enzymatic and mechanical dissociation of the cerebral tissue followed by filtration through nylon meshes of 230- and 130-μ pore sizes. Single cell suspension was plated at 1–10 × 106 cells per ml in DMEM (Cellgro: Dulbecco’s modified Eagle medium) supplemented with 5% fetal calf serum (FCS: Gemini Bio-products, Woodland, CA), penicillin (100U/ml), streptomycin (100μg/ml) and fungizone (0.25μg/ml) (Gibco) for 2 weeks, and then microglial cells were collected by aspiration of the culture medium. Monolayers of microglia were prepared in 60-mm or 100mm tissue culture dishes at 1 × 106 cells per 10 ml medium or in 96-well tissue culture plates at 104 per 0.1 ml medium. Two to four hours later, cultures were washed twice to remove non-adherent cells (neurons and astrocytes). Microglial cultures were highly pure consisting of > 98% CD68+ cells. Highly enriched human astrocyte cultures were generated by repeated passage of the mixed CNS cultures, as described previously (Lee et al., 1992; Liu et al., 1996). Cells were kept as monolayers in culture media that consisted of DMEM with 5% FCS and antibiotics.
Cell treatment with stimulants and inhibitors
PIC and LPS were purchased from Sigma (St. Louis, MO) and was diluted in endotoxin-free sterile phosphate buffered saline (PBS) and used at 20μg/ml or 100 ng/ml, respectively. IFN-s was purchased from PBL Biomedical Laboratories (Piscataway, NJ; 1 ng = 80 U) and used at 10ng/ml. TSA, VA, and SB216763 were purchased from Sigma. TSA was diluted in DMSO at 2 mg/ml (stock solution). Further dilutions were made in culture medium containing FCS to final concentrations of 50–500 ng/ml. VA was diluted in sterile H2O and used at 1–8 mM final concentrations. SB216763 was diluted in DMSO, and used at 10 μM. Microglia or astrocytes were grown to confluence in 6 cm plastic Petri dish for protein or RNA extraction or in 96-well tissue culture plates for ELISA. Cells were pretreated with inhibitors for 1 h, and then treated with IFNβ, LPS or PIC for an addition 6 hour (real-time PCR or microarray analysis) or 24 h (Western blot or ELISA).
Western blot analysis
Briefly, cell cultures in 60 mm dishes were scraped into lysis buffer (10 mM Tris-HCl [pH 8.8], 50 mM NaCl, 0.5 mM Na3VO4, 30 mM Na4P2O7, 50 mM NaF, 2 mM EDTA, 1% Triton X-100) at various time points. Thirty to seventy micrograms of protein was separated by 10% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membrane. The blots were blocked in Tris-buffered saline-0.1% Tween-20 containing5% nonfat milk and then incubated with antibodies at 4°C for 16 h (Suh et al., 2007). Primary Antibody, IDO was used at 1/3000 (gift of Osamu Takikawa)(Suh et al., 2007). The secondary antibody was horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG and was used at 1:2,000 to 1:10,000for 1 h at room temperature. Signals were developed using enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL). All blots were reprobed for vinculin (117 kDa) or β–actin using antibodies from Santa Cruz to control for protein loading. Densitometric analysis was performed using Scion NIH Image software (Scion, Frederick, MD).
Real time PCR
Quantitative real-time reverse transcription-PCR (Q-PCR) was performed as described previously (Rivieccio et al., 2005; Rivieccio et al., 2006; Suh et al., 2007), using porphobilinogen deaminase (PBDA) or GAPDH as internal controls. Primer sequences for TLR3, IFN-s, IDO, viperin, IRF3, TNFα, IP-10 were published previously (Rivieccio et al., 2006; Suh et al., 2007; Suh et al., 2009b). Briefly, total RNA was extracted from microglia in culture with TRIzol (Invitrogen Life Technologies), following the manufacturer’s instructions. PCR was performed using a SYBR green PCR mix and conducted with the ABI Prism 7900HT (Applied Biosystems). All values were expressed as the increase relative to the expression of PBDA. The mean value of the replicates for each sample was calculated and expressed as the cycle threshold (CT; cycle number at which each PCR reaches a predetermined fluorescence threshold, set within the line arrange of all reactions). The amount of gene expression was then calculated as the difference between the CT of the sample for the target gene and the mean CT of that sample for the endogenous control.
ELISA
Cell culture supernatants or lysates (for IL-1β only) were harvested and subjected to protein assay using commercial ELISA kits for cytokines and chemokines as previously described (Lee et al., 1992; McManus et al., 2000). Standard curves were generated with known concentrations of recombinant cytokines and chemokines, and the samples were diluted until the optical reading (OD) values fall within the range of the ELISA detection.
Human cDNA 28k microarrays
Astrocyte-enriched cultures plated at 2 × 106 cells/100 mm dish were pretreated with 400 ng/ml TSA for 1h, then with 20 μg/ml PIC for an additional 6h in DMEM containing 5% FCS. Cell harvest, RNA isolation, and microarrays (obtained from the Albert Einstein College of Medicine cDNA Microarray Facility: http://microarray1k.aecom.yu.edu) were performed as described previously (John et al., 2005; Rivieccio et al., 2005; Rivieccio et al., 2006). Approximately 5 μg of total RNA was used for T7 linear amplification. Linear amplification of total RNA and subsequent fluorescent labeling of corresponding cDNA was carried out using the Message Amp T7 linear amplification kit (Ambion) and cDNA labeling protocols developed at the AECOM Microarray Facility. Hybridization to cDNA arrays was carried out overnight at 50 C in a buffer containing 30% formamide, 3X SSC, 0.75% SDS and 100 ng of human Cot-1 DNA. Following hybridization, slides were briefly washed with a solution of 1X SSC, 0.1% SDS, then washed for 20 min at room temperature in 0.2X SSC, 0.1% SDS and 20 min at room temperature in 0.1X SSC (without SDS). Independent measurements of Cy5 and Cy3 signal intensity and background were generated for each cDNA element using a Genepix 4000B scanner (Axon Instruments) and Scanalyze software (http://rana.lbl.gov/EisenSoftware.htm). The ratio of the fluorescence intensities of the two dyes represented a measure of differential gene expression between the individual treatments and their corresponding controls. Cutoff values were established as Cy5:Cy3 ratio >1.5 or <0.67and as net signal intensity more than three times background in one or both channels. All elements not satisfying both criteria were discarded. Controls included reversing the fluorochromes and labeling the same sample using both Cy3 and Cy5.
Statistical Analysis
Results shown are pooled data from two to four independent experiments using cells from different brain cases. Values (protein levels by ELISA or mRNA levels by Q-PCR) were normalized to those induced by LPS or PIC within the same experiment, then all values from multiple different experiments were pooled. One-way analysis of variance (ANOVA) with Dunnett’s post test was performed to determine which drug treatment conditions significantly modulated the gene expression, using the GraphPad Prism 4.0 software.
RESULTS
Effects of HDACi on microglial cytokine protein production
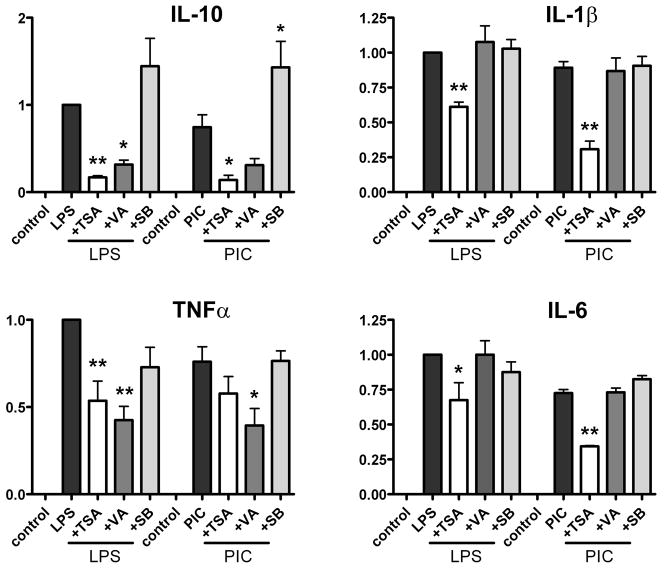
We examined the role of TSA and VA on LPS- and PIC-induced microglial proinflammatory and anti-inflammatory cytokine production by ELISA (Figure 1). Since VA (in addition to HDACi activity) has been shown to inhibit GSK3 (Dewhurst et al., 2007; Dou et al., 2003; Maggirwar et al., 1999), we tested a cell-permeable inhibitor of GSK3 (SB216763) in parallel. The inhibitor concentrations (TSA at 400 ng/ml, VA at 4 mM, and SB216763 at 10 μM) were chosen based on known IC50 (Dokmanovic and Marks, 2005; Minucci and Pelicci, 2006) and previous reports. Microglial cultures were pretreated with the inhibitors for 1 h, and then treated with the TLR ligands for an additional 24h. ELISA was performed for IL-10, IL-1β, TNFα and IL-6. Figure 1 shows pooled data from four separate microglial cases for IL-10, IL-1β and TNFα, and two separate cases for IL-6.
Figure 1. The effect of HDAC inhibitors on cytokine protein production from human microglia.
Primary human fetal microglial cultures were prepared in triplicates in DMEM 5% FCS with antibiotics, as described. Cells were treated with the inhibitors (TSA 400 ng/ml, VA 4 mM or SB216763 10 μg/ml) for 1 h, then with the TLR ligands (LPS at 100 ng/ml or PIC at 20 μg/ml) for 24 h and ELISA was performed as described in the Materials and Methods. All values (ng/ml) were normalized to LPS-induced amounts (LPS = 1) and data from multiple experiments were pooled. Each experiment was derived from different microglial case (n= 4 for IL-10, IL-1β and TNFα, and n = 2 for IL-6). Data are Mean ± SEM, with one way ANOVA with Dunnett’s post test using LPS or PIC as control. * = p <0.05, ** = p < 0.01.
Microglial responses were generally similar regardless whether they were stimulated with LPS (left of the graph) or PIC (right of the graph, all values were normalized to those induced by LPS). The results with the non-selective HDACi TSA showed that it inhibited the production of all four microglial cytokines but with a stronger inhibition of IL-10 than IL-1β, TNFα or IL-6. Again, the TSA effects on PIC-induced cytokines were similar to those on LPS-induced cytokine production. The effects of another non-selective HDACi VA were more variable showing inhibition of IL-10 and TNFα but not IL-1β or IL-6 (Figure 1). Finally, the GSK3 inhibitor SB216763 also had more variable effects, ranging from mild inhibition to no change to even an increase (IL-10).
Effects of HDACi on microglial cytokine, chemokine and antiviral gene expression
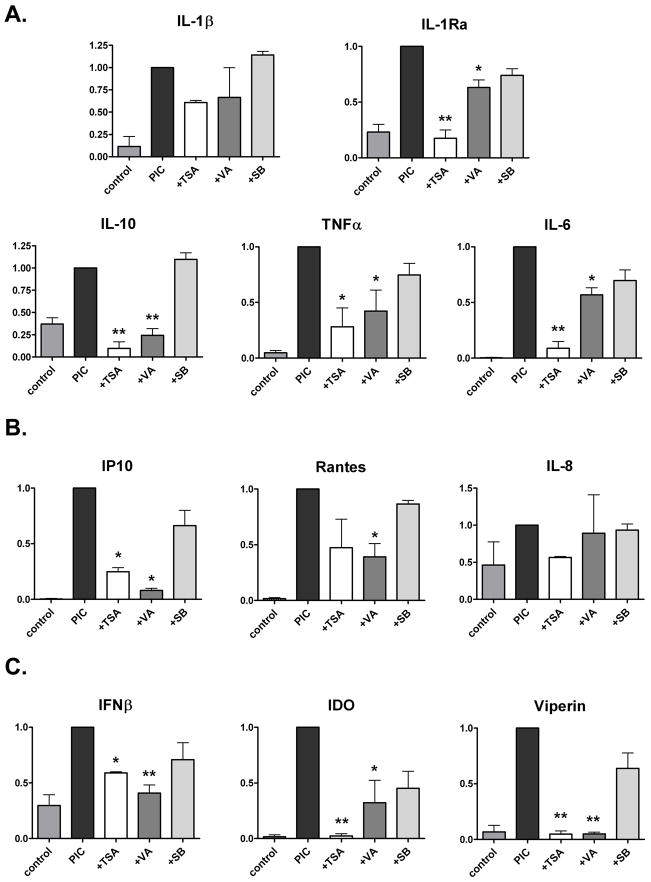
We next examined the effects of TSA, VA, and SB216763 on microglial cytokine, chemokine and antiviral gene expression, using real-time PCR (Figures 2 and 3: A, B and C, respectively). Microglial cultures were treated with LPS, PIC, and the inhibitors, as shown in Figure 1, and RNA was harvested at 6h. The results of experiments with LPS stimulation are shown in Figure 2 and those with PIC are shown in Figure 3. Data are pooled from two separate experiments using different microglial cases and represent normalized (LPS or PIC = 1) values.
Figure 2. The effect of HDAC inhibitors on microglial cytokine, chemokine and antiviral gene mRNA expression induced by LPS.
Microglial cultures were treated with the inhibitors for 1h, then with LPS as described in Figure 1 legend. Total RNA was harvested 6 h after LPS treatment and Q-PCR was performed for cytokines (A), chemokines (B), and innate immune molecules (C), as described in the Materials and Methods using GAPDH as control. The mRNA levels were normalized to the values induced by LPS alone (LPS = 1) and data from two different experiments were pooled. Mean ± SEM. ANOVA with Dunnett’s post test with LPS as control. * = p <0.05, ** = p < 0.01.
Figure 3. The effect of HDAC inhibitors on microglial mRNA expression induced by PIC.
Microglial cultures were treated in an identical manner to that described in Figure 2 legend, except that PIC, instead of LPS, was used to stimulated microglial gene induction. Q-PCR with RNA harvested 6h after PIC treatment. Pooled, normalized (PIC = 1) data for cytokines (A), chemokines (B) and innate immune molecules (C) from two separate microglial cases. ANOVA with Dunnett’s post test: * = p <0.05, ** = p < 0.01.
Once again, microglial mRNA responses were very similar regardless whether they were stimulated with LPS (Figure 2) or PIC (Figure 3). Similar to protein production shown in Figure 1, TSA inhibited mRNA production for all cytokines measured, but with strong inhibition shown for IL-10, IL-6, IL-1ra, TNFα mRNA and smaller (not significant) inhibition for IL-1β mRNA (Figures 2A and 3A). VA also inhibited cytokine mRNA production, but less potently than TSA. SB216763 had smaller and variable effects, with the only significant effect being the increase of (LPS-induced) IL-10 mRNA (see Discussion).
Examination of additional microglial chemokine and antiviral genes (Figures 2B and 3B, and Figure 2C and 3C, respectively) demonstrated that IP-10, IFNβ, IDO and viperin mRNA were all strongly inhibited by TSA and VA, whereas SB216763 had much less and insignificant effects. The HDACi suppressed chemokines differentially with a rank order IP-10 > Rantes > IL-8. The responses were again similar between LPS- and PIC-stimulated microglia.
Effects of HDACi on astrocyte inflammatory and antiviral gene expression
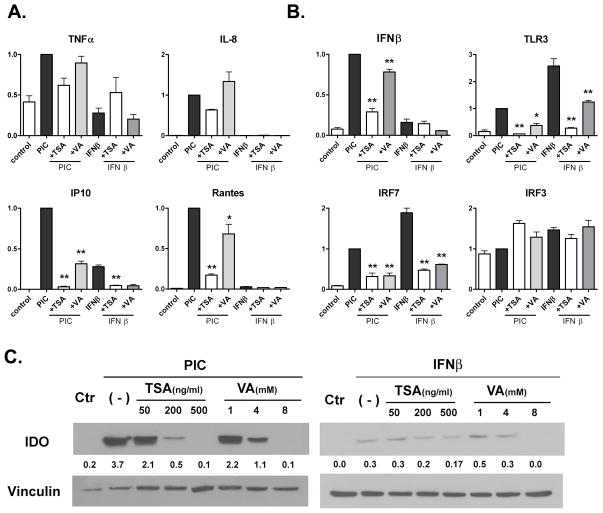
We next examined the expression of cytokines, chemokines and IFN-related molecules in astrocytes stimulated with PIC or IFNβ, as human astrocytes do not respond to LPS. Figure 4(A and B) shows the Q-PCR results of astrocytes pretreated with HDACi for 1 h, then stimulated with PIC or IFNβ for 6 h (n = 2, data are normalized to PIC = 1, then pooled). Note the small amount of astrocyte TNFα induced by PIC as reported, while the induction of IL-8, IP-10 and Rantes was robust (John et al., 2005; Rivieccio et al., 2005; Rivieccio et al., 2006; Suh et al., 2007). Human astrocytes produce little or no IL-1β, IL-1ra or IL-10 (Liu et al., 1998). The effects of HDACi (TSA 400 ng/ml or VA 4 mM) were similar to those observed in microglia in that they inhibited IP-10 the strongest and IL-8 the weakest (IP-10 > Rantes > IL-8). In IFNβ-stimulated astrocytes, IP-10 mRNA alone was induced (as expected) and this was inhibited by HDACi. The protein levels measured by ELISA in astrocyte cultures demonstrated results similar to mRNA (data not shown).
Figure 4. The effect of HDAC inhibitors on astrocyte inflammatory and innate immune gene expression.
Primary human fetal astrocytes were pretreated with TSA at 400 mg/ml or VA at 4 mM for 1h, and then stimulated with PIC at 20 μg/ml or IFNβ 10 ng/ml for an additional 6 h. Q-PCR was performed as described in Figure 2 and Figure 3 legend. Data are shown for cytokines and chemokines (A) and molecules involved in TLR3 signaling (B). Pooled, normalized (PIC = 1) data from two separate astrocyte cases are shown. Mean ± SEM. ANOVA with Dunnett’s post: * = p <0.05, ** = p < 0.01. (C) Western blot analysis of astrocytes for IDO protein expression. Astrocytes were treated with varying concentrations of TSA or VA as indicated for 1 h, then with IFNβ or PIC for 24h. The numbers are densitometric ratios to vinculin, a protein loading control. The data are representative of two separate experiments with similar results.
Astrocyte mRNA expression for several molecules involved in TLR3 signaling (the receptor itself, primary response gene IFNβ, and the transcription factors involved in TLR3 signaling, IRF3 and IRF7) were also examined by real-time PCR. As shown in Figure 4B, IFNβ induced larger amounts of TLR3 and IRF7 than PIC, suggesting that IFNβ was probably the mediator of PIC action. IFNβ did not induce IFNβ. IRF3, the constitutive transcription factor present in all cells, showed a small increase by IFNβ and not by PIC. HDACi suppressed the expression of all with the exception of IRF3.
We next examined astrocyte IDO protein expression by Western blot analysis (Figure 4C). PIC was a much more potent inducer of IDO than IFNβ, and TSA and VA dose-dependently inhibited IDO induced by either stimulus. For PIC-induced IDO expression, IC50 was ~100 ng/ml for TSA and ~4 mM for VA. The generally stronger inhibition shown for TSA than for VA in our study might be related to the drug concentrations chosen (TSA 400 ng/ml and VA 4 mM). Taken together, these results show that HDACi inhibit astrocyte cytokines, chemokines and innate antiviral molecule expression.
Microarray analysis of the TSA effect on astrocyte PIC-induced genes
Because of the potent down-regulatory effect of TSA on gene expression that we observed, we next examined the effect of TSA on astrocyte global gene expression induced by PIC (Table 1). Highly enriched fetal astrocyte cultures were pre-treated with TSA (400 ng/ml) for 1 h, then treated with PIC (20 μg/ml) for an additional 6 h. Microarray analysis was performed using human 28K cDNA microarray chips from the AECOM Microarray Facility (http://microarray1k.aecom.yu.edu/) as described in the Materials and Methods. There were ~2500 genes upregulated and ~2000 genes downregulated by TSA using 2X and 0.5X as cutoffs. Table I shows examples of some of the downregulated and upregulated genes by TSA in these cultures. The full array results are provided as Supplemental Data. The results corroborate those obtained by Q-PCR in Figure 4. Importantly, the interferon-inducible genes were among the most profoundly downregulated genes (Table 1).
Table 1.
TLR3-induced genes altered by HDACi (TSA) treatment1
| Decreased genes | Gene Symbol | Fold Change |
|---|---|---|
| Chemokine (CXC) ligand 11 | CXC11 | 0.01 |
| Chemokine (CXC) ligand 9 | CXCL9 | 0.02 |
| Myxovirus (influenza virus) resistance 1 | Mx1 | 0.02 |
| 2′,5′-oligoadenylate synthetase 1 | OAS1 | 0.03 |
| 2′,5′-oligoadenylate synthetase 2 | OAS2 | 0.03 |
| TNT (ligand) superfamily, member 10 (TRAIL) | TNFSF10 | 0.03 |
| IFN-induced protein with tetratricopeptide repeat 1 | IFIT1 | 0.03 |
| Indoleamine-pyrrole 2,3 dioxygenase | IDO | 0.04 |
| Myeloid differentiation primary response gene 88 | MyD88 | 0.07 |
| IFN-stimulated exonuclease gene 20 kDa | ISG20 | 0.10 |
| IFN-stimulated transcription factor 3 gamma 48 kDa | ISGF3G | 0.11 |
| ISG15 ubiquitin-like modifier | ISG15 | 0.11 |
| Transforming growth factor, beta 1 | TGFB1 | 0.13 |
| Interleukin 15 receptor, alpha | IL15R | 0.16 |
| Vascular endothelial growth factor C | VEGFC | 0.17 |
| Interferon regulatory factor 7 | IRF7 | 0.19 |
| Colony stimulating factor 1 | CSF1 | 0.19 |
| Activating transcription factor 3 | ATF3 | 0.20 |
| caspase 1 (IL1 converting enzyme) | CASP1 | 0.21 |
| Toll-like receptor 3 | TLR3 | 0.23 |
| Interleukin 13 | IL13 | 0.24 |
| Fibroblast growth factor 2 | FGF2 | 0.24 |
| CD38 molecule | CD38 | 0.25 |
| Interferon beta (IFNβ) | IFNB1 | 0.28 |
| Claudin 1 | CLDN1 | 0.31 |
| Chemokine (CC) ligand 5 (Rantes) | CCL5 | 0.33 |
| fms-related tyrosine kinase 1 (VEGF receptor) | FLT1 | 0.36 |
| Interleukin 1B (IL-1β) | IL1B | 0.44 |
| Interleukin 8 | IL8 | 0.46 |
|
| ||
| Increased genes | Gene Symbol | Fold Change |
| Proliferating cell nuclear antigen | PCNA | 19.3 |
| Mitogen-activated kinase kinase kinase 3 | MAP3K3 | 18.4 |
| Interleukin 13 receptor, alpha 1 | IL13RA1 | 11.2 |
| Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CDKN1B | 9.83 |
| Caspase 9 | CASP9 | 9.75 |
| Chemokine (CXC) ligand 1 (GROα) | CXCL1 | 8.28 |
Highly enriched human fetal astrocyte cultures (~1% microglia, ~2% neurons) were pretreated with 400 ng/ml TSA for 1h, then with 20 μg/ml TLR3 ligand PIC for an additional 6h. Microarray was performed as described in the Materials and Methods. Fold changes are the ratios as calculated by values in TSA+PIC culture divided by those in PIC alone culture. (1= no change; >1 increase; <1 decrease)
DISCUSSION
Previous studies in the laboratory have shown that PIC induces a wide array of inflammatory and antiviral genes, as well as upregulation of the receptor TLR3 in human astrocytes and microglia (Rivieccio et al., 2006; Suh et al., 2007; Suh et al., 2009b). In addition to a broad spgectrum of antiviral response genes, PIC also induced an antiviral state in astrocytes and microglia. Among the most highly induced genes in PIC-activated glia were viperin/cig5, a cytomegalovirus-inducible gene that is also known be induced by IFN α/β, as well as indoleamine 2, 3-deoxygenase (IDO), a pleiotropic molecule involved in the induction of immune tolerance. The involvement of these proteins in anti-HIV activity has also been demonstrated, supporting that IDO and viperin are components of the CNS innate immune response.
In this study, we found that HDACi TSA and VA modulate TLR-induced cytokine, chemokine and antiviral gene expression in human glia. In microglia, analysis of protein and mRNA showed that TSA and VA suppressed the expression of all genes examined. By ELISA, TSA suppressed all four cytokines (IL-10, IL-1α, TNFα and IL-6), with the anti-inflammatory cytokine IL-10 being the most strongly suppressed. VA, on the other hand, generally produced more modest suppression compared to TSA, with no suppression observed for IL-1β or IL-6. Microglial mRNA analysis showed that similar to protein, IL-10 mRNA also showed strong suppression by TSA and VA, and this pattern was also shown for IP-10, IFNβ, IDO and viperin (all genes that we previously determined to be IFN-inducible) (Rivieccio et al., 2006; Suh et al., 2007; Suh et al., 2009b). On the other hand, the expression of other cytokine/chemokine genes was less severely suppressed. This was particularly true for IL-1β and IL-8.
In astrocytes, we find that TSA and VA also suppressed the expression of IFNβ and the molecules involved in TLR signaling (TLR3 and IRF7), as well as IDO and viperin, all IFN- or interferon regulatory factor 3- (IRF3) dependent genes. Microarray analysis remarkably showed that the proteins belonging to the IFN-inducible gene family were globally inhibited and were among the most severely downregulated genes. Among the least inhibited genes were IL-1β, IL-8 as well as GROα, all IFN-independent inflammatory genes. In addition to downregulated genes, ~2000 genes were also upregulated by TSA, demonstrating that HDACi were not global inhibitors of transcription.
We noted that in both microglia and astrocytes, VA (4 mM) tended to be less inhibitory than TSA (400 ng/ml), with some exceptions. Several possible explanations exist for this observation. First, although we chose the drug concentrations based on the literature, the IC50 of the two inhibitors might have been different in glial cells (see Figure 4C). Alternatively, the effect of VA might have reflected more than one pharmacological action. For example, VA has been shown to exert GSK3β inhibitory activity, in addition to HDACi. In this regard, it is worthy of note that the GSK3β inhibitor SB216763 did not significantly modulate microglial inflammatory and antiviral gene expression, leaving the possibility that in VA-treated cells, the anti-GSK3 activity negated the HDACi-induced inhibition. The only significant effect that SB216763 showed was on IL-10, which was enhanced by SB216763, similar to that reported in human monocytes (Martin et al., 2005). The opposing effects of VA and SB216763 for microglial IL-10 expression make it unlikely that VA functions as a (pure) GSK3β inhibitor in microglia.
In our study, a less than perfect correspondence between microglial protein and mRNA expression (Figures 1–3) was observed. Several possible explanations include different time points for RNA and protein examination, the different microglial cases used for RNA and protein examination, and limited numbers of microglial samples available for study (i.e., low statistical power). Although not actively explored, there is also the possibility that the protein expression was inhibited at the level of protein translation involving mechanisms such as microRNAs.
The results of our study have several implications. First, they suggest that the use of HDACi could suppress the innate antiviral immune response. Several earlier studies have shown that HDACi have surprising suppressive effects on the transcription of type I interferons and IFN-inducible genes (Chang et al., 2004; Genin et al., 2003; Nusinzon and Horvath, 2003; Sakamoto et al., 2004; Vlasakova et al., 2007). Moreover, a study using siRNA for specific HDACs has found that HDAC1 and HDAC8 repress and HDAC6 enhances IFNβ expression and that HDAC6 acts as a coactivator of IRF3-dependent gene transcription (Nusinzon and Horvath, 2006). These studies indicate that HDAC6 is an essential component of the innate antiviral immune response, and that the use of non-selective HDACi could compromise the host innate antiviral immune response by inhibiting HDAC6. These results are not restricted to tissue culture studies, as HDACi have recently been shown to enhance the replication and spread of (oncolytic) viruses in vivo, offering a therapeutic opportunity in this case (Nguyen et al., 2008).
Second, our results suggest that HDACi could suppress inflammation by suppressing inflammatory cytokine expression from microglia and astrocytes. However, given the differential effects shown for certain cytokines, for example, a much stronger inhibition of IL-10 and IL-1 receptor antagonist vs. IL-1, and IP-10 vs. IL-8, we believe that the results might not be simply a suppression but a change in balance. These results may also reflect the underlying mechanisms of HDACi action, since IL-1 and IL-8 are two genes with no known IFN-stimulated response elements (ISRE) in their promoters. Indeed, our results may indicate that the primary target of HDACi in the TLR3/4 signaling is IRF3, its primary response gene IFNβ and downstream IFN-stimulated genes (ISGs). The strikingly similar effects of HDACi on either LPS- or PIC- activated microglia provide additional support for the notion that the common TLR3/4 signaling components (the TRIF-IRF3-IFNβ axis) might be the target. Additional mechanisms for the differential gene regulation by HDACi may include the induction of activating transcription factor 3 (ATF3). ATF3 is a member of the CREB/ATF family transcription factor induced by TLR3/4 that negatively regulates inflammatory cytokine gene expression (Gilchrist et al., 2006; Whitmore et al., 2007). In human glia, ATF3 is induced by TLR3/4 activation and is suppressed by TSA (Suh et al., 2009b) (see Table 1). This opens the possibility that HDACi-mediated inhibition of ATF3 de-represses inflammatory cytokine gene expression. These findings hint at complex mechanisms by which HDACi modulate cytokine gene expression.
Our investigation has implications for the potential utility as well as the danger associated with the use of HDACi for treatment of HIV-associated diseases. For example, VA has been promoted as an agent to purge latent HIV reservoir through its HDACi activity (Lehrman et al., 2005). VA has also been studied extensively for its possible neuroprotective activity against HIV infection. Specifically, VA has been shown to inhibit neuronal apoptosis and tau phosphorylation in in vivo and in vitro experimental models of HIV encephalitis and the neuroprotection conferred by VA has been attributed to its anti-GSK3β activity (Dewhurst et al., 2007; Dou et al., 2003; Maggirwar et al., 1999; Sui et al., 2006). More recently, GSK3β inhibitors including VA and lithium chloride have been promoted as an adjunct therapy for HIV-associated neuropsychiatric conditions (Dewhurst et al., 2007; Schifitto et al., 2006; Schifitto et al., 2009). In spite of these data, our study suggests that the role of HDACi and GSK3β inhibitors in glial inflammatory gene expression might be quite different and that in microglia VA might act predominantly as an HDACi rather than a GSK3 inhibitor.
Supplementary Material
Acknowledgments
This study was supported by NIH RO1 MH55477, KO1 MH084705, Molecular Neuropathology Training grant NIH T32 NS007098 and Einstein CFAR grant P30 AI051519. We thank the Einstein Human Fetal Tissue Repository for the tissue and Meng-Liang Zhao for tissue culture.
Abbreviations
- HDAC
histone deacetylase
- HDACi
HDAC inhibitor
- IFNAR
type I interferon receptor
- ISRE
IFN-stimulated response element
- PIC (poly I:C)
polyinosinic-polycytidylic acid
- PBDA
porphobilinogen deaminase
- Q-PCR
real time RT-PCR
- VA
valproic acid
- IDO
indoleamine 2, 3- dioxygenase
- IRF
interferon regulatory factor
- ISGs
IFN-stimulated genes
- LPS
lipopolysaccharide
- TLR
toll-like receptor
- TRIF
toll/interleukin-1 receptor domain-containing adaptor protein inducing IFNβ
- TSA
trichostatin A
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. Expression of latent human immunodeficiency type 1 is induced by novel and selective histone deacetylase inhibitors. AIDS. 2009 doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camelo S, Iglesias AH, Hwang D, Due B, Ryu H, Smith K, Gray SG, Imitola J, Duran G, Assaf B, Langley B, Khoury SJ, Stephanopoulos G, De GU, Ratan RR, Ferrante RJ, Dangond F. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005;164:10–21. doi: 10.1016/j.jneuroim.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Carpentier PA, Duncan DS, Miller SD. Glial toll-like receptor signaling in central nervous system infection and autoimmunity. Brain Behav Immun. 2008;22:140–147. doi: 10.1016/j.bbi.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I, Levy DE. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 7.Dewhurst S, Maggirwar SB, Schifitto G, Gendelman HE, Gelbard HA. Glycogen synthase kinase 3 Beta (GSK-3beta) as a therapeutic target in neuroAIDS. J Neuroimmune Pharmacol. 2007;2:93–96. doi: 10.1007/s11481-006-9051-1. [DOI] [PubMed] [Google Scholar]
- 8.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 9.Dou H, Birusingh K, Faraci J, Gorantla S, Poluektova LY, Maggirwar SB, Dewhurst S, Gelbard HA, Gendelman HE. Neuroprotective activities of sodium valproate in a murine model of human immunodeficiency virus-1 encephalitis. J Neurosci. 2003;23:9162–9170. doi: 10.1523/JNEUROSCI.23-27-09162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genin P, Morin P, Civas A. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J Virol. 2003;77:7113–7119. doi: 10.1128/JVI.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 12.Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: a priceless link to innate immunity. Trends Mol Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 13.John GR, Lee SC, Song X, Rivieccio M, Brosnan CF. IL-1-regulated responses in astrocytes: relevance to injury and recovery. Glia. 2005;49:161–176. doi: 10.1002/glia.20109. [DOI] [PubMed] [Google Scholar]
- 14.Kiernan R, Bres V, Ng RW, Coudart MP, El MS, Sardet C, Jin DY, Emiliani S, Benkirane M. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 15.Kim HJ, Rowe M, Ren M, Hong JS, Chen PS, Chuang DM. Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: multiple mechanisms of action. J Pharmacol Exp Ther. 2007;321:892–901. doi: 10.1124/jpet.107.120188. [DOI] [PubMed] [Google Scholar]
- 16.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 17.Lee SC, Liu W, Brosnan CF, Dickson DW. Characterization of primary human fetal dissociated central nervous system cultures with an emphasis on microglia. Lab Invest. 1992;67:465–476. [PubMed] [Google Scholar]
- 18.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, Modena D, Moras ML, Pozzi P, Reznikov LL, Siegmund B, Fantuzzi G, Dinarello CA, Mascagni P. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Zhao M-L, Brosnan CF, Lee SC. Expression of type II nitric oxide synthase in primary human astrocytes and microglia: role of IL-1b and IL-1 receptor antagonist. J Immunol. 1996;157:3569–3576. [PubMed] [Google Scholar]
- 21.Liu JS, Amaral TD, Brosnan CF, Lee SC. IFNs are critical regulators of IL-1 receptor antagonist and IL-1 expression in human microglia. J Immunol. 1998;161:1989–1996. [PubMed] [Google Scholar]
- 22.Maggirwar SB, Tong N, Ramirez S, Gelbard HA, Dewhurst S. HIV-1 Tat-mediated activation of glycogen synthase kinase-3beta contributes to Tat-mediated neurotoxicity. J Neurochem. 1999;73:578–586. doi: 10.1046/j.1471-4159.1999.0730578.x. [DOI] [PubMed] [Google Scholar]
- 23.Mahlknecht U, Will J, Varin A, Hoelzer D, Herbein G. Histone deacetylase 3, a class I histone deacetylase, suppresses MAPK11-mediated activating transcription factor-2 activation and represses TNF gene expression. J Immunol. 2004;173:3979–3990. doi: 10.4049/jimmunol.173.6.3979. [DOI] [PubMed] [Google Scholar]
- 24.Marks PA. Discovery and development of SAHA as an anticancer agent. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]
- 25.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McManus CM, Liu JS, Hahn MT, Hua LL, Brosnan CF, Berman JW, Lee SC. Differential induction of chemokines in human microglia by type I and II interferons. Glia. 2000;29:273–280. [PubMed] [Google Scholar]
- 28.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 29.Monneret C. Histone deacetylase inhibitors. Eur J Med Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TL, Abdelbary H, Arguello M, Breitbach C, Leveille S, Diallo JS, Yasmeen A, Bismar TA, Kirn D, Falls T, Snoulten VE, Vanderhyden BC, Werier J, Atkins H, Vaha-Koskela MJ, Stojdl DF, Bell JC, Hiscott J. Chemical targeting of the innate antiviral response by histone deacetylase inhibitors renders refractory cancers sensitive to viral oncolysis. Proc Natl Acad Sci U S A. 2008;105:14981–14986. doi: 10.1073/pnas.0803988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci U S A. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nusinzon I, Horvath CM. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol. 2006;26:3106–3113. doi: 10.1128/MCB.26.8.3106-3113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng GS, Li G, Tzeng NS, Chen PS, Chuang DM, Hsu YD, Yang S, Hong JS. Valproate pretreatment protects dopaminergic neurons from LPS-induced neurotoxicity in rat primary midbrain cultures: role of microglia. Brain Res Mol Brain Res. 2005;134:162–169. doi: 10.1016/j.molbrainres.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol Cell Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivieccio MA, John GR, Song X, Suh HS, Zhao Y, Lee SC, Brosnan CF. The cytokine IL-1beta activates IFN response factor 3 in human fetal astrocytes in culture. J Immunol. 2005;174:3719–3726. doi: 10.4049/jimmunol.174.6.3719. [DOI] [PubMed] [Google Scholar]
- 36.Rivieccio MA, Suh HS, Zhao Y, Zhao ML, Chin KC, Lee SC, Brosnan CF. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]
- 37.Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J Biol Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 38.Schifitto G, Peterson DR, Zhong J, Ni H, Cruttenden K, Gaugh M, Gendelman HE, Boska M, Gelbard H. Valproic acid adjunctive therapy for HIV-associated cognitive impairment: a first report. Neurology. 2006;66:919–921. doi: 10.1212/01.wnl.0000204294.28189.03. [DOI] [PubMed] [Google Scholar]
- 39.Schifitto G, Zhong J, Gill D, Peterson DR, Gaugh MD, Zhu T, Tivarus M, Cruttenden K, Maggirwar SB, Gendelman HE, Dewhurst S, Gelbard HA. Lithium therapy for human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol. 2009;15:176–186. doi: 10.1080/13550280902758973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suh HS, Brosnan CF, Lee SC. Chapter 4: TLRs in CNS viral infections. [Current Topics in Microbiology and Immunology] In: Kielian T, editor. Toll-like Receptors: Roles in Infection and Neuropathology. 2009a. Ref Type: Serial (Book, Monograph) [DOI] [PubMed] [Google Scholar]
- 41.Suh HS, Kim MO, Lee SC. Inhibition of granulocyte-macrophage colony-stimulating factor signaling and microglial proliferation by anti-CD45RO: role of Hck tyrosine kinase and phosphatidylinositol 3-kinase/Akt. J Immunol. 2005;174:2712–2719. doi: 10.4049/jimmunol.174.5.2712. [DOI] [PubMed] [Google Scholar]
- 42.Suh HS, Zhao ML, Choi N, Belbin TJ, Brosnan CF, Lee SC. TLR3 and TLR4 are innate antiviral immune receptors in human microglia: role of IRF3 in modulating antiviral and inflammatory response in the CNS. Virology. 2009b doi: 10.1016/j.virol.2009.07.001. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh HS, Zhao ML, Rivieccio M, Choi S, Connolly E, Zhao Y, Takikawa O, Brosnan CF, Lee SC. Astrocyte indoleamine 2, 3 dioxygenase (IDO) is induced by the TLR3 ligand poly IC: mechanism of induction and role in anti-viral response. J Virol. 2007;81:9838–9850. doi: 10.1128/JVI.00792-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui Z, Sniderhan LF, Fan S, Kazmierczak K, Reisinger E, Kovacs AD, Potash MJ, Dewhurst S, Gelbard HA, Maggirwar SB. Human immunodeficiency virus-encoded Tat activates glycogen synthase kinase-3beta to antagonize nuclear factor-kappaB survival pathway in neurons. Eur J Neurosci. 2006;23:2623–2634. doi: 10.1111/j.1460-9568.2006.04813.x. [DOI] [PubMed] [Google Scholar]
- 45.Suuronen T, Huuskonen J, Pihlaja R, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by histone deacetylase inhibitors. J Neurochem. 2003;87:407–416. doi: 10.1046/j.1471-4159.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- 46.Vlasakova J, Novakova Z, Rossmeislova L, Kahle M, Hozak P, Hodny Z. Histone deacetylase inhibitors suppress IFNalpha-induced up-regulation of promyelocytic leukemia protein. Blood. 2007;109:1373–1380. doi: 10.1182/blood-2006-02-003418. [DOI] [PubMed] [Google Scholar]
- 47.Whitmore MM, Iparraguirre A, Kubelka L, Weninger W, Hai T, Williams BR. Negative regulation of TLR-signaling pathways by activating transcription factor-3. J Immunol. 2007;179:3622–3630. doi: 10.4049/jimmunol.179.6.3622. [DOI] [PubMed] [Google Scholar]
- 48.Zhang B, West EJ, Van KC, Gurkoff GG, Zhou J, Zhang XM, Kozikowski AP, Lyeth BG. HDAC inhibitor increases histone H3 acetylation and reduces microglia inflammatory response following traumatic brain injury in rats. Brain Res. 2008;1226:181–191. doi: 10.1016/j.brainres.2008.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.