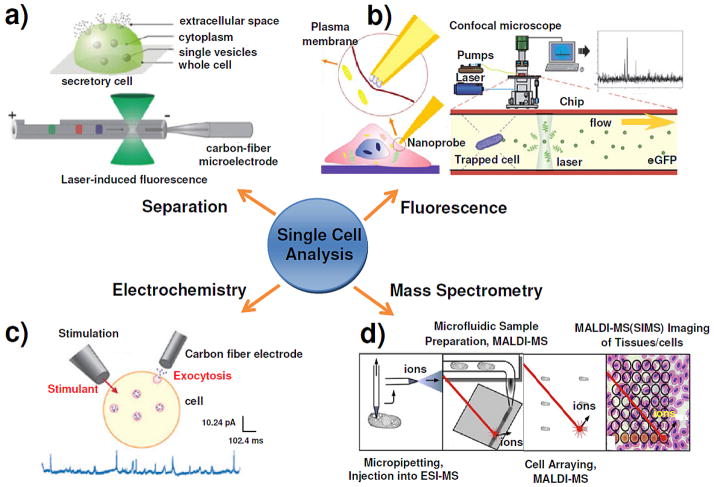
Single Cell Analysis
In this review we attempt to give an overview of methods developed for single cell analysis. This is an important topic for several reasons. Many biological systems have heterogeneous make up of cells and it is important therefore to analyze them one at a time to determine these differences. Additionally, in detection of cell-based diseases, it is important to develop methods of extreme sensitivity down to single cells to gain the best ability for diagnoses. These are some highly exciting times for single cell analysis. The area began for all practical purposes with enzyme activity measurements in the 1940s and slowly gained speed until the 1980s and 90s when microcolumn liquid chromatography, capillary electrophoresis, amperometry, and new fluorescence reagents all seemed to converge on the subject at once. The development of total internal reflectance fluorescence and subsequent rise of mass spectrometry imaging have all made this an analytical chemists’ playground. The biologists have simultaneously developed genetic technologies to label compounds in cells with molecules like green fluorescent protein and the repertoire of techniques is huge.
We have made an attempt to be comprehensive, but have only covered the last three years. There are many great works in related areas and it was difficult to draw a specific line between quantitative cell analysis and qualitative analysis. For example, we have not covered a great deal of studies that are truly at the single cell level concerning biological applications of fluorescence imaging with agents like FURA II, FM1-43, and green fluorescent protein (GFP). There are simply too many applications as these tools become more highly used in biology. We have chosen to aim mostly at new techniques or variations of techniques. The review is the first on single cell analysis in this specific series and, hence, we have covered the slightly more than three-year period from 2007 through the beginning of 2011.
We have split the topics discussed into sections more or less by technique, these ranging from separations, direct fluorescence, electrochemistry, mass spectrometry, and other methods (Figure 1). Microfluidic devices show up in several places as well, as these seem to cross all the barriers. This just exemplifies that there is a great deal of cross-fertilization in an area such as single cell analysis where most methods are hybrid methods. Thus, fluorescence is used heavily in detection with liquid separations, and is also used directly on cells for quantitative analysis. We sincerely hope we have paid attention to all the work done in this field and would be happy to hear from anyone whose work we omitted.
Figure 1.
Diagram showing the four major approaches to chemical analysis in, at, and of single cells, with an emphasis on exocytosis measurements. (a) Single cell analysis of exocytosis with capillary electrophoretic separation, which is capable of selectively sampling from localized regions within a single secretory cell (upper panel); Two of the most popular separation-based single cell detection schemes are LIF detection and electrochemical detection (lower panel). Reproduced from reference 51 with permission. (b) Fluorescence detection of extracellular lactate by an optical fiber based nanobiosensor (left panel); the general setup for the real time fluorescence analysis of single secreted proteins from a single yeast cell which is trapped in the chip by an electrical field is shown; enhanced green fluorescence protein molecules in the cell secretes can be detected by confocal microscopy (right panel). Reproduced from reference 60 and reference 282 with permission. (c) The typical setup for amperometry and typical amperometric data of a single cell in which exocytosis is stimulated by a pipette containing a stimulant. Reproduced from reference 90 with permission. (d) Schematics of the four mass spectrometry-based approaches for single cell metabolomics. Reproduced from reference 170 with permission.
Separation-based analysis of single cells
Separation-based techniques have historically played an important role in single cell studies. One area where this has been used is the understanding of exocytosis at the single-cell level. Investigation of the exocytotic behavior at the single-cell level is difficult for separation-based techniques, principally because of the small volume of a single cell, where the total volume is only of the order of a picoliter.1, 2 Capillary electrophoresis (CE) separation is based on the differential electrophoretic mobilities of charged molecules in an electrical field, typically within a narrow-bore capillary. CE has advantages in nanoliter sample consumption, high efficiency separations, the ability for online analyte concentration, and can be combined with detectors that have very mass sensitivity compared with other conventional separation methods, for example liquid chromatography and gas chromatography.3, 4 CE has been successfully applied to acquire chemical information from single cell metabolites of gaseous nitric oxide (NO) to small amines and amino acids to neuropeptides and larger proteins. Several reviews focusing either fully or partially on the applications of CE to single-cell analysis in general are available.5–8
Traditionally, detection methods employed for CE include absorbance, fluorescence, and electrochemistry. By far, the two most widely used detection methods for single cell work are laser-induced fluorescence (LIF) and microelectrode-based electrochemical methods. Both detection methods are capable of measuring quantities down to the zeptomole level, and even yoctomole capability in some LIF schemes. Recent trends suggest that new sampling, separation, and detection strategies with CE are being developed for obtaining an ever-increasing quality of temporal, spatial and chemical information from increasingly minuscule structures. This section highlights CE-based separation methods for understanding and quantifying of chemicals at the single cell level collected based on different detection scheme used.
CE-laser-induced fluorescence (LIF) separations of single cell components
LIF uses a laser to promote the analyte molecule to its excited electronic state, and the resulting fluorescence emission is collected by suitable optics and focused onto an appropriate detector, such as a charge coupled device or a photomultiplier tube. CE-LIF provides the lowest reported limits of detection among the detection methods available allowing measurement of trace levels of signaling molecules in complex microenvironments and minimizes the amount of scattered excitation radiation detected which otherwise would degrade analyte detection. Due to the high sensitivity, With the development of derivatization agents and better optical detection instruments, a variety of components including amino acids, peptide, protein, nucleic acid have been detected inside a single cell at increasingly lower concentrations. Generally, there are two classes of molecules that have been detected by LIF. The first group is chemical messenger species with native fluorescence, typically bearing aromatic rings. Catecholamine, indolamine, and aromatic amino acids are representative examples. The other group of target molecules is those which are not natively fluorescent but can be tagged with a fluorophore via proper derivatization chemistry.
For the analysis of proteins extracted from single cells, Dovichi’s group reported two innovations that allow capillary isoelectric focusing (CIEF) to be performed on minute amounts of labeled proteins.9 Using the Chromeo P540 dye with excitation at 532 nm and the high-power photodiodes to photobleach ampholytes resulted in an extremely low background signal, producing femtomolar concentration detection limits, zeptomole mass detection limits, and the ability to analyze a protein homogenate corresponding to the content of three cells. Additionally, Dovichi’s group demonstrated that CE-LIF can be used to detect GFP, in a cellular supernatant and that the amount of GFP in the supernatant correlates strongly with the classic lactate dehydrogenase (LDH) assay.10 In the case that the macrophages express GFP, CE-LIF can be used to measure increasing levels of GFP in the supernatant of a cell population as more cells expire. They also used a standard LDH assay to measure the release of LDH, a standard marker in supernatants. They observed the rate of cell death quantified by release of GFP and LDH into supernatant to be essentially identical, demonstrating that GFP release is at least as good as an indicator of macrophage cell death as the established LDH release method. They also coupled CIEF with an LIF detector that is based on a post-column sheath flow cuvette and employed Chromeo P503 as a fluorogenic reagent to label proteins before analysis. Detection limits for Chromeo P503-labeled β-lactoglobulin were 5 amol injected into the capillary.11 The results demonstrated sub-zeptomole detection limits, high efficiency, and high-resolution separation generated by Chromeo P503-labeled proteins in sieving and sub-micellar electrophoresis.
Jiao’s group described a versatile and selective CE-LIF method to measure NO release in single cell.12 This work was based on 8-(3,4-diaminophenyl)-2,6-bis(2-carboxyethyl)-4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene (DAMBO-PH) trapping. CE-LIF was used for the sensitive determination of NO release in individual neurons and mammalian cells with DAMBO-PH as the fluorescent probe. The method was validated by successful application to the measurements of NO release from four single cell study models.
Yeung’ group developed a CE method based on an in-capillary enzymatic cycling reaction to determine both NAD+ and NADH in a single cell in a single run.13 The detection limit was as low as 0.2 amol of NAD+ and 1 amol of NADH with a homemade CE-LIF setup. Cellular NAD+ and NADH levels of a rat myoblast cell line were determined using this method. Both NAD+ and NADH levels decreased when the cells were exposed to oxidative stress induced by H2O2.
Brown et al. developed a generally applicable method to overcome the challenges presented by the measurement of enzyme activity in single cells.14 The applicability of this approach was demonstrated by examining the intracellular fate of a protease substrate derived from the β-amyloid precursor protein (β-APP). A combination of single-cell CE-LIF, CE-LIF, and liquid chromatography (LC)-MS has been used to examine the intracellular fate of a protease substrate derived from the β-APP. This study demonstrated how single-cell CE, MS, and peptide substrates can be combined to identify and measure enzyme activities in single live cells.
Separation of cell components in microfabricated devices
Since many biomolecules or their derivatization species show fluorescent properties, detection can be adopted to investigate these molecules directly. Martin’s group described the fabrication and evaluation of a multilayer microchip device that can be used to quantitatively measure the amount of catecholamines released from pheochromocytoma (PC12) cells of the rat adrenal medulla cells immobilized within the same device.16 To enable detection of catecholamines released from these cells, a post-column derivatization scheme using naphthalene-2,3-dicarboxaldehyde and 2-β-mercaptoethanol was described and optimized in terms mixing/reaction time. Off-chip PC12 cell stimulation verified the ability of this system to monitor the stimulated release of dopamine. This is the first report of a fully integrated microchip device that couples cell immobilization and hydrodynamic flow over the cells to microchip electrophoresis.
Kennedy’s group developed a microfluidic device for high-throughput, automated, and online monitoring of insulin secretion from individual islets in parallel.17 This chip consists of a 15-channel network with each capable of superfusing a single islet and mixing superfusate online with fluorescein isothiocyanate labeled insulin and anti-insulin antibody for a competitive immunoassay. Serial immunoassays were performed at 10-s intervals on all 15 channels, corresponding to 5400 immunoassays per hour, to create temporally resolved insulin release profiles that captured single islet secretion dynamics. The chip was used to demonstrate that free fatty acid induced lipotoxicity in islets eliminates pulsatile insulin secretion. They also developed a microfluidic chip capable of long-term unattended electrophoresis operation.28 With the automated device, insulin secretion from individual beta cell islets was monitored for 24 h resulting in the collection of 14400 immunoassays for a single experiment. The immunoassay had detection limits of 0.4 nM for insulin and generated ~4% relative standard deviation over an entire 24 h period with no evidence of signal drift. The combined system was used to monitor insulin secretion from single islets of Langerhans for 6–39 h. The device resulted in considerable time and cost savings compared with the conventional insulin assay techniques.
Jiang et al. developed an automated microelectrophoresis system with buffer switching and cell lysis to rapidly analyze the contents of adherent cells in a serial manner.18 In this system, an array of cell microwells was used to position the cells in a channel at known addresses so that a capillary could be repeatedly and accurately positioned above a cell. By regulating the flow rate, electrophoresis buffer is excluded from the cells that reside in microfabricated cell microwells within the channel filled with physiologic buffer. A laser rapidly lysed individual cells and 28 cells loaded with Oregon green and fluorescein were serially analyzed in under 16 min, a rate of 1.8 cell min−1.
Phillips et al. descrived an air-stable and storable membrane-coated polydimethylsiloxane (PDMS) device for separations and single-cell cytometry.19 Protein-reinforced supported bilayer membranes (rSBMs) composed of phosphatidylcholine, biotin-PE and Neutravidin were used to coat hybrid microchips composed of PDMS and glass, which demonstrated unique properties such as efficiencies of up to 700 000 plates m−1 for fluorescent dyes and peptides, no significant change in the fluorescein efficiency, and it is more stable for electrophoresis. The performance of single-cell cytometry, a stringent test, yielded nearly identical results (after a 1 month storage time) compared with devices that were used immediately after fabrication and coating. These results demonstrate that rSBMs are an excellent functional coating for PDMS devices and possess unique properties for electrophoresis and emerging live cell analysis strategies.
Marc et al. demonstrated the feasibility of combining the electrical lysis of adherent cells held in micro-wells with capillary-based chemical cytometry.20 Electrodes composed of indium tin oxide (ITO) were patterned on a glass surface followed by formation of topographical cell traps using 1002F photoresist. Cell lysis is fast, occurring in 33–66 ms, so that rapidly changing analytes in cells could be measured with this method. The maximal cellular analysis rate is 120 cells h−1. This compares favorably to the current analysis rate of 20 cells day−1 using capillary electrophoresis. Thus, full integration and automation of cell lysis with electrophoretic separation could permit greatly increased rates of analysis of single adherent cells.
The Dovichi group has pioneered developments in single-cell proteomics and has developed a range of methods and microfabricated devices for chemical analysis of single cells, normally using CE with LIF detection. Recently, many attempts have been made by that group to study metabolite molecules including amino acids and proteins extracted from single cells. They reported a microfluidic system that can capture a cell, lyse the cell, derivatize the lysate, and separate the labeled components with fluorescence detection for amino acids from single cells.21 This system is applied to the analysis of ganglioside metabolism in AtT-20 cells. Cells are incubated with a fluorescent substrate, tetramethylrhodamine (TMR)-GM1. These cells take up the substrate and create a series of metabolites that retain the fluorescent tag. By employing capillary electrophoresis with laser-induced fluorescence detection, they were able to employ chemical cytometry for the analysis of metabolism at the single-cell level. In another microfabrication study, they described a microfabricated device for the capture and injection of a single mammalian cell into a fused silica capillary for subsequent analysis by chemical cytometry.22 The device consists of a 500 μm diameter well made from PDMS on an ITO coated microscope slide, which is both electrically conductive and transparent, and allows the visualization of the cell and the subsequent application of a voltage to assist injection of the cell into the grounded analysis capillary. A cellular suspension was allowed to settle on the device, and aspirated through the aperture after applying a voltage to the ITO, thus trapping a single NG-108 cell.
Greif has developed novel chip architectures for single cell analysis based on full body quartz glass microfluidic chips (QG chips) that extend their previous studies in PDMS chips, and enhance the detection sensitivity of native UV laser-induced fluorescence (UV-LIF), close to the pM range for the aromatic amino acid tryptophan, which is the lowest amount of this amino acid detected in a microfluidic chip by native UV-LIF.23 With these optimization steps the three proteins α- chymotrypsinogen A, ovalbumin and catalase each at a concentration of 100 μg mL−1 (equal to 4 μM, 0.4 μM and 2.2 μM) were injected electrokinetically and could be separated with nearly baseline resolution. Finally they exploited the improved sensitivity for single cell electropherograms of Spodoptera frugiperda (Sf9) insect cells.
Lai et al. demonstrated the use of a pulsed laser microbeam for cell lysis followed by electrophoretic separation of cellular analytes in a microfluidic device.24 They performed a detailed examination of the system including the pulse energy of the laser, the position of the laser focal point relative to the cell and its position relative to the floor of the microchannel to establish the optimal parameters for laser based single-cell lysis in a PDMS separation channel. Finally, the fluorescent cytosolic contents released from a single cell lysed in the channel were electrophoretically separated to demonstrate the feasibility of chemical cytometry employing laser-based lysis on-chip.
Toriello and coworkers developed an integrated microdevice for the analysis of gene expression in single cells.25 The system captures a single cell, transcribes and amplifies the mRNA, and quantitatively analyzes the products of interest. Efficient microchip integration of these processes enables the sensitive and quantitative examination of gene expression variation at the single-cell level. This microdevice was used to measure siRNA knockdown of the GAPDH gene in individual Jurkat cells.
The group of Lin demonstrated a simple way to rapidly lyse cells in a microfabricated device with sodium dodecyl sulfate (SDS) followed by analysis of the cell contents by LIF.26 Each cell can be lysed as rapidly as 500 ms owing to the high concentration of SDS used at a mixing cross section resulting from the absence of electroosmosis after surface coating in a microchannel. In each run, approximately 100 cells could be analyzed in about 10 min, which demonstrated comparatively high throughput. The reliability and quality of the analysis was confirmed by analysis of glutathione and rhodamine 123 in single K562 cells.
Zhu et al. reported a microchip electrophoresis method for the determination of intracellular superoxide (O2•−) in individual HepG2 cells.27 Dihydroethidium (DHE) was used as the specific fluorescent probe to react with intracellular O2•− to form the fluorescent 2-hydroxyethidium. An extremely low detection limit of 2.0 amol was achieved for the determination of O2•− in single HepG2 cells by microfluidic chip electrophoresis combined with LIF detection after examination of the electrophoretic behavior of 2-hydroxyethidium and E+ owing to the minute sample volume and insignificant dispersion effect during microfluidic chip-based electrophoretic separation. Different from the HPLC analysis results, only 2-hydroxyethidium but not E+ was detected, indicating that photooxidation of DHE to E+ was avoided by using the suggested single-cell analysis method.
Tang’s group described the application of microchip electrophoresis with laser-induced fluorescence detection to simultaneously determine glutathione (GSH) and hydrogen peroxide (H2O2) in mitochondria.29 The results demonstrated the feasibility of sampling, simultaneous detection, and quantification of GSH and H2O2 in subcellular organelles, and the levels of GSH and H2O2 in mitochondria isolated from HepG2 cells were found to be 2.01 ± 0.21 mM and 5.36 ± 0.45 μm, respectively. The method was further extended to observe situations of the two species in mitochondria of HepG2 cells experiencing cell apoptosis that were induced by doxorubicin and photodynamic therapy.
Xu and Yin described a chip-based microfluidic system integrated with continuous introduction of individual cells, rapid dynamic lysis, capillary electrophoretic separation and laser induced fluorescence detection for high-throughput single-cell analysis on a cross microfluidic chip with one sheath-flow channel located on each side of the sampling channel.30 The introduction of sheath-flow streams not only helps to align individual cells to sequentially enter the separation channel, but also ensures rapid dynamic lysis of the moving cells at the entry of the separation channel. In addition, single cell analysis is automatic and does not require a microscope to observe cell positioning. This simple and robust method is another that shows great potential in high-throughput single-cell analysis.
Capillary electrophoresis with electrochemical detection in single cell analysis
CE combined with electrochemical detection (CE-ED) has been used to obtain a wealth of information about the catecholamine content of neurons from a number of important neurobiological models, as well as mechanistic details of the storage and release of catecholamines within and from these neurons. Amperometry is by far the most common detection mode employed in CE-ED. This detection approach has been a popular choice owing largely to the high sensitivity achieved for electroactive molecules of neurochemical importance and facile compatibility.
Zhang’s group describe a method for the direct determination of ascorbic acid (AA) in individual rat hepatocytes based on CE-ED using a new kind of homemade carbon fiber micro-disk bundle electrode.31 The recovery was between 91% and 97%, and the amount of AA in single rat hepatocyte ranged from 28 to 63 fmol.
Sun’s group also used carbon fiber bundle electrodes to determine the amount of AA in single rat peritoneal mast cells by CE-ED.32 A single cell was lysed completely within 5 s using 0.1% SDS as the cell lysis solution together with a lysis voltage of 2 kV. Quantitative analysis of the amount of AA in single rat peritoneal mast cells was found to range from 2.4 to 7.1 fmol. The method is a simple, sensitive and reliable technique for the analysis of chemical species in single cells.
Ewing’s group developed a hybrid capillary-microfluidic device for the separation of vesicles that are chemically lysed and subsequently detected using end-column carbon-fiber amperometry.33 As vesicles exit the separation capillary, the microfluidic device provides an interface for the chemical lysis of the membrane in a sheath-flow format and subsequent electrochemical detection of the vesicle contents in the analyte flow stream. They call this electrochemical cytometry. Dopamine vesicle size was estimated by electrochemically measuring the amount of internalized analyte and relating this to the volume. The characterized hybrid capillary-microfluidic device can be used to investigate the amount of neurotransmitter present in individual synaptic vesicles isolated from the cell environment. Based on the hybrid capillary-microfluidic device, the total catecholamine content of vesicles extracted from secretory cells was directly compared with the more conventional amperometric release experiments performed on intact single cells from matched populations revealing that the average vesicle releases only 40% of its total transmitter load.34
Capillary electrophoresis with chemiluminescence detection for single cells
Chemiluminescence (CL) is characterized by simple and low-cost optical systems requiring no external light source, thus avoiding the effects of stray light and the instability of light sources. This provides low backgrounds with excellent sensitivity. Liu’s group developed a method based on microchip electrophoresis with CL detection for quantitative analysis of AA and amino acids including tryptophan (Trp), glycine (Gly) and alanine (Ala) present in single cells.35 A single cell was loaded in the cross section of two channels by electrophoretic means by applying a set of potentials at the reservoirs. This immobilized cell was then lysed rapidly under a direct electric field. The intracellular contents were separated within 130 s. CL detection was based on the enhancing effects of AA and amino acids on the CL reaction of luminol with K3[Fe(CN)6]. The average intracellular concentrations of AA, Trp, Gly and Ala in single rat hepatocytes were found to be 38.3, 5.15, 3.78 and 3.84 fmol, respectively.
Based on the oxidation reaction of luminol-labeled GSH with NaBrO, Liu’s group described an application of microchip electrophoresis with chemiluminescence detection for intracellular content of glutathione in single human red blood cells.36 Intracellular glutathione was first labeled by incubating cells with diazoluminol, and then individual cells were injected in the chip, lysed, and separated. The average content of glutathione in individual human red blood cells was found to be 64.9 amol (n=17). When compared to microchip methods with LIF detection, the CL assay described for glutathione is simple and about 100 times more sensitive.
Ye et al. reported on a method based on microchip separations coupled with CL detection to determine taurine (Tau) and amino acids including Ala, Gly, Trp, glutamic acid (Glu) and aspartic acid (Asp) present in mice single fibrosarcoma (S180) cells.37 The CL detection was based on the enhancement effects of Tau and amino acids on the CL reaction of luminol with H2O2 and Cu2+. The average amounts of Tau, Trp, Gly, Ala, Glu and Asp in per S180 cell from a population were 4.73, 1.23, 2.65, 1.94, 1.61 and 1.99 fmol. These results demonstrate that microchip separations coupled with CL detection is a simple, quick and highly sensitive analytical tool for single-cell analysis.
Single cell analysis with capillary electrophoresis and UV-VIS detection
Ren et al. developed a method for continuous intact cell detection and viability determination of individual trypan blue stained cells by CE with ultraviolet-visible dual-wavelength detection. For cell viability determination, only two parameters were needed: the range of absorbance of single cells at 214 nm and a cut-off point for living and dead cells at 590 nm.38 The results were in good agreement with conventional cell counting methods. In this CE method, only a diode array detector was needed. These are more common and cheaper than LIF detectors.
Ding and Zhang et al. used CE to determine the activity Hela cells treated with 0–46 mM methylmercury an apoptosis model.39 With induction of cell apoptosis by methylmercury, cell shrinkage and component leakage were indicated at 214 nm. Mitochondrial activity, lysosome phagocytosis ability and cell membrane integrity were detected at maximum absorption wavelengths of the Janus Green B, Rhodamine 123, Neutral Red and Trypan Blue dyes, respectively. Cell activity was described by corrected peak area ratios between apoptosis and control cells.
Capillary electrophoresis and electrogenerated chemiluminescence detection
Zhang’s group has reported a rapid, high-sensitivity, convenient preparation CE method coupled with electrogenerated chemiluminescence (ECL) detection of AA at a carbon fiber microdisk bundle electrode.40 Tris(2,20-bipyridine) ruthenium(II) (Ru(bpy)32+)was injected into the capillary tip after a cell was lysed. AA in individual rat hepatocytes reacted with Ru(bpy)32+ at the working electrode resulting in ECL. This method has been successfully applied to determine AA in single rat hepatocytes and the amount of AA in seven rat hepatocytes ranged from 16 to 62 fmol.
Micellar electrokinetic capillary chromatography (MECC) with LIF detection
Xu and Arriaga recently demonstrated that chemical cytometry can be used to quantitate superoxide levels in the mitochondrial matrix of single myoblasts by including rhodamine 123 (R123) as an internal mitochondrial membrane potential calibrant in chemical cytometry experiments.41 After loading with triphenylphosphonium hydroethidine and R123, a single cell was lysed within a separation capillary and its contents were separated and detected by MECC with LIF detection. They applied this method to single skeletal muscle myoblasts and determined that the steady state superoxide levels in the mitochondrial matrix were ~ (0.29 ± 0.10) × 10−12 M. Similarly, cultured muscle fibers in individual nanoliter-volume wells were treated with triphenylphosphonium hydroethidine, which forms the superoxide specific reporter hydroxytriphenylphosphonium ethidium (OH-TPP-E+).15 After lysis of each fiber in their corresponding nanowell, the contents of each well were processed and analyzed by MECC with laser-induced fluorescence detection making it possible to detect superoxide found in single fibers. Detection of the superoxide specific product, OH-TPP-E+, from each individual soleus skeletal muscle fiber demonstrated the feasibility of superoxide detection. Further single cell experiments validated the suitability of this approach for superoxide detection from individual fibers.
Other separation-based analyses of single cells
Using high-performance liquid chromatography with electrochemical and fluorescence detection, Yamboliev et al. simultaneously evaluated secretion of dopamine, ATP, adenosine 5′-diphosphate, adenosine 5′-monophosphate, adenosine, β-NAD and its immediate metabolites ADP-ribose and cyclic ADP-ribose in superfused nerve growth factor-differentiated rat pheochromocytoma PC12 cells.42 They demonstrated that β-NAD, a putative neurotransmitter and a neuromodulator, is subject to constitutive and regulated release in NGF-differentiated PC12 cells.
Fercher et al. reported an end-to-end differential contactless conductivity sensor for on-chip capillary electrophoresis.43 This was apparently the first successful CE separation using the end-to-end differential capacitively coupled contactless conductivity measurement approach on a miniaturized device. They fabricated this with low-temperature cofired ceramics multilayer technology. The working principle is based on the placement of two distinct detector areas near both ends of the fluid inlet and outlet of the separation channel. Both output signals are subtracted from each other, and the resulting differential signal is amplified and measured. Electrophoretic separation experiments of inorganic ions showed sensitivity enhancements by about a factor of 30–60 compared to the single-end measurement scheme.
Liu’s group observed that an efficient chemiluminescence resonance energy transfer (CRET) between a luminol donor and a CdTe QD acceptor was suppressed by the presence of certain organic compounds of biological interest.44 These allowed the development of sensitive microchip assays with CRET-based detection. Five categories of organic compounds were selected as model analytes including biogenic amines and thiols, amino acids, organic acids, and steroids. The proposed microchip assays were 10–1000 times more sensitive than those previously reported microchip methods with CL, LIF, or electrochemical detection for quantifying the corresponding compounds. Amino acids in individual red blood cells were determined by using the present method.
Single cell analysis by direct fluorescence
Studies of living cells often involve optical methods, particularly fluorescence spectroscopy and microscopy readily adaptable to microchip technology.45,66 Fluorometric assays are generally based on the presence of fluorescent tags or probes.46 Only a few metabolites can be analyzed directly in single cells by native fluorescence. In many cases, fluorometric assays can be applied in targeted analysis of small-molecule compounds in cells and cell extracts. In addition, fluorescence methods also can provide a real-time view into the conformation of proteins and multiprotein complexes over a wide range of timescales (picoseconds to hours).47–49 Research in this field has focused on novel sensing strategies and has been supported by a large number of publications on the enhancement of specificity, sensitivity, and response time.50 A specific area where fluorescence has had major impact is in monitoring exocytosis. In general, there are three major strategies to monitor exocytosis that include loading cells with fluorescent molecules, membrane-based fluorescent tracers, and engineered green fluorescent proteins (GFPs) fused to selected vesicle/granule resident proteins to label secretory vesicles/granules in single cells.51
Fluorescence methods for single cell analysis have grown rapidly in scope and applications in the past few years because of their fast, simple, and often quantitative capabilities for chemicals of living cells events. Direct fluorescence monitoring of live cells, in some cases, avoids the permeabilization or the lysis of the target cell widely involved in the electrochemical and separation methods developed for single cell analysis. The main advantages of fluorescence detection of intracellular metabolites include high sensitivity, capabilities for revealing the real-time concentration dynamics of secretory vesicles/granules, its nondestructive nature, and high-throughput capabilities in living cell.52 Compared with patch clamp or carbon-fiber microelectrode amperometry, direct fluorescence is more suited to revealing the spatial distribution of the release active zones, but it is not as quantitative concerning what is released.
For a general overview of different strategies to study metabolism in single cells using fluorescence methods, there are several reviews in the past few years.53–56 Since there are so many and widely applied fluorescence methods and devices including fluorescence spectroscopy and fluorescence microscopy for single-cell, a comprehensive review of all fluorescence analysis techniques is not realistic for this article. Thus, in this review we will only focus on the available fluorescence quantification methods for single cell analysis by showing some illustrative examples and discussing applications and important aspects, rather than aiming for full coverage of the field.
Direct fluorescence in single cell analysis
Lu etc. used confocal fluorescence microscopy imaging, fluorescence resonance energy transfer, and fluorescence recovery after photobleaching techniques to explore the roles of dihydroartemisinin- (DHA, a front-line antimalarial herbal compound with anticancer activity and low toxicity) elicited reactive oxygen species in the DHA induced Bcl-2 family proteins activation, mitochondrial dysfunction, caspase cascade, and cell death.57 Their findings demonstrate for the first time that DHA induces cell apoptosis by triggering reactive oxygen species- (ROS) mediated caspase-8/Bid activation and the mitochondrial pathway, which might function as a novel therapeutic strategy for the treatment of lung adenocarcinoma.
With a proteasome-specific cleavage motif fused to the TAT sequence and linked to the fluorophores 4-(4′-dimethylaminophenylazo) benzoic acid and 5-(2′-aminoethyl)aminonaphthalene-1-sulfonic acid, Urru et al. engineered an internally quenched fluorogenic peptide, which can penetrate cell membranes and is rapidly cleaved by the proteasomal chymotrypsin-like activity, generating a quantitative fluorescent reporter of in vivo proteasome activity in living cells as assessed by time-lapse or flow cytometry fluorescence analysis.58 This approach is an innovative tool for monitoring proteosomal proteolytic activities in physiological and pathological conditions.
Nunez-Milland etc. tested the ability of synchrotron X-ray fluorescence (SXRF) using empirical phosphorus (P) conversion factors derived by calibration versus other elements in National Institute of Standards and Technology (NIST) thin-film standards to accurately quantify P in individual cells of a model organism using analytical conversion factors for P empirically derived from the other elements in the NIST thin-film standards.59 It shows SXRF is a useful tool for studying spatial and taxonomic variations in the P quotas of individual cells, and this method will lead to advances in understanding of element dynamics within cellular systems.
Zheng et al. developed a unique nanoscale optical fiber lactate sensor to monitor the extracellular lactate concentrations of cancer cells by modifying its nanotip with LDH, which could then catalyze lactate conversion to generate NADH for sensitive fluorescence detection.60 It was demonstrated that the fabricated nanosensor can successfully detect the extracellular lactate concentrations for single HeLa, MCF-7, and human fetal osteoblast (hFOB) cells, as well as to investigate the effect of a monocarboxylate transporter inhibitor on the lactate efflux from cancer cells. This work demonstrates that the nanosensor has potential for evaluating the effect of metabolic agents on cancer metabolism and survival.
Imamura and Nhat et al. reported on a method to measure intracellular ATP levels by using genetically encoded fluorescence resonance energy transfer (FRET) based indicators for ATP, they call these ATeams.61 These probes show high selectivity to ATP over other nucleotides and have apparent dissociation constants for ATP ranging from 7.4 μM to 3.3 mM. By targeting ATeams to different subcellular compartments, they unexpectedly found that ATP levels in the mitochondrial matrix of HeLa cells are significantly lower than those of the cell cytoplasm and nucleus. The ATP levels in different cellular compartments and the dynamics of ATP in real-time can be monitored at the single cell level. In principle, it is possible to measure ATP levels at levels ranging from 2 μM to 8 mM by modulating the affinity of the ATeam. One possible explanation is that adenine nucleotide translocator pumps ATP from the mitochondria and thus maintains a high ADP/ATP ratio.
Based on dielectrophoresis and microfluidics in a lab-on-chip system, Kirschbaum et al. presented an approach that allowed the initiation of cell-cell or cell-particle interactions and the analysis of cellular reactions within various regimes while the identity of each individual cell was preserved.62 They contacted single T cells with functionalized microbeads and monitored their immediate cytosolic Ca2+ fluorescence response. The cytosolic Ca2+ level in the cells obtained by averaging all fluorescence image pixels of a cell was monitored before, during and after the contact formation procedure and correlated to the bead stimulation with sub-second resolution.
Palomero and Pye et al. measured intracellular ROS generation in real-time in mature skeletal muscle fibers by loading muscle fibers with 5- (and 6-) chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate and quantifying the fluorescence of 5- (and 6-) chloromethyl-2′,7′- dichlorofluorescein (CM-DCF) from individual fibers obtained by microscopy.63 The sensitivity of this approach was demonstrated by addition of 1 μM H2O2 to the extracellular medium. Contractions of isolated fibers induced by field electrical stimulation caused a significant increase in CM-DCF signal that was abolished by pre-treatment of fibers with GSH ethyl ester, indicating CM-DCF fluorescence microscopy can detect physiologically relevant changes in intracellular ROS activity in single isolated mature skeletal muscle fibers in real time.
Philippe’s group identified tyrosine kinase substrates by imaging their phosphorylation levels after inhibition of protein tyrosine phosphatases.64 With the analysis of the correlation between protein phosphorylation and expression levels at single cell resolution, they identified positive feedback motifs. Using fluorescence lifetime imaging microscopy on cell arrays, they discovered components that transduce signals from epidermal growth factor receptor and quantified the dynamic phosphorylation response of protein tyrosine kinase and phosphatase substrates to epidermal growth factor (EGF) to identify components that relay signals from EGF receptors and classified them according to their biological functionality.
Camacho and Machado et al. observed that disruption of the vesicular gradient of pH in turn causes the leakage of Ca2+ from vesicles in the cell, as measured with fura-2 through single cell fluorescence. Using the dye Oregon green BAPTA-2, it was shown with fluorimetric measurements that bafilomycin, an inhibitor of V-ATPase inducing alkalinization of vesicular pH, directly released Ca2+ from freshly isolated vesicles.65 The Ca2+ released from vesicles to the cytosol dramatically increased the granule motion of chromaffin- or PC12-derived granules and triggered exocytosis (measured by amperometry). They concluded that the gradient of pH of secretory vesicles might be involved in the homeostatic regulation of cytosolic Ca2+ and in two of the major functions of secretory cells, vesicle motion and exocytosis.
Matsunaga et al. described a microfluidic device equipped with black polyethylene terephthalate (PET) micromesh for the entrapment of mammalian cells and high-throughput measurements of specific mRNA expression by fluorescence. Cell adsorption was prevented by treating the PDMS surface of the microchannel with air plasma and Pluronic F-127 while each microcavity was fabricated to ensure that only one cell is trapped in a single microcavity.67 Thus, cells introduced into the microfluidic device were trapped onto the black PET micromesh with high efficiency. In addition, on-chip fluorescence in situ hybridization could be directly performed on cells that have been trapped onto the black PET micromesh, and they could successfully identify cell-to-cell variations in β-actin mRNA expression in individual Raji cells. Differences in the levels of β-actin mRNA expression were observed in serum-supplied or serum starved cell populations.
Lu’s group reported the use of a variation of total internal reflection fluorescence to carry out flow cytometry (TIRF-FC) to examine the region of the cell membrane with a throughput of ~100–150 cells s−1 and single cell resolution using an elastomeric valve. This valve is partially closed to force cells flowing in contact with the glass surface where the evanescent field resides.68 They demonstrate that TIRF-FC is capable of detecting the differences in the subcellular location of an intracellular fluorescent protein. Combined with proper data processing and analysis, TIRF-FC could be used for quantification.
Yamada et al. demonstrated a novel method for applying different solutions to a part of or all of a cell at high spatiotemporal resolution.69 They fabricated a microfluidic device using PDMS, and the sharp interface between the two solution streams flowing in the channel was used for the application of different solutions. Using a computer-controlled system to control the interface movement precisely, rapidly and reproducibly during positioning, the spatial and temporal resolution were found to be 1.6 μm and 189 ms, respectively. They applied the present system to measure [Ca2+]i increases by fluorescence and with times of 500 ms. This method can be used as a generic platform to investigate responses to drugs at the single cell level.
Berg et al. constructed a fluorescent sensor of adenylate nucleotides by combining a circularly permuted variant of GFP with a bacterial regulatory protein, GlnK1.70 The sensor affinity for Mg-ATP was <100 nM, a surprisingly high affinity considering that normal intracellular ATP concentration is in the millimolar range. ADP is also bound the same site of the sensor as Mg-ATP and produces a smaller change in fluorescence. At physiological ATP and ADP concentrations, the binding site is saturated, but competition between the two substrates makes the sensor behave as a nearly ideal reporter of the ATP/ADP concentration ratio, which can be used to monitor the ATP/ADP ratio during live-cell imaging. This cellular ATP/ADP ratio reporter is particularly valuable because it determines (along with free inorganic phosphate concentration) the actual free energy of ATP hydrolysis available for cellular reactions.
Taniguchi and Kajiyama et al. reported a method for quantitative polymerase chain reaction (qPCR) featuring a reusable single-cell cDNA library immobilized on beads.71 This was used to measure the expression of multiple genes in a single cell. Using this method, they analyzed multiple cDNA targets (from several copies to several hundred thousand copies) with an experimental error of 15.9% or less. This method is sufficiently accurate to investigate the heterogeneity of single cells.
Shim et al. described a microfluidic system that can simultaneously monitor the time-dependence of protein expression of monomeric red fluorescent protein (mRFP1)28 and the enzymatic activity of coexpressed alkaline phosphatase (AP) in compartmentalized E. coli cells.72 The device was built in multilayered PDMS to incorporate microfluidic valves and wells in which the droplets were arrayed to allow automated readout of the fluorescence corresponding to protein expression and product formation. This is capable of storing picoliter droplets containing single bacteria at constant volumes. By measuring the fluorescence intensity of both the mRFP1 inside the cells and a fluorescent product formed as a result of the enzymatic activity outside the cells, gene expression and enzymatic activity can be simultaneously and continuously monitored. Based on the microfluidic system described in this work, it was possible to maintain thousands of droplets in a constant environment that allows quantitative measurements of each droplet and enabled the concurrent study of the kinetics of protein expression and enzymatic activity in individual cells.
By using an in vitro cell-SELEX procedure, the group of Tan developed an aptamer, sgc8, towards the T-cell ALL CCRF-CEM cell line and successfully elucidated the target protein, human protein tyrosine kinase-7 (PTK7), which was discovered to be highly expressed on the cell membrane in a series of leukemia cell lines and recognized as a potential cancer biomarker.73 They set out to demonstrate fluorescence correlation spectroscopy as an effective approach for mapping receptor densities on live cells with this aptamer. They chose two different cell types with different expression levels of PTK7 on the cell membrane for a proof of principle. This appears to be the first study that has used fluorescence correlation spectroscopy to estimate the density of the membrane receptor PTK7 on different cell types with the fluorophore-labeled aptamers as receptor recognition through aptamer-receptor interactions.
To determine the phosphorylation status of many proteins involved in cell signaling at the single-cell level, Firaguay and Nunes presented a protocol for using state-specific antibodies to detect target phosphoproteins.74 They used fluorescence measurements with flow cytometry. They improved the signal intensity by using a sandwich-labeling method for the analysis of signaling proteins. By comparing the phosphorylation state of proteins in the presence and absence of nonspecific tyrosine phosphatase inhibitor, the relative amount of tyrosine-phosphorylated protein, which reflects the activity of the signaling pathway, in the samples was determined. This dynamic approach, combined with the signal amplification through a sandwich-labeling method, produces accurate and reproducible measurements of the activity of signaling pathways.
Li et al. demonstrated that the zinc (II) probe FluoZin-3AM possesses sensitive properties to distinguish different stages of apoptotic cells according to trace intracellular zinc (II) fluorescence flux.75 When apoptosis in HeLa or K562 cells was artificially induced, FluoZin-3AM selectively and strongly stained apoptotic cells only at early and middle stages, which was attributed to significantly increased free zinc (II) flux during these stages. FluoZin-3AM work was thus confirmed to be a robust and versatile tool for illuminating intracellular zinc (II) uptake, mobilization, and quantification.
Combining a semi-automated fluorometric method with an experimental model, the group of Viveiros quantified ethidium bromide transport in E. coli strains that differ in their overall efflux activity.76 The method can be used for the early detection of differences in the drug efflux capacity in bacteria accounting for antibiotic resistance, as well as to expedite screening of new drug efflux inhibitor libraries and transport studies across the bacterial cell wall. This information can be used to interpret different phenotypes resulting from this efflux activity, including multidrug resistance in clinical bacterial strains.
Weekley et al. investigated the speciation and distribution of Se in A549 human lung carcinoma cells treated with the selenoamino acids, SeMet and MeSeCys, using X-ray absorption near edge structure spectroscopy, extended X-ray absorption fine structure spectroscopy, and synchrotron radiation X-ray fluorescence microscopy.77 X-ray absorption and X-ray fluorescence studies both showed that the selenium content of MeSeCys-treated cells was much lower than that of SeMet-treated cells. Selenium was distributed homogeneously throughout the MeSeCys-treated cells.
Han and Nakamura et al. described a sensor system for a hormonal drug effect at the single cell level using a novel low invasive single cell DNA delivery technology using a nanoneedle.78 An estrogen responsive GFP reporter vector (pEREGFP9) delivered to a single breast cancer MCF-7 cell using a nanoneedle was constructed and its estrogenic response activity was confirmed using lipofection as the means of transferring the vector to the cells. By treatment with ICI 182,780 (an antagonist of estrogen) the fluorescence intensity of the GFP was decreased by 30–50% within 24 h. This technique could be applied for precise cell manipulations and single cell diagnostics.
The group of Lakowicz prepared Cy5-avidin conjugate-bound silver nanoparticles as a fluorescence molecular reagent for single cell imaging using scanning confocal microscopy.79 Compared with the metal-free avidin conjugate, the avidin-metal complex was observed to display a stronger emission intensity, shorter lifetime, and better photostability. The emission intensity over the cell image was increased with an increase of the number of avidin-metal complexes conjugated with the biotin-sites on the surfaces of PM1 cell lines, but the lifetime was decreased. A quantitative regression curve was made to deal with the correlation between the amount of avidin-metal complexes on the cell surface and the emission intensity or lifetime maxima over the entire cell. This was a useful approach that can be used to quantify the amount of target molecules on the cell surfaces using the cell intensity and lifetime images at the single cell level.
Using a fluorescent Mg2+ indicator (mag-fluo-4-AM), Lee et al. examined the propagation and dynamics of Mg2+ signaling across the cell membrane by employing real-time visualization of intracellular Mg2+ waves in living ventricular myocytes by TIRF microscopy.80 They were able to directly observe the intracellular sites where Mg2+ “sparks” occur. The majority of the Mg2+ ions are located within the matrix (41%) and intramembranous space (50%), with some also existing outside (4%) and inside (5%) of the mitochondrial membrane. The so called Mg2+ sparks and waves showed random temporal propagation patterns in nonhomogeneous substructures. This system provided better temporal resolution and real-time detection during Mg2+ sparks for individual living cells than conventional fluorescence microscopic techniques have shown in the past.
Sasuga developed a novel platform for single cell lysis by a combination of an array of micrometer-sized wells of 10–30 pL volume with a commercially available cell lysis reagent.81 Based on the flow cell constructed with a PDMS microwell array, they succeeded in trapping and lysing single cells in each of the subnanoliter-scaled wells. They were able to confirm that the resulting single-cell lysates could be successfully used for measuring the quantity of intracellular proteins and the activities of endogenous enzymes by fluorescent substrates. Since this system obtained several unique features: (1) simple protocols consisting of merely two steps for lysis, (2) minimized dilution of cell lysate during lysis, and (3) parallel processing of an ensemble of single cells, together with highly sensitive fluorescent detection techniques, it points another possible way towards analysis of the heterogeneity of a cell population at the single-cell level.
Li’s group for the first time constructed a nanobiosensor on an optical fiber to successfully detect a general cancer biomarker in the form of a telomerase at the single cell level with its nanoscale tip.82 The nanotip with a specific antibody immobilized on it was inserted into a MCF-7 breast cancer cell nucleus to capture telomerases directly. Following this, an in vitro enzymatic sandwich immunoassay was performed to achieve sensitive single living cell detection. The nanotip inserted into a MCF-7 cell nucleus provides significantly higher average (F F0)/F0 fluorescent response than when placed in a human mesenchymal stem cell (hMSC) nucleus. This demonstrates the successful detection of the telomerase over-expression in cancer cells as compared to normal cells, and provides an approach to investigate telomerase regulation in various cell types at the single cell level.
Duhamel et al. reported a simple protocol that enables the detection of enzyme-labelled fluorescence (ELF) alcohol, the product of ELF97-phosphate hydrolysis, allowing the detection of phosphatase positive bacteria, using fluorescence and flow cytometry.83 The bacterial cells need to be concentrated, the cell clumps disaggregated, and the subsequent pellet incubated with ELF97-phosphate in the liquid phase so that the sample can be fully analyzed using flow cytometry. The analysis of such samples by flow cytometry allowed quantitative assessment of the phosphatase activity for heterotrophic bacteria in both oligotrophic marine and mesotrophic lake samples.
Picazo et al. developed biosensors based on the estrogen receptor α-ligand binding domain and fluorescent proteins to detect estrogenic compounds.84 Estrogens altered the fluorescence signal of cells transfected with the indicators in a dose-dependent manner. They imaged local estrogen production in adrenocortical H295 cells expressing aromatase and transfected with the fluorescent sensors. In addition, paracrine detection was observed in HeLa cells showing the indicators and co-cultured with H295 cells. These cell-based assays allow detection of estrogen receptor ligands and can detect the local production of estrogen in single living mammalian cells.
Gaggia et al. reported use of carboxyfluorescein diacetate succinimidyl ester as pH indicator to assess changes in intracellular pH in one strain of Mycobacterium avium subsp. paratuberculosis (MAP). This was accomplished with fluorescence ratio imaging microscopy after exposure to nisin and neutralized cell-free supernatants from five bacteriocin-producing lactic acid bacteria.85 This study represents a new means to investigate intracellular pH of individual cells of slow growing pathogenic microorganisms such as MAP.
Using a novel optical technique, called triplet imaging, Geissbuehler et al. exploited oxygen-induced triplet lifetime fluorescence changes and showed how this is compatible with a variety of fluorophores.86 A modulated excitation with varying pulse widths allows the extraction of the lifetime of the essentially dark triplet state using a high fluorescence signal intensity. This enables the monitoring of kinetics of oxygen concentration changes in living cells with high temporal and spatial resolution. Oxygen consumption in single smooth muscle cells was measured. The results indicated a consumption leading to an intracellular oxygen concentration that decays exponentially with time. The proposed triplet-state imaging method has the potential to investigate oxygen metabolism at the single cell and the subcellular level.
Yu and Heikal reported a non-invasive two-photon fluorescence dynamics assay that was used to determine the intracellular NADH concentration as well as molecular conformation (i.e., free and enzyme-bound fluorescence fractions) in living cells.87 Two-photon fluorescence lifetime imaging of intracellular NADH showed sensitivity to both cell pathology and inhibition of the respiratory chain activities using potassium cyanide (KCN). Using this newly developed fluorescence dynamics imaging assay, they quantitatively estimated the average NADH concentration in cancer cells (168 ± 49 μM) to be ~1.8-fold higher than in breast normal cells (99 ± 37 μM), demonstrating the significance of intracellular NADH level dynamics (rather than intensity) imaging for probing some degenerative diseases.
Electrochemical methods at single cells
Electrochemical systems have been used since the late 1930s88 as bioanalytical tools. Electrochemical sensors offer the possibility to achieve quantitative in-situ measurements, are cheap, can be easily fabricated with lab bench techniques, are prone to miniaturization and generally require simple instrumentation. Recently, publications using electrochemical approaches to single cell analysis have mostly included carbon microfiber electrodes, microfabricated devices, scanning electrochemical microscopy (SECM) and ion selective electrodes. Interestingly, steady state amperometry is still the most popular electrochemical methods for biological assays, because of its excellent, submillisecond, time resolution. However, other techniques, such as chronoamperometry or fast scan cyclic voltammetry have been recently reported. More background information can be found in several recent reviews.51, 89–91
Carbon microfiber electrodes
Cylindrical carbon microfiber electrodes were initially reported by Gonon et al. in 197892 and disk-shaped carbon fiber electrodes by Wightman and coworkers in 1981, for the study of dopamine release in the rat brain.93 However, they were not applied to single cell analysis until the early 1990s when they were used to monitor single exocytosis events.94 These sensors are built by threading a carbon microfiber into a glass capillary, pulling the capillary and beveling the tip to obtain a clean and flat surface. Their fabrication has been well described and detailed in the papers referenced below.
Exocytosis measurements at adrenal or adrenal-derived cells
Cells from adrenal glands, and in particular chromaffin cells and PC12 cells, can release catecholamines and have been reviewed recently as popular models for exocytosis measurements.95 It has been recently shown that the electrochemical sensing of these amperometric exocytotic events and the characteristic features of the obtained spikes do not depend on the size of the electrode, and that data obtained with 5–7 μm diameter electrodes are therefore biologically relevant.96
Most of the recent studies focus on the effect of pharmacology and the intracellular physiology. Resveratrol, an antioxidant polyphenol showing cardiovascular protecting effects, was shown to inhibit catecholamine exocytosis in bovine chromaffin cells, for concentrations down to 30 nM.97 This effect was shown to be Ca2+ independent and was attributed to interactions with nitric oxide mediation. The phosphorylation of two actin associated proteins, myosin II and myristoylated alanine-rich C-kinase substrate has been demonstrated to be critical for tuning the exocytotic mode in bovine chromaffin cells (ie. ‘kiss and run’ vs full exocytosis), supporting its regulation by the rearrangement of the cell actin cortex.98 Similarly, disrupting dynamin I activity hinders both exocytotic modes, by blocking re-internalization of the membrane in the ‘kiss and run’ mode, and by limiting fusion pore dilation in full exocytosis mode.99 This was reported to be an evidence of dynamin I regulation of the fusion pore dilation or contraction. In another study, 17-β-estradiol at PC12 cells was reported to inhibit depolarization induced calcium influx in PC12 cells.100 The subsequent effect on exocytosis was found to be rather complex, as exposure to 17-β-estradiol decreased the number of exocytotic events for some cells, but induced exocytosis for others. Overall, it was indicated that 17-β-estradiol has an effect on N-type voltage-gated calcium channels, as 17-β-estradiol induced inhibition of exocytosis was abolished by ω-conotoxin.
In work related to vesicle mobility and dynamics, several single cell studies have been important. Myosin II inhibition was shown to decrease granule (vesicle) mobility near the plasma membrane.101 Inhibition of actin polymerization decreased the fusion pore expansion rates, without changes in quantal release. A study reports that protein kinase Cε facilitates the recovery of the extent of exocytosis after a long stimulation in a phosphatidylinositol biphosphate-dependent manner.102 The authors postulate that this might be used to enhance the rate of vesicle delivery and faster actin network reorganization. A quantitative investigation of the characteristics of spikes produced by chromaffin cells has led to the hypothesis that the duration and size of the fusion pore are determined by the features (size and topology at least) of the vesicle, and that the duration of the spike is characterized by the molecular factors of the cell membrane.103
Sulzer et al. showed, using amperometry and intracellular patch-clamp electrochemistry, that prolonged administration of methamphetamine, contrary to acute exposure to this drug, increased the quantal release of dopamine in rat chromaffin cells and induced hyperacidification.104 Similarly, it has been reported that beta-blockers can accumulate into the vesicles of bovine chromaffin cells, thus decreasing the concentration of released catecholamines and inducing a decrease in sympathetic tone.105 This result could explain, in part, the delay in the hypotensive effect of beta-blockers.
Adrenal cells are also a widely used model for the study of the role of lipids in exocytosis.106 Reduction of cellular cholesterol and extraction of cholesterol from the cytosol both decrease the pre spike features, usually referred to as the foot, and reduced the probability of ‘stand alone foot’, i.e. exocytotic events where there is no full fusion and recording of a spike, and increase in cellular cholesterol has the opposite effect.107 Similar results, and the fact that free cholesterol can induce exocytosis, have been reported by Zhang et al.108 Exocytosis in PC12 could also be triggered by external infusion of lysophosphatidylinositol, probably through increased intracellular Ca2+ levels, but not by other lysophospholipids.109 Depletion of cholesterol attenuated this response.
Measurements of serotonin in platelets, immune cells, and neurons
Measurements at single cells with carbon fiber electrodes have also been used to study platelet physiology. It has been shown that these cells release serotonin, highly associated with an aggregate, from their dense-core granules, and that the concentration of serotonin in these granules is thought to be about 0.5 M.110 The role of lipids in this process has been investigated.111 The results show that these cells can be used as a model for exocytosis, simpler than PC12 cells, and that cholesterol has a biophysical effect on the characteristics of exocytosis.
The toxicity of nanoparticles and immunotoxicants has been investigated using murine mast cells as a model for the exocytotic response. Exposing these cells to 28 nm gold nanoparticles altered the exocytotic events by decreasing the frequency of these events and by increasing the rate of intravesicular matrix expansion.112 These effects were found to be dose and time dependent, as 72-h exposure to the nanoparticles decreased the amount of granular release.113 Bisphenol A and mono-2-ethylhexyl phthalate also decreased the quantal level of released serotonin and the spike frequency in this model.114
Finally, amperometric measurements at MN9D cells, obtained from the somatic fusion of murine primary neurons with neuroblastoma cells, following butyric acid induced differentiation showed that the amount of dopamine released and the half-width of exocytosis events both increased.115 This is important as is shows plasticity as the nerve cells differentiate. Exocytosis from neurons and PC12 cells has also been investigated with carbon nanofiber electrodes, produced by electrochemical etching of a carbon microfiber.116 When compared to a conventional carbon microfiber electrode, this sensor shows better signal to noise ratio, and lower hindrance of the signal due to diffusion.
Modified carbon fiber microelectrodes
Modifying the carbon fiber electrode used is a method commonly used to improve its chemical selectivity and its spatial resolution. Some recent developments are described below.
Platinized electrodes have been used for the study of oxidative stress. Platinizing the tip of the electrodes can improve sensing of reactive nitrogen and oxygen species released by macrophages or fibroblasts. This has been used to investigate the mechanisms of nitrosative and oxidative stress. Triangulation of the exocytosis of reactive species released by a single fibroblast after mechanical depolarization of the membrane showed that the active area is constrained to a 15 μm radius disc centered over the point of contact.117 The composition of the chemical cocktail released by immunostimulated macrophages was also investigated, revealing the presence of nitric oxide, nitrite and peroxynitrite.118 Complete abolition of the measured signal after iNOS inhibition by 1400W, a selective inhibitor of iNOS, indicates that this isoform of nitric oxide synthase co-releases NO and hydrogen peroxide. The same study was repeated on mechanically stimulated single MG63 osteosarcoma cells.119 It was found that these cells release a high quantity of nitric oxide, compared to the amount of released hydrogen peroxide, thus explaining their malignity. As an extension to this work, triple potential step chronamperometry was applied to immunostimulated macrophages, thus allowing the simultaneous detection and quantification of the co-released species.120 In particular, temporal variations in the release of different reactive species have been reported.
Electrodes modified with a mixture of Nafion and platinum nanoparticles have also been used to demonstrate oxidative stress in plant protoplasts after triggering by a pathogen analogue.121 Genetically modified plants showed a sustained and longer-lasting response to this pathogen.
This type of sensor was also applied to investigate pharmacological interactions with oxidative stress. Vitamin C, or AA, has been shown to be an anti-oxidant in phagocytes (PLB-985), but a pro-oxidant in macrophages (RAW-264.7).122 α-Lapachone, a drug used for its anti-tumor properties, has been shown to be initially an anti-oxidant in macrophages, but a pro-oxidant after 4 h of incubation.123 This was attributed to calcium chelation leading to decreased enzyme activity and then to redox cycling in electron transfer mediation to dioxygen, inducing high concentration of reactive species and apoptosis. Similarly, some lead-based compounds, found in ancient Egyptian make-ups, have reported to increase nitric oxide release and oxidative stress in human keratinocytes.124
Azidothymidine, a drug used in HIV treatment, was also found to increase oxidative stress in macrophages.125 In particular, this property was attributed to the azido moiety of this compound. MnII pentaazamacrocyclic complexes are known to act as superoxide dismutase mimics. It has been demonstrated that one of these molecules, [MnII(pyane)Cl2], also acts as a nitric oxide dismutase, without hindering the upstream kinase pathway, and could be used as a powerful anti-inflammatory drug.126
There have been several recent developments in nitric oxide sensing. Selective sensing of nitric oxide released by a single cell has been performed recently. Using Nafion and a single-walled carbon nanotube modified carbon microfiber electrode, Cheng et al. have developed a sensor with a limit of detection of 4.3 nM.127 This electrode was used to sense L-arginine- or acetylcholine-evoked nitric oxide release from single human umbilical vein endothelial cells. Another approach used Nafion and a poly-eugenol coated microfiber to study nitric oxide released from a single identified neuron in the intact brain of the pond snail.128 This study showed that levels of nitric oxide are unaltered by aging, contrary to serotonin levels, which are altered.
Electrochemical arrays for sensing exocytosis at single cells
A microelectrode array, made of 7 individually addressable carbon microfiber electrodes, has been used to investigate the spatial and temporal resolution of exocytosis at the surface of a single PC12 cell.129 It has been use to show that some events happen simultaneously at different locations of the cell surface and to clearly show the presence of active zones of release across the cell membrane. This array has been recently used to compare amperometry and fast scan cyclic voltammetry, again for the imaging of exocytosis for a single PC12 cell.130 In comparison to amperometry, fast scan cyclic voltammetry offered excellent chemical, but poor temporal and spatial resolutions.
Models and data analysis for monitoring single cell exocytosis
The shape of the exocytotic peaks measured by electrochemistry is directly related to the dynamics of the vesicular release process. Analyzing amperometric peaks can be a challenging task, and extensive comments can be found in the literature.131 Amatore et al. report that 30% of the detected spikes feature a foot prior to the full exocytosis measurement. These can be classified into two categories: feet showing a simple ramp, and feet showing a ramp followed by a plateau.103 The amount of transmitter released during an open and stagnant fusion pore, e.g. during the foot, is correlated to the amount released during the spike, thus indicating that the maximum pore size may be correlated to the size of the vesicle. Tse et al. have also considered the probabilities of a pre spike foot, but have studied the case where the pore does not dilate to complete the vesicle fusion, resulting in an exocytotic event without a spike.107 This phenomenon was found to be dependent on the lipid composition of the membrane.
Several models have been recently proposed to explain the biochemical and biophysical events regulating exocytosis. The pH drop due to neurotransmitter oxidation occurring during the electrochemical sensing, and important analytical parameter, was modeled.132 This phenomenon is usually ignored, but can lead to a pH that is below 6 in the vicinity of the electrode. Another model was used to establish a procedure to obtain the aperture function describing the fusion of the vesicle with the membrane.133 This method does not allow direct calculation of the pore angle, but statistical analysis indicates that this angle should be smaller than 10°, and that full opening of the fusion pore is therefore very unlikely.
Microfabricated devices in the measurement of exocytosis at single cells
Recent advances in microfabrication have opened new possibilities for bioelectrochemical devices. Most of these electrochemical microsensors offer the opportunity to study single cells without having to position precisely an electrode on top of the cell, but are usually designed to study the chemistry of several cells. However, an increasing number of recent microfabricated devices for single cell analysis have been recently reported.
Electrode arrays
Most lithographically fabricated or printed microelectrode arrays feature metal or carbon based electrodes. Lactate microsensors were fabricated from screen-printed carbon ink containing cobalt phthalocyanine redox mediator and lactate oxidase enzyme.134 This sensor was used to demonstrate that m-dinitrobenzene induces a decrease in extracellular lactate in HepG2 single liver cells.
Mathies et al. reported a DNA barcode chip, where iridium oxide pH sensitive electrodes where functionalized with a single DNA strand.135 Cells (in this case, primary T cells and Jurkat T lymphoma cells) modified with membrane bound complementary single stranded DNA were then injected into the system. This allowed selective binding of different cell types and the measurement of their acidification rate.
An addressable microelectrode/microwell array, made of a basal layer of horizontally aligned electrodes, an intermediate layer of microwells, and a top layer of vertically aligned electrodes, allowed identification of secreted AP modified HeLa cells by redox cycling at the electrode intersection.136 Exocytosis from chromaffin and mast cells has been monitored with a platinum based electrode array.137 Fused silica was used as an insulator and poly-D-lysine as a biocompatible layer, allowing good adhesion of the cells to the electrode. A nanocrystalline array has also been successfully used as a transparent electrode material for the sensing of exocytosis from chromaffin cells.138 A fully automated device, allowing measurement of dopamine release with mercaptopropionic acid gold electrodes, has been reported.139 This chip allowed measurement of spike characteristics consistent with the results obtained with the conventional microfiber methods.
The optically transparent ITO is another good candidate for electrode material in microfabricated devices as it allows the simultaneous use of optical microscopy. Caged Ca2+ was used to elicit exocytosis from chromaffin cells grown on a device featuring 20 20 μm × 20 μm ITO electrodes.140 Ca2+ was released by UV illumination, and the amount of released Ca2+ was measured by fluorescence through the ITO layer while using the ITO electrodes to monitor release by exocytosis. Optical tweezers were also used to bring a Shewanella bacteria in contact with an ITO electrode to measure directly the extracellular electron transfer from its outer membrane c-type cytochrome.141 This method was expected to reduce the effect of fouling and to provide direct access to membrane molecules.
More recently, the complementary metal oxide semi-conductor technology, or CMOS, has been used to develop fully integrated electrochemical devices for these measurements at single cells.142 One device features several gold 12 μm × 20 μm electrodes connected to an amplifier. This architecture offers low noise and multiple parallel measurements, as this sensor can potentially be scaled to a large array. As a proof of concept, exocytosis from chromaffin cells was detected, without any significant differences from previous results obtained with carbon microfiber electrodes.
Field effect transistors used to monitor exocytosis at cells
Chen et al. have used a single-walled carbon-nanotube field-effect transistor modified with anti-chromogranin A antibody to study exocytosis at a single bovine adrenal chromaffin cell, triggered by different levels of histamine stimulation.143 This method, coupled with atomic force microscopy (AFM) imaging, allowed determination of the average size of the vesicles. Another single-walled carbon-nanotube field-effect transistor, modified with IGF1R-specific and Her2-specific antibodies was used to identify breast tumor cells.144 Binding of these markers, expressed at specific concentrations in breast tumors, induced a decrease in conductivity across the nanotube. A device coupling gold microelectrode arrays and field effect transistors was also used to monitor anionic membrane currents from modified HEK293 cells.145 These cells were used as transducers for glycine sensing through glycine evoked anionic currents.
Impedance measurements
Impedance has been commonly used as measurement method in cell-based microdevices. Hydrodynamic flows can be used to position a HeLa cell between two electrodes.146 Surfactant Tween and streptolysin-O have then been added, and have been shown to decrease the measured impedance, probably because of membrane permeation.
Another sensor using fluidic trapping was made by producing a bottle-neck shaped channel, with a 10-μm width at its narrowest point, allowing the capture of a single hepatocellular liver carcinoma cell.147 Impedance sensing was then performed by use of highly doped silicon electrodes, and the cell was then released by increasing the hydrodynamic pressure in the system.
A biosensor aimed at the diagnosis of HIV has been developed by modifying an array of 7 μm × 7 μm electrodes with anti-CD4+ antibodies.148 The electrodes were considered as individual pixels, and impedance monitoring was performed. Adhesion of cells on the electrode allowed rapid counting of CD4+ cells. Zhang et al. have also demonstrated, using different sizes of electrodes and different types of surface modification, that variations up to 50% can be measured for different types of sensors.149 Similarly, another article addressed the problems of the cell sensor interface, showing that biomodified electrodes offer better results, and suggesting an electrochemical model for cell based impedance assays.150
Scanning electrochemical microscopy (SECM) measurements at single cells
SECM is frequently used for topological cell study because of its improved spatial resolution and its ability to scan the sample surface.151 It has been recently applied to several components of cell physiology, such as morphology, metabolism, release and uptake.
Morphology
The most common use of SECM is in mapping a cell monolayer with a nanoelectrode to obtain high resolution images of single cells along a surface.152 It has also been suggested that the electrode can be used to induce an “electrochemical attack”, thus killing cells specifically with electrochemical products.153 Again, the spatial resolution of SECM allows the time course of this treatment to be monitored. SECM has been recently used to detect specific molecules on the cell surface, such as epidermal growth factor receptors,154 or membrane glycans.155 This was achieved by AP, via an antibody, labeling in the case of epidermal growth factor receptors, and with lectins labeled with horseradish peroxidase for the glycans. Finally, this method has been applied to the study of changes in cell morphology and membrane permeation after exposure to different concentrations of Triton X-100.156 A similar study focused on the swelling rate and rupture of cells in cold (4°C) media, and the effect of anti-freeze proteins on this hypothermic process.157 Anti-freeze proteins were shown to slow down the swelling and to delay the cell rupture.
Metabolism
The production of p-aminophenol from HeLa cells transfected with vectors encoding secreted AP was sensed by chronoamperometry.158 This method, allowing the secreted AP vector expression to be detected, was improved by growing the cells into PDMS microwells, to allow better single cell resolution. In another setup, the same cells were trapped into microwells by dielectrophoresis.159 By modifying the secreted AP vectors to add a κB element control, higher expression of p-aminophenol was detected after exposure to TNFα. This method of detection has also been coupled with optical microscopy and shear-force-based probe-sample distance regulation to simultaneously obtain electrochemical, topographical and fluorescence data.160 Extracellular and intracellular electrochemical recordings have been obtained for human breast epithelial cells.161 This allowed mass transport across the membrane to be measured, and imaging of the cell surface with nanometer resolution.
Release
Constant distance SECM was applied to the topological detection of reactive oxygen species released by macrophages.162 The main finding was that the nucleus region releases more reactive oxygen species than any other site. These molecules were also studied in single human bladder and kidney epithelial cells after exposure to heat-killed uropathogenic E. coli GR-12.163 It was found that this treatment induced release of reactive species in bladder cells, probably through toll-like receptors, but not in kidney cells.
Uptake
Respiratory activity of single neutrophils was quantified using two successive SECM scans.164 The first scan contained currents corresponding to cell topography and oxygen consumption. The cell was then killed with cyanide, and a second scan, measuring the cell topography only, was performed. The difference of these two scans allows the determination of respiratory activity. Enzyme modified electrodes were used to map glucose consumption at the surface of a single fibroblast.165 Respiration profiles for PC12 cells have also been investigated, showing higher oxygen uptake at the points where the cell touches the Petri dish. Interactions between HeLa cells immobilized on a cell array and silver nanoparticles have been studied with iridium chloride as the redox mediator.166 The IrCl62− produced in-situ can indeed be reduced by silver nanoparticles and can be used as a probe to detect silver nanoparticle uptake.
Ion selective electrodes
Potentiometric methods and ion selective electrodes are attractive technologies to investigate ionic variations, for instance regulated by potassium channel activity, 167 at the surface of single cells. Self-referencing ion-selective (H+, Ca2+, K+ and Cl−) potentiometric microelectrodes were applied to the measurement of membrane fluxes in the algal cell Eremosphaera viridis, and to the elucidation of the biochemical pathways controlling respiration and chloroplast translocation in photosynthesis.168 Measurements of extracellular K+, released by Ca2+ controlled potassium channels, were performed with K+ selective electrodes in Chinese hamster ovary cells.169 These electrodes created an ion trap, because of diffusion hindrance, thus enhancing the measured signal.
Mass spectrometric analysis of single cells
Mass spectrometry (MS) is emerging as a powerful tool for measurements of single cell components. This technique can provide both qualitative and quantitative information about a specific analyte or analytes in a cell, be used to determine their elemental composition, chemical structure, and concentration. This powerful technique can provide information about the nature of biocompounds (e.g. proteins, peptides, glycoproteins etc) in complex mixtures such as cells, cell lysates etc. Certain approaches of MS also have the capability to study the dynamic processes in cells. Several reviews of this area have been published recently.170–172
In this part of the single cell analysis summary we provide an overview of the application of different MS techniques for the analysis of biomolecules at single-cell level. We have chosen to divide these techniques by the ionization source used as this often relates to the state of the cell and overall sampling procedure.
Mass spectrometric analysis of single cells with electrospray ionization
Electrospray ionization (ESI) operates at atmospheric pressure with the molecule to be analyzed dispersed by an electric-field-generated spray of column effluent into a fine aerosol. A sample solution is sprayed from a small tube into a strong electric field in the presence of a nitrogen flow to assist desolvation. The droplets formed evaporate and ejected charged particles then enter the mass analyzer. ESI can be easily coupled to high-performance liquid chromatography (LC-ESI MS) and capillary electrophoresis (CE-ESI MS) to analyze complex mixtures.
LC-MS is a powerful tandem technique used for many applications and provides high sensitivity and selectivity. Generally its application is oriented towards the specific detection and potential identification of chemicals in a complex mixture. Tandem LC-MS/MS was recently demonstrated for quantification and characterization of shellfish poisoning toxins in single-cell isolates,173 quantitation of protein expression levels in single islets of Langerhans174 and of human tumor proteomes.175
The proteolysis of the β-APP in single live cells was examined using CE coupled to LIF for single-cell analysis and LC-MS for identification of peptide modification.14 Initially, single cells from a human erythroleukemia nonadherent cell line were loaded with a fluorescently labeled peptide and analyzed using CE-LIF. Then the cells were lysed, peptide fragments were isolated (purified) by use of antibodies, and the masses of the peptide fragments in the lysate were determined using LC coupled to ESI MS. Three distinct fragments, which differed by a single uncharged amino acid, were generated from the β-APP. This approach has enabled the profiling multiple proteolytic activities acting on a single substrate and the identification of cleavage sites in single cells. Hence, it might find application in profiling a diverse array of enzyme activities (kinases and phosphatases) at the single-cell level.
Williamson et al. described a method of microarray analysis for relative quantification of nuclear extracts of embryonic stem cells by two-dimensional LCLC-MS/MS.176 Stem cell signatures have been defined at the mRNA level using microarrays. Cells were fractionated, enriching for nuclei. The nuclear fractions from the four populations were each trypsinized and labeled with a specific iTRAQ reagent. The samples were then mixed and subjected to two-dimensional LC-MS/MS. Proteins were identified and relatively quantified by integrating the peptide data for each specific protein.
Relative protein content in astrocytes was quantified using multidimensional LC MS/MS analysis.177 Initially, samples were separated using SDS-PAGE. Then proteins in each segment of the gel lane are reduced and digested. An LC-MS/MS experiment is then performed to obtain peptide sequence information and identify the proteins in each segment. A total of 516 proteins in astrocyte conditioned media and cell lysate were identified and relatively quantified.
Separation and identification of 2243 proteins digested from immune-captured prolactin cells were performed by two-dimensional-nanoscale liquid chromatography (2D-nanoLC/MS) and characterized by tandem MS.178, 179 The 2D-nanoLC/MS approach seems to be a suitable method to perform sensitive proteomic analysis on limited protein quantities.
The secretome of isolated endothelial progenitor cells from the lungs of adult mice cells were studied using nanoflow LC–MS. As many as 133 proteins categorized as membrane-bound or secreted were found. More than 500 proteins were identified in total.180
A shotgun analysis method for protein identification in a small number of breast cancer cells was described by Wang et al.181 The sample preparation included cell lysis, protein precipitation and tryptic digestion. The digest was analyzed using nanoflow liquid chromatography coupled to quadrupole time-of-flight (Q-TOF) MS. A human blood sample was spiked with MCF-7 tumor cells to demonstrate the potential use of this method. Then the cells were isolated by antibodies and collected using flow cytometry. It was shown that this method could identify an average quantity of proteins from 500–5000 MCF-7 breast cancer cells.
Mass spectrometry has been combined with laser capture microdissection (LCM) in order to examine solid tumor heterogeneity on a cellular basis.182 Decomposition of tumor cells by LCM enables a more direct assessment of tumor heterogeneity. Six specimens 50000 cells each were collected by LCM, lysed, and the digested proteins were extracted. Prepared samples were analyzed by LC-MS/MS resulting in more than 500 unique protein identifications and revealing the content of the cell. Further optimization of this method including analysis of additional clinical specimens might help to improve the understanding of tumor heterogeneity, and serve as a platform for solid tumor biomarker discovery.
A metabolomic analysis method using a combination of capillary electrochromatography and ESI-MS to analyze the metabolome of a human hepatocellular carcinoma cell line (HepG2) was described by Katoa et al.183 The method was used to separate simultaneously more than 100 charged and neutral compounds from standard samples. So far eighteen compounds (mainly amino acids, nicotinamide, creatinine) were identified from the cell extract, including marker compounds related to hepatocellular cell activity. The authors claim that further improvements in this detection method will make it possible to measure hundreds or thousands of biological compounds in a cell in a single measurement. This would make the system useful both for metabolome analysis and for diagnostic measurements of cell function.
Lapanis et al. have described a highly efficient tandem method based on coupling the electrophoretic separation with ESI time-of-flight (TOF) MS detection for single-cell metabolomic profiling of the Aplysia californicas R2 neuron and metacerebral cell.184 Detection limits are in the low nanomolar range (<50 nM, <300 amol) for a number of cell-to-cell signaling molecules, metabolites, neurotransmitters (e.g. acetylcholine, histamine, dopamine, serotonin). The instrument yields high-efficiency separations, e.g. ~600 000 theoretical plates for eluting acetylcholine bands. A nebulizer-free coaxial sheath-flow interface provides a stable electrospray, yielding a highly reproducible signal. Due to the improved CE sampling interfaces (optical trap, smaller nanovials) a larger fraction of individual cell contents can be used for these measurements. This enables characterization of smaller structures. The authors claim that the described method provides better mass and concentration limits of detection (LOD) for neurotransmitters and other small molecules than reported before.185, 186
A fully integrated microfabricated fluidic device was developed for the automated real-time analysis of human erythrocytes using CE and ESI-MS.187 The microfluidic structure comprised a means for rapid lysis of single cells, electrophoresis channel, where cellular constituents were separated, and an integrated electrospray emitter for ionization of separated components in eluent. The dissociated heme group and the α and β subunits of hemoglobin from individual erythrocytes were detected using this method. The average analysis throughput was approximately 12 cells per minute.
Another tandem method combining microscale electrophoretic separation and Q-TOF MS detection was developed for rapid identification of cytochrome C, β-lactoglobulin, ovalbumin and BSA from human muscle cell lysate.188 The separation was performed using a monolithically integrated, polymer microchip comprising an electrophoretic separation unit, a sheath flow interface and an ESI emitter. Good analytical performance of the microchips and good repeatibility was shown by performing both peptide mass-fingerprinting of complex cell lysates and protein identification based on single peptide sequencing. In contrast to HPLC-MS, the separation performed with the developed microchips is fast (all peptides migrated within 98 s), requires much smaller amount of sample (<3 μL), and provides better resolution (plate numbers approaching 106). Protein sequence coverage can be obtained without any pretreatment of the proteolytic samples. Peptides from digested proteins can be detected with reasonably high sensitivity (at the femtomole level). The total analysis of the proteins from sampling to detection step required less than ten minutes.
Nano-ESI MS
Direct and quick molecular analysis of live plant, mouse-embryonic fibroblasts, and mast single cells has been developed using nano-electrospray ionization (nano-ESI) tip and mass spectrometers.189–191 A nano-ESI tip was inserted into the single plant cell acting as a micropipette. The process was viewed under a video-microscope. The cell content was sucked into the tip, electrosprayed, and the compounds were detected and identified by Q-TOF-MS or MS/MS. This method was also applied to the cell classification of seven types of cell lines at the single-cellular level by principal component analysis.
Laser ablation electrospray ionization mass spectrometry-MS
A relatively recent invention, laser ablation electrospray ionization mass spectrometry (LAESI-MS), is an ambient ionization technique to acquire mass spectra under normal atmospheric conditions for analysis of biological samples with sufficient water content. This ionization technique combines laser ablation of the sample with a mid-infrared laser and a secondary ESI process. The mid-infra red laser excites the water molecules of the sample, generates gas phase particles that are then ionized through interactions with charged droplets from the ESI source. The ionized molecules are then swept into the mass spectrometer, which analyzes and records the composition of the released ions. This ionization approach allows analytical investigations to be performed directly on a tissue or biofilm under native-like experimental conditions.192, 193
Applicability of LAESI-MS was demonstrated for the in situ analysis of the compounds and metabolites in individual plant cells (Allium cepa, Narcissus pseudonarcissus and Aphelandra squarrosa) to explore metabolic variations in cell populations.194–196 The molecular images produced by LAESI in combination with optical and scanning electron microscopy (SEM) allowed identification of metabolic pathways with ~50 μm depth resolution. Over 40 primary and secondary cell metabolites were detected (36 of them assigned). The limit of detection was decreased to only a few femtomoles.
LAESI-MS also allows the rapid, direct and sensitive identification of peptides, proteins, metabolites, and other biomolecules with molecular weight up to 66 kDa in tissue sections and cells, with minimal invasion and destruction of the sample. Thus this technique is suitable for two-dimensional molecular imaging and for depth profiling, for the investigation of biochemical changes in live organisms. The current lateral and depth resolution is ~100 μm and ~40 μm, respectively. Although at this stage LAESI cannot compete with vacuum imaging methods such as SIMS, MALDI, and graphite-assisted laser desorption/ionization imaging in terms of spatial resolution, its ability to work with live specimens in situ without sample preparation and high-throughput enable unique applications and appears to open new perspectives for application in the fields of pharmaceutical and biological research, molecular pathology, clinical diagnostics, chemical and biological defense, forensics, agriculture, food process monitoring.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)
Matrix-assisted laser desorption/ionization mass spectrometry is a widespread analytical tool for analysis of peptides, proteins and many other biomolecules. MALDI provides nondestructive vaporization and ionization of biomolecules. In MALDI analysis, the analyte is first co-crystallized with an excess of a matrix compound (e.g. UV absorbing organic acid). After laser radiation of analyte–matrix mixture, matrix absorbs the laser light energy, vaporizes and carries the analyte with it. The matrix also serves as a proton donor and receptor, which allows ionization of the analyte in both positive and negative ionization modes, respectively. There are three main types of mass analyzers used with the MALDI ionization source: a linear time-of-flight (TOF), a TOF reflectron, and a Fourier transform mass analyzer.197
MALDI-MS combined with microscale sample preparation, thin layer matrix deposition, negative-ion mode detection with the aminoacridine as a matrix was recently exploited for the detection of endogenous metabolites (ADP, GDP, ATP, GTP, acetyl-CoA) in individual yeast cells.198 The method is extremely fast, highly throughput and sensitive. The calculated LOD varied from 5 to 12 attomoles, which is three orders of magnitude lower than for other MALDI methods. The matrix deposition approach is compatible with any microfluidic platform, including lab-on-a-chip, lab-on-capillary and can be used in nano-liquid chromatography and capillary electrophoresis.199
A high-throughput and nontargeted metabolomic technique using MALDI-MS in negative ionization mode was developed for the rapid analysis of metabolites from human acute lymphoblastic leukemia Jurkat cells.200 A detection limit lower than 10 fmol well−1, and a high linearity, at low concentrations was achieved. Up to 150 metabolite peaks were detected from a single analysis within 90 s. For multivariate analysis of Jurkat cells against drug-treatment, three anticancer drugs were utilized. Using principal component analysis of metabolites, clear independent clusters for cells treated with anticancer drugs was demonstrated.
Quantitative microanalysis of peptides in individual Aplysia californica neurons and small pieces of tissue was described using MALDI MS.201 A combination of appropriate sample preparation, analyte labeling with stable isotope, and advanced MALDI MS approaches allowed relative and absolute quantification of the physiologically active neuropeptide cerebrin from nerves and neuronal clusters with a limit of detection of 19 fmol in nanoliter volume samples.
Rapid bacterial typing of whole cells of Salmonella strains has been carried out using MADI-TOF MS.202 More than 200 biomarker peaks were found and identified as ribosomal or nucleic acid binding proteins. The MS-based bacterial classification was in good correlation with the results of DNA sequence-based methods.
MALDI-TOF MS and tandem MS were utilized for the determination of the peptide content in single dissected neurons from the nematode Ascaris sum.203 Six novel neuronal peptides were discovered and sequenced. The results of the peptide content measured were compared favorably with immunocytochemistry data.
An interesting approach combining a novel neuronal affinity capture technique followed by MALDI-TOF MS profiling has been developed to characterize the localization of neuropeptides in the brain of adult Drosophila melanogaster.204 Fruit fly brains, genetically labeled with the surface marker mCD8-GFP, were dissociated into single cell preparations and incubated with paramagnetic beads conjugated to a monoclonal anti-mCD8 antibody for purification. The mixture was then placed in a magnetic column, and the cells captured by antibodies were retained in the column. Then, the cells were eluted from the column and analyzed by MALDI-TOF MS. A total of 42 neuropeptides from 18 peptide families were sequenced and extensively characterized.
To identify cell lines in culture MALDI-TOF MS was used to provide rapid but exact characterization of 34 different species in 66 various cell cultures.205 A reference spectra library was generated that allows unambiguously the identification of all 66 cell lines. Depending on the availability of reference spectra from the same or related species, unknown samples could be classified taxonomically to the level of species identification. The procedure is rapid, results for single sample can be obtained within one hour, cost effective, construction of laboratory specific spectrum libraries is feasible and scheduled MALDI-TOF identification of the authenticity of the cell lines is straightforward. The cell-specific mass lists created might serve as a characteristic bar-code to complement cytological, biochemical, genetic or virological data.
Urban et al. have described the application of functional micro-arrays for high throughput analysis of unlabeled molecules in single cells and picoliter-volume samples by mass spectrometry.206 High-density micro-arrays for mass spectrometry (MAMS) were fabricated by picosecond-laser ablation patterning of a hydrophobic layer coated on a conductive support, such as steel or ITO. The high-density microscale pattern on the MAMS surface enabled unsupervised aliquoting of very small volumes of solutions or suspensions. Various compound classes (metabolites, peptides, a drug and a protein) could readily be analyzed with limits of detection in the low attomole range. Detection limits for a few metabolites in yeast cells were as low as ~500 zeptomoles. Although in this work micro-arrays were implemented with MALDI-MS, it is also possible to imagine that MAMS could be coupled to matrix-free ionization methods (e.g. NIMS). This sample preparation strategy completely eliminates the need for using microscale dispensing tools, such as microspotters, to digitize samples prior to MS analysis. The versatility of MAMS has great potential to facilitate high-throughput proteomic analysis and screening of biomolecular complexes with minute amounts of sample.
A mass-spectrometry-based strategy to determine the absolute quantity, the average number of protein copies per cell in a cell population for a large fraction of the proteome in genetically unperturbed human pathogen Leptospira interrogans cells has been reported.207 Initially, LC-MALDI and LC-ESI tandem MS were used to isolate and identify the peptides contained in the fractions of the digested protein extract from whole cells. Out of the 2221 identified proteins, 1864 proteins were provided with estimated copy per cell numbers, ranging from 40 to 15000 copies per cell. The validated method was applied to study the reorganization of the Leptospira interrogans proteome upon exposure of the antibiotics Ciprofloxacin. It was indicated that the cells react by expressing massive amounts of a small number of normally unexpressed proteins of unknown function while keeping the total cellular protein constant. The technique described is fast, efficient, versatile, and can be applied to various biological systems of low and medium complexity in future studies.
Aerni et al. have described the rapid analysis of hundreds of specimens in human kidney tissue with MS.208 The strategy combined parallel sample processing and on-chip electrophoresis with automated MALDI MS analysis. The method was optimized for small quantities of tissues allowing detection of 700 peptides in 60000 cells in a tissue microarray section. The method was high throughput with several hundred samples per day possible.
MALDI imaging mass spectrometry (MALDI-IMS) is a powerful tool for investigating of the spatial distribution of proteins, peptides, metabolites, biomarkers species within tissue section through visualizing it. Feasibilities and limitations of MALDI-IMS, for its application in clinical analysis and for molecular studies of complex biological samples such as tissue sections are described in detail in a number of reviews.209–213
The abilities of MALDI-IMS were recently demonstrated to visualize phospholipids in cultured mouse neuronal cells.214 First, the parameters which may influence the identification of lipids in cultured neurons were optimized in order to get an optimum detection of lipid molecules. The obtained results showed that the phosphatidylcholine molecules at m/z 734 and 826 form characteristic distribution patterns depending on the location of neurons. Authors claim that the obtained information will offer an understanding of aspects of neural lipid metabolism within the subcellular components of the neurons.
It can be concluded that in contrast to other techniques, for instance, LC-MS, MALDI-MS has advantages for metabolite analysis because of its high sensitivity, high-throughput ability (thousands of samples can be analyzed within a day), low sample-consuming (~ 1 μL). Although the present levels of sensitivity allow the detection of a small group of cells but they are not sufficient to detect discrete modification at a single cell level.
Matrix-free laser induced desorption/ionization MS
The use of the laser desorption/ionisation mass spectrometric imaging (LDI-MSI) technique with no matrix required has been demonstrated on animal and plant cells for analysis of cellular species at single-cell level.215, 216
Single-cell localization of individual astrocytes was achieved by monitoring the distribution of cholesterol in a cell population using LDI imaging mass spectrometry with colloidal silver as an additive.215 No labeling or sophisticated sample preparation was required. Good correlation between LDI-MS and conventional enzymatic fluorometric assay was observed.
Another application of LDI-MS was demonstrated for the detection of cellular components, such as phospholipids at individual single-cell algae Euglena gracilis and Chlamydomonas reinhardtii.216 First, algae was seeded on a bare stainless steel plate and analyzed by microspectrophotometer to obtain the information about the distribution of photoactive species (e.g., β-carotene, chlorophyll). Then cells were analyzed by LDI-MS in the negative ion mode. The content and arrangement of proplastids, photosystem components, and chemical composition of various phospholipids in individual algal cells were studied.
LDI-MS allows the analysis of cell components with higher resolution and sensitivity than that can be achieved by commonly used MALDI techniques (10 μm versus approximately 50 μm).217 A review by Russel highlights the recent progress made in the application of single particle mass spectrometry (MALDI and LDI) to the analysis of microbial cells.218
Nanostructure-initiator mass spectrometry (NIMS) uses a matrix-free desorption/ionization approach for sample introduction. A liquid initiator is trapped in a nanostructured surface to facilitate desorption and ionization of intact molecules adsorbed on the surface. NIMS initiators, in contrast to MALDI matrices, do not absorb UV energy, most do not ionize, and the analytes are not co-crystallized with the initiator. During NIMS desorption/ionization, the porous silicon absorbs laser energy that results in rapid surface heating, vaporization of the trapped initiator, and desorption/ionization of the adsorbed analyte without fragmentation. An overview of NIMS technology and its application in the detection of biofluids and tissue metabolites was recently presented by Greving et al.219
NIMS produces minimal analyte fragmentation, provides enhanced sensitivity, requires no sample preparation and enables of direct analysis of biotissues, offers the flexibility of different initiators, various surface modifications and choice of positive- or negative-ion mode. These characteristics of NIMS have been used to analyze metabolic profiles and detect specific endogenous and xenobiotic metabolites in biofluids and biological tissues. Coupling NIMS to high-resolution instruments such as a Fourier transform mass spectrometer or a Q-TOF mass spectrometer may increase specificity and improve the measurement of complex biological samples. Also NIMS is a powerful platform for MS-based tissue imaging. In NIMS the laser can be focused to a much smaller diameter (15–20 μm) than is typically used in MALDI imaging (50–300 μm). This improves spatial resolution significantly compared to MALDI imaging. However, despite all flexibilities and enhancements provided by NIMS, further improvements of ionization efficiency and ion suppression are clearly needed for the analysis of biological samples.
NIMS in negative ion mode with 3-aminopropyldimethylethoxysilane as initiator was applied for the detection of small phosphorylated molecules in a yeast cell extract such as phosphoenolpyruvate and nucleotides like ATP. Using this technique, a limit of detection in the tens of femtomoles was reached.220
Laser-induced acoustic desorption
A novel approach for investigating the amount of nano-/microparticle uptake into mammalian cells in the gas phase has been presented using a technique the authors call cell mass spectrometry (CMS).221 The experimental setup of CMS includes matrix-free laser-induced acoustic desorption of microparticles, a low-frequency quadrupole ion trap for the measurement of ultralarge m/z values, a pressure-controlled corona discharge to enhance the number of charges on a cell or microparticle, and a low-noise charge detector for total-charge measurement. Cells were incubated with gold-nanoparticles (NPs) and the kinetics of the uptake of the gold NPs by the cells was measured by investigating their mass spectra. This approach can be used for rapid and accurate determination of the number of NPs taken up into each individual cell (in contrast to ICP-MS, which measures only a mean uptake for all cells). CMS can be used to measure the cellular uptake not only of metal nanoparticles but also of nonmetal nano- and microparticles. CMS may also be useful for the measurement of the quantity of liposomes taken up by cancer cells, in drug delivery experiments.
Secondary ion mass spectrometry of cells
Time of flight secondary ion mass spectrometry (TOF-SIMS) is useful for the compositional analysis of small samples and molecules of low molecular weight. It is based on bombardment of a sample surface with a primary ion beam followed by mass spectrometry of the emitted (sputtered) secondary ions. The secondary ions are sorted by mass and energy and counted in an ion detector. Since the size of the sputtered area depends only on the primary ion beam diameter (typically this is between 100 nm and 2 μm), SIMS analysis has a relatively high lateral resolution. In addition, new methodology is making it possible to carry out depth profiling to obtain 3-D images.
A review of the current state of the art in biological imaging mass spectrometry using TOF–SIMS has been published recently.222 Recent and remarkable improvements in terms of sensitivity of TOF–SIMS imaging methods have allowed many biological applications to be successfully tested in the last few years. The diverse application of various formats of SIMS for identifying the specific role of micro-organisms in biogeochemical cycles was highlighted by Orphan et al.223 SIMS imaging of single cells was applied for quantifying the relative difference in the amount of lipid distribution between cellular sections using 40 keV C60+ ion source.224–226
Three surface analysis techniques with different sensitivity, probing depth, and lateral resolution, such as in situ AFM, X-ray photoelectron spectroscopy, and SIMS were exploited to gain insight into the surface properties of the conidia of the human fungal pathogen Aspergillus fumigates.227 Due to its high surface sensitivity, SIMS provides semi-quantitative chemical analysis of the outermost molecular layers of the sample surface. The amino acid composition of the surface of fungal rodlets was identified. A good agreement between the three methods was demonstrated. XPS and SIMS spectra were used to demonstrate that the surfaces of the wild-type A. fumigatus conidia are fairly rich in proteins, reflecting essentially the rodlet structures seen in the AFM images. Furthermore, the SIMS data confirmed that rodlets are rich in glycine, alanine, proline, and leucine, in agreement with the known amino acid composition of fungal proteins RodAp hydrophobins.
A novel TOF-SIMS MS instrument was developed and validated by analyzing prostatic hyperplasia cells, HeLa cells, and human cheek cells.228 The instrument that was developed overcomes many of the drawbacks of current TOF-SIMS spectrometers by removing the need to pulse the primary ion beam. The instrument uses a continuous primary ion beam and then samples the secondary ions using a buncher that feeds into a specially designed TOF analyzer. This novel instrument was applied for molecular depth profiling on a subcellular scale with high resolution and throughput and obtaining 3D molecular imaging.
Malm et al. have used TOF-SIMS, SEM, and interference reflection microscopy (IRM) to study the effects of different fixation and drying methods on the morphology and chemical structure of human fibroblast cells adhered to a silicon surface.229 Two fixation techniques (chemical fixation with glutaraldehyde and cryofixation by plunge freezing) and two drying techniques (freeze drying and alcohol substitution drying) were applied in order to investigate lipid distributions and sodium/potassium ion gradients.
TOF SIMS analysis of freeze fractured PC12 cells using a Bi cluster ion source was reported in this time period.230 The analyzed cells were prepared for the experiment in a newly designed in situ freeze fracture device. The work again showed the ability to obtain high-resolution SIMS imaging of single cells with subcellular lateral resolution from cells preserved in an ice matrix. The signal from lipid fragment ions rarely identified in freeze fried single cells is better observed in the freeze-fractured samples for these experiments. The freeze fracture device was coupled to high-resolution cluster SIMS imaging system to provide the sensitivity and resolution required to carry out biologically relevant SIMS experiments.
Chemical species within intact mammalian cells were imaged using SIMS including the simultaneous mapping of subcellular elemental and molecular species along with intrinsic membrane-specific cellular markers.231 Appropriate sample preparation protocols to maintain the physical and chemical integrity of treated mammalian cells and well-developed MS imaging procedures were combined. Spatial resolution of approximately 400–500 nm was achieved. The results from mapping the cell surface for phosphatidylcholine and several other molecular ions present in the cells established that spatially resolved chemical signatures of individual cells can be derived from novel multivariate analysis and classification of the molecular images obtained at different m/z ratios. These developments can in principle be combined with new SIMS instruments that employ tandem mass spectrometry (MS/MS) for chemical speciation of unknown mass peaks and dynamic SIMS beam currents for enhanced molecule detection.
NanoSIMS is a version of secondary ionization mass-spectrometry using dynamic ablation of the sample surface. This approach provides better spatial resolution than conventional SIMS for cases where specific element isotope labeling and elemental analysis can be used. NanoSIMS can provide an image of the elemental and isotopic composition of a sample with a spatial resolution of about 50 nanometers.
The distribution, uptake and fixation of CO2 and N2 by cyanobacterium Trichodesmium IMS-101 cells were directly quantified and characterized by imaging of 13C and 15N isotopes with high-resolution NanoSIMS.232 A few cells at once were scanned with 150-nm spatial resolution to capture intercellular variability. After performing correlated transmission electron microscopy and NanoSIMS analysis on trichome thin-sections, transient inclusion of the isotopes 15N and 13C into discrete subcellular granules was observed. The dynamics of the uptake of newly fixed CO2 and N2 within all cells along a Trichodesmium trichome was also observed and investigated.
Li et al. described the application of NanoSIMS for visualization of oligonucleotide probe-conferred hybridization signals and isotopic measurements at the single cell level on microbial cells.233 An oligonucleotide containing iodized cytidine was hybridized on fixed cells of Escherichia coli cultured on media containing different levels of isotopes 13C or 15N. It was observed that iodine signals could be localized on targeted cells and the isotopic enrichment could be monitored at the single-cell level.
Behrens et al. developed a method combining element labeling fluorescence in situ hybridization (EL-FISH) and NanoSIMS based on the fluorine or bromine isotope imaging to examine phylogenetic identity and metabolic activity of individual bacterial cells in complex microbial communities.234 The FISH technique was modified by using halogen containing, fluorescently labeled tyramides in order to overcome the natural fluorine and bromine backgrounds. This resulted in enhanced sensitivity and allowed them to distinguish the target from non-target cells in NanoSIMS fluorine or bromine images. The method was optimized on single cells of axenic E. coli and Vibrio cholerae cultures; and then applied to study interrelationships of cyanobacterium and alphaproteobacterium and evaluation of microbial aggregates obtained from human oral biofilms. EL-FISH/NanoSMS was shown to be a suitable method to study metabolic interactions within cells by visualizing the fate of substrates labeled with 13C and 15N and identification of individual cells by halogen labeling via EL-FISH. The aim is to help to understand the distribution of microbial activities in and among individual cells of probe-identified populations.
Imaging mass spectrometry (MALDI-IMS and SIMS) is currently receiving a significant amount of attention because of its capability to provide an efficient means to locally observe complex biological phenomena and to directly identify biomolecules in cells, tissue sections with high spatial resolution (MALDI 50 or even 20 μm or SIMS 100 nm). This makes these methods powerful tools for biomedical diagnostics and molecular histology. The challenges, opportunities, possibilities and difficulties associated with the application of imaging MS to biomedical tissue studies have been comprehensively discussed in a series of reviews.235–238
Inductively coupled plasma mass spectroscopy of single cells
Inductively coupled plasma (ICP) MS combines the easy sample introduction and quick analysis of ICP technology with the accurate and low detection limits of a mass spectrometer. The sample being analyzed is converted into an aerosol and then is swept into the plasma to create the ions. A less common method to introduce a sample is via the use of laser ablation (LA). In this method, a laser is focused on the sample and creates a stream of ablated material that can be swept into the plasma. This method is particularly useful for solid samples.
A novel mass-cytometric technique for real-time immunochemical analysis of multiple antigens in single-cells of a leukemia cell line based on ICP-TOF MS was described by Bandura et al.239 and Tanner et al.240 The equipment for the analysis included a plasma-vacuum interface, DC quadrupole turning optics for decoupling ions from neutral components, an RF quadrupole ion guide discriminating against low-mass dominant plasma ions, a point-to-parallel focusing DC quadrupole doublet, an orthogonal acceleration reflectron analyzer, a discrete dynode fast ion detector. A high spectrum generation frequency provides capability to collect multiple spectra from each particle-induced ion cloud. 1100 cells per second can be resolved and characterized individually. The technique was applied for the real time simultaneous assay of 20 antigens (biomarkers) at the single cell level and for the assay of leukemia patient samples immuno-labeled with lanthanide-tagged antibodies. Mass cytometry based on ICP-TOF MS overcomes existing limitations in flow cytometry resulting from overlap of fluorescent tag emission spectra.
A method of simultaneous cell counting and determination of constituent metals in alga Chlorella vulgaris single cells using time-resolved ICP-MS was reported by Ho et al.241 Rapid measurement of sorbed analyte ions on the biological cells without separation of the cells from the original suspension was demonstrated in a kinetic study of the interactions of metal ions and algae in aqueous standards. The ICP-MS spike intensity of an element is related to its quantity in the algal cell. Quantitative and semi-quantitative determination of the metal contents of the algal cells is feasible. Major elements, for example Mg, which was present at the level of 108 atoms cell−1, and trace elements, for example Mn and Cu which was present at the level of 5 × 106 atoms cell−1, could be detected.
Additional techniques for single cell analysis
Several other techniques can be used as a complement to the methods presented so far in this review. Here, we do not attend to be exhaustive, but rather we present a selection of interesting examples of non chemical methods (imaging, microscopy and labs on a chip) and of single cell ‘omics’.
Imaging and microscopy
Directly imaging a cell is the most obvious way to obtain significant insights about its physiology.52 AFM is a commonly used method for cell imaging, as chemically modified tips can provide access to membrane receptors, such as cadherins.242, 243 Adding electrode systems to the tip also allows impedance, potentiometric, or amperometric measurements to be made in a wide range of samples (plant or animal cells, microfabricated substrates).244 Using the cantilever to compress the cell is also a popular way to investigate cellular mechanics.245, 246 SEM offers better resolution, and some methods for liquid phase imaging have been proposed.247, 248
The recently developed stimulated emission depletion (STED) microscopy method, where a secondary laser depletes the halo of the excitatory laser thus improving its resolution, is attracting increasing attention in the neurobiological field, as it allows, for instance, detection of dynamics of dendritic spines of pyramidal neurons with a resolution better than 100 nm.249 Recent reports with this technique include mapping of Na+-K+-ATPase in dendritic spines, showing that this enzyme is mostly located in the post-synaptic part of the spine,250 and description of vesicle mobility in the axons of cultured neurons.251
Coherent anti-Stokes Raman scattering (CARS)252 has been recently utilized to image glucose in living yeast cells.253 A similar method, stimulated Raman scattering (SRS) microscopy, has been proposed for biomedical applications, such as measurement of fatty acid uptake in human lung cancer cells.254
Lab on a chip
Development of lab on chip devices for biological applications is a very popular and active field. Most of the reported systems are based on microfluidics.7, 255, 256 These systems can allow co-culture of two specific cells, thus making possible the study of close interactions,257 or manipulation and stimulation of these cells by optical tweezers258 or hydrodynamic flow.259 Cytometry based systems260 using dielectrophoresis261 or hydrodynamic flows262, 263 have been reported and used for rapid screening of a cell population. These methods have been successfully applied to the study of mitogen activated protein kinase signaling in Saccharomyces cerevisiae,264 the measurement of antigen-antibody reaction kinetics,265 the identification of a possible signaling signature in human myeloproliferative disorders266 and the temporal analysis of protozoan lysis.267 Culture of cells in microwells also facilitates single cell analysis by enabling the quick identification of a specific cell and by allowing the study of interactions between a small number of cells.268, 269 An original sorting method involves the production of PDMS microwells, with a detachable base, termed as microraft.270 Cells can be grown individually on this microraft, and selected and isolated by detaching the microraft from the rest of the chip with a needle.
Other methods, such as optical stretching,271 chemiluminescence based detection on an array of thin film transistor photosensors272 and the real monitoring of protein translation using a fusion polypeptide containing a streptavidin modified moiety and a streptavidin modified quartz crystal microbalance.273
Single cell ‘Omics’
‘Omics’ refers to genomics, metabolomics and proteomics, and is the main field in molecular biology. Its application to single cell analysis has recently gained considerable importance following the development of single molecule methods reducing the amount of sample and allowing on chip analysis.171 Single cell genomics were applied to analysis of developmental fate of daughter cells,274 single DNA molecule sequencing using a nanopore,275 single cell qPCR,71 single molecule real-time sequencing data from a single DNA polymerase276 and from a viral genome277 and the genome haplotyping of single cells.278
Transcriptomics applied to single cells include reverse transcriptase polymerase chain reaction (RT-PCR) on a single cell fluidic chip,25 real time PCR for different types of cells isolated from hematopoietic tissues showing the presence of rare progenitor and stem cells,279 and a single-cell mRNA digital sequencing assay which, when applied to a single mouse blastomere, identified 75% more genes than the conventional microarray technique.280 In the field of proteomics, a fluidic chip performing microcavitation induced cell rupture, capture of the released proteins on antibody printed spots and measurement by total internal reflection microscopy.281,282 This method was tested on GFP transfected cells and showed a relative precision of 88%.
‘Omics’ is therefore a very powerful technique for modern biology and the next generation of these techniques is expected to provide a significant contribution to the field of single cell analysis.
Subcellular measurements of singlet oxygen as an analytical probe
The last topic we wish to introduce in this section is the laser-based generation of singlet oxygen in cellular regions. This has recently been reviewed.283 Singlet oxygen has been selectively produced in subcellular volume elements upon irradiation with a focused laser using one- or two-photon excitation, and this singlet oxygen phosphorescence was detected in time-resolved experiments from these sub-cellular volume elements.284 This was then used to make qualitative and quantitative measurements of the effects of this on cell function in the part of the cell where the singlet oxygen was selectively created. Using these tools, unique information can be provided to help further elucidate how singlet oxygen perturbs cell function. This has been used to investigate the viscosity of the intracellular matrix upon cell death.285
Acknowledgments
We acknowledge the many coworkers that have come before and who’s works we cite in this review. The European Research Council (Advanced Grant), Knut and Alice Wallenberg Foundation, the Swedish Research Council (VR), and the National Institutes of Health are all thankfully acknowledged for financial support.
Biographies
Yuqing Lin is currently a postdoctoral researcher in the Department of Chemistry at the University of Gothenburg. She received her B.S. in 2003 from Zhengzhou University (China) and her PhD in Analytical Chemistry from Institute of Chemistry, Chinese Academy of Science (ICCAS) in 2008. She worked as an Assistant Professor in the field of in vivo and on line electroanalytical chemistry in ICCAS from 2008 to 2010. Her research interests include micro/nano structured electroanalytical methods for probing neurochemistry both in in vivo and in single cells.
Raphaël Trouillon is currently a postdoctoral researcher in the Department of Chemistry at the University of Gothenburg. He graduated with his B.Eng and M.Eng. from the Ecole Polytechnique (France) in 2006 and with his M.Sc from the Department of Bioengineering of the Imperial College London (UK) in 2007. He obtained his Ph.D degree in Biomedical Engineering from the Imperial College in 2010. His research interests include electrochemistry applied to life sciences and cells-on-a-chip.
Gulnara Safina is currently Assistant Professor in the Department of Chemistry at the University of Gothenburg (Sweden). She received her B.S in 2002 and PhD in 2006 from the Department of Chemistry at Kazan State University (Russia). Previously she was a postdoc in Department of Pure and Applied Biochemistry and Department of Biochemistry at Lund University (Sweden) for two years. Her research interests include bio-, enzyme- and immunosensors, development of analytical methods and tools for multicompound assay, bioanalytical and single-cell electrochemistry.
Andrew G. Ewing is currently Professor of Analytical Chemistry at both Chalmers University of Technology and the University of Gothenburg in Sweden. He is also Director of the newly formed Chalmers/GU Center for Bioanalytical Chemistry. He was previously Professor at Penn State University. He received his B.S. from Saint Lawrence University in Canton, NY and his PhD at Indiana University in Bloomington, IN with R. Mark Wightman as supervisor. He was a postdoc in the laboratory of Royce Murray at the University of North Carolina for 13 months. His research interests have been in single cell analysis, and neuroanalytical chemistry, chemical imaging, and methods in electrochemistry, separations, and mass spectrometry.
Literature Cited
- 1.Lindstrom S, Andersson-Svahn H. Lab Chip. 2010;10:3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- 2.Kortmann H, Blank LM, Schmid A. Advances in Biochemical Engineering/Biotechnologies. Springer; Berlin/Heidelberg: 2010. [DOI] [Google Scholar]
- 3.Omiatek DM, Cans AS, Heien ML, Ewing AG. Anal Bioanal Chem. 2010;397:3269–3279. doi: 10.1007/s00216-010-3698-4. [DOI] [PubMed] [Google Scholar]
- 4.Frost NW, Jing M, Bowser MT. Anal Chem. 2010;82:4682–4698. doi: 10.1021/ac101151k. [DOI] [PubMed] [Google Scholar]
- 5.Stuart JN, Sweedler JV. Anal Bioanal Chem. 2003;375:28–29. doi: 10.1007/s00216-002-1609-z. [DOI] [PubMed] [Google Scholar]
- 6.Arcibal IG, Santillo MF, Ewing AG. Anal Bioanal Chem. 2007;387:51–57. doi: 10.1007/s00216-006-0690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WH, Ai F, Wang ZL, Cheng JK. J Chromatogr B. 2008;866:104–122. doi: 10.1016/j.jchromb.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Lapainis T, Sweedler JV. J Chromatogr A. 2008;1184:144–158. doi: 10.1016/j.chroma.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 9.Ramsay LM, Dickerson JA, Dada O, Dovichi NJ. Anal Chem. 2009;81:1741–1746. doi: 10.1021/ac8025948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swearingen KE, Loomis WP, Kehimkar B, Cookson BT, Dovichi NJ. Talanta. 2010;81:48–953. doi: 10.1016/j.talanta.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsay LM, Dickerson JA, Dovichi NJ. Electrophoresis. 2009;30:297–302. doi: 10.1002/elps.200800498. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Zhang X, Bao X, Lu H, Zhang W, Wu W, Miao H, Jiao B. J Chromatogr A. 2008;1201:120–127. doi: 10.1016/j.chroma.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Xie W, Xu A, Yeung ES. Anal Chem. 2009;81:1280–1284. doi: 10.1021/ac802249m. [DOI] [PubMed] [Google Scholar]
- 14.Brown RB, Hewel JA, Emili A, Audet J. Cytometry A. 2010;77:347–355. doi: 10.1002/cyto.a.20880. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Thompson LV, Navratil M, Arriaga EA. Anal Chem. 2010;82:4570–4576. doi: 10.1021/ac100577q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MW, Martin RS. Analyst. 2008;133:1358–1366. doi: 10.1039/b807093h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dishinger JF, Reid KR, Kennedy RT. Anal Chem. 2009;81:3119–3127. doi: 10.1021/ac900109t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D, Sims CE, Allbritton NL. Electrophoresis. 2010;31:2558–2565. doi: 10.1002/elps.201000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips KS, Kang KM, Licata L, Allbritton NL. Lab Chip. 2010;10:864–870. doi: 10.1039/b921817c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marc PJ, Sims CE, Bachman M, Li GP, Allbritton NL. Lab Chip. 2008;8:710–716. doi: 10.1039/b719301g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boardman AK, McQuaide SC, Zhu C, Whitmore CD, Lidstrom ME, Dovichi NJ. Anal Chem. 2008;80:7631–7634. doi: 10.1021/ac800890b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boardman A, Chang T, Folch A, Dovichi NJ. Anal Chem. 2010;82:9959–9961. doi: 10.1021/ac1022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greif D, Galla L, Ros A, Anselmetti D. J Chromatogr A. 2008;1206:83–88. doi: 10.1016/j.chroma.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Lai HH, Quinto-Su PA, Sims CE, Bachman M, Li G, Venugopalan V, Allbritton NL. J R Soc Interface. 2008;5:S113–S121. doi: 10.1098/rsif.2008.0177.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. Proc Natl Acad Sci USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L, Huang H, Dong X, Wu D, Qin J, Lin B. Electrophoresis. 2008;29:5055–5060. doi: 10.1002/elps.200800331. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Lu M, Yin X. Talana. 2008;75:1227–1233. doi: 10.1016/j.talanta.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Reid KR, Kennedy RT. Anal Chem. 2009;81:6837–6842. doi: 10.1021/ac901114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Li Q, Wang X, Wang Z, Zhang R, Yin M, Yin L, Xu K, Tang B. Anal Chem. 2010;82:2006–2012. doi: 10.1021/ac902741r. [DOI] [PubMed] [Google Scholar]
- 30.Xu CX, Yin XF. J Chromatogr A. 2011;1218:726–732. doi: 10.1016/j.chroma.2010.11.049. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, Niu Y, Bi S, Zhang S. J Chromatogr B. 2008;870:46–50. doi: 10.1016/j.jchromb.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 32.He P, Niu Y, Mei Z, Bao J, Sun X. J Chromatogr B. 2010;878:1093–1097. doi: 10.1016/j.jchromb.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Omiatek DM, Santillo MF, Heien ML, Ewing AG. Anal Chem. 2009;81:2294–2302. doi: 10.1021/ac802466g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omiatek DM, Dong Y, Heien ML, Ewing AG. ACS Chem Neurosci. 2010;1:234–245. doi: 10.1021/cn900040e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, Huang Y, Liu YM. J Chromatogr A. 2009;1216:6746–6751. doi: 10.1016/j.chroma.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S, Li X, Liu YM. Anal Chem. 2009;81:3873–3878. doi: 10.1021/ac900391u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye F, Huang Y, Xu Q, Shi M, Zhao S. Electrophoresis. 2010;31:1630–1636. doi: 10.1002/elps.200900665. [DOI] [PubMed] [Google Scholar]
- 38.Ren X, Qu F, Zhang L, Ding J, Liu Q. Electrophoresis. 2010;31:324–330. doi: 10.1002/elps.200900417. [DOI] [PubMed] [Google Scholar]
- 39.Ding J, Zhang L, Qu F, Ren X, Zhao X, Liu Q. Electrophoresis. 2011;32:455–463. doi: 10.1002/elps.201000324. [DOI] [PubMed] [Google Scholar]
- 40.Sun X, Niu Y, Bi S, Zhang S. Electrophoresis. 2008;29:2918–2924. doi: 10.1002/elps.200700792. [DOI] [PubMed] [Google Scholar]
- 41.Xu X, Arriaga EA. Anal Chem. 2010;82:6745–6750. doi: 10.1021/ac101509d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamboliev IA, Smyth LM, Durnin L, Dai Y, Mutafova-Yambolieva VN. Eur J Neurosci. 2009;30:756–768. doi: 10.1111/j.1460-9568.2009.06869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fercher G, Haller A, Smetana W, Vellekoop MJ. Anal Chem. 2010;82:3270–3275. doi: 10.1021/ac100041p. [DOI] [PubMed] [Google Scholar]
- 44.Zhao S, Huang Y, Shi M, Liu R, Liu YM. Anal Chem. 2010;82:2036–2041. doi: 10.1021/ac9027643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmid A, Kortmann H, Dittrich PS, Blank LM. Curr Opin Biotechnol. 2010;21:12–20. doi: 10.1016/j.copbio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Vo-Dinh T. Spectrochim Acta B. 2008;63:95–103. doi: 10.1016/j.sab.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Carlo D, Lee LP. Anal Chem. 2006;78:7918–7925. doi: 10.1021/ac069490p. [DOI] [PubMed] [Google Scholar]
- 48.Taraskaz JW, Zagotta WN. Neuron. 2010;66:170–189. doi: 10.1016/j.neuron.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiller DG, Wood CD, Rand DA, White MRH. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 50.Czechowska K, Johnson DR, van der Meer JR. Curr Opin Microbiol. 2008;11:205–212. doi: 10.1016/j.mib.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Ge S, Koseoglu S, Haynes CL. Anal Bioanal Chem. 2010;397:3281–3304. doi: 10.1007/s00216-010-3843-0. [DOI] [PubMed] [Google Scholar]
- 52.Petibois C. Anal Bioanal Chem. 2010;397:2051–2065. doi: 10.1007/s00216-010-3618-7. [DOI] [PubMed] [Google Scholar]
- 53.Cohen D, Dickerson JA, Whitmore CD, Turner EH, Palcic MM, Hindsgaul O, Dovichi NJ. Annu Rev Anal Chem. 2008;1:165–190. doi: 10.1146/annurev.anchem.1.031207.113104. [DOI] [PubMed] [Google Scholar]
- 54.Rigler R. Biochem Biophys Res Commun. 2010;396:170–175. doi: 10.1016/j.bbrc.2010.04.070. [DOI] [PubMed] [Google Scholar]
- 55.Chiu DT, Lorenz RM. Acc Chem Res. 2009;42:649–658. doi: 10.1021/ar8002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinaud F, Dahan M. Science. 2008;320:187–188. doi: 10.1126/science.1156510. [DOI] [PubMed] [Google Scholar]
- 57.Lu YY, Chen TS, Wang XP, Li L. J Biomed Opt. 2010;15:046-028. doi: 10.1117/1.3481141. [DOI] [PubMed] [Google Scholar]
- 58.Urru SA, Veglianese P, De Luigi A, Fumagalli E, Erba E, Gonella Diaza R, Carra A, Davoli E, Borsello T, Forloni G, Pengo N, Monzani E, Cascio P, Cenci S, Sitia R, Salmona M. J Med Chem. 2010;53:452–7460. doi: 10.1021/jm100362x. [DOI] [PubMed] [Google Scholar]
- 59.Nunez-Milland DR, Baines SB, Vogt S, Twining BS. J Synchrotron Radiat. 2010;17:560–566. doi: 10.1107/S0909049510014020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng XT, Yang HB, Li CM. Anal Chem. 2010;82:5082–5087. doi: 10.1021/ac100074n. [DOI] [PubMed] [Google Scholar]
- 61.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Proc Natl Acad Sci USA. 2009;106:5651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirschbaum M, Jaeger MS, Duschl C. Lab Chip. 2009;9:3517–3525. doi: 10.1039/b911865a. [DOI] [PubMed] [Google Scholar]
- 63.Palomero J, Pye D, Kabayo T, Spiller DG, Jackson MJ. Antioxid Redox Signal. 2008;10:1463–1474. doi: 10.1089/ars.2007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grecco HE, Roda-Navarro P, Girod A, Hou J, Frahm T, Truxius DC, Pepperkok R, Squire A, Bastiaens PI. Nat Methods. 2010;7:467–472. doi: 10.1038/nmeth.1458. [DOI] [PubMed] [Google Scholar]
- 65.Camacho M, Machado JD, Alvarez J, Borges R. J Biol Chem. 2008;283:22383–22389. doi: 10.1074/jbc.M800552200. [DOI] [PubMed] [Google Scholar]
- 66.Urbanski JP, Johnson MT, Craig DD, Potter DL, Gardner DK, Thorsen T. Anal Chem. 2008;80:6500–6507. doi: 10.1021/ac8010473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsunaga T, Hosokawa M, Arakaki A, Taguchi T, Mori T, Tanaka T, Takeyama H. Anal Chem. 2008;80:5139–5145. doi: 10.1021/ac800352j. [DOI] [PubMed] [Google Scholar]
- 68.Wang J, Bao N, Paris LL, Geahlen RL, Lu C. Anal Chem. 2008;80:9840–9844. doi: 10.1021/ac801940w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamada A, Katanosaka Y, Mohri S, Naruse KA. IEEE T Nanobiosci. 2009;8:306–311. doi: 10.1109/TNB.2009.2035253. [DOI] [PubMed] [Google Scholar]
- 70.Berg J, Hung YP, Yellen G. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taniguchi K, Kajiyama T, Kambara H. Nat Methods. 2009;6:503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- 72.Shim JU, Olguin LF, Whyte G, Scott D, Babtie A, Abell C, Huck WT, Hollfelder F. J Am Chem Soc. 2009;131:15251–15256. doi: 10.1021/ja904823z. [DOI] [PubMed] [Google Scholar]
- 73.Chen Y, Munteanu AC, Huang YF, Phillips J, Zhu Z, Mavros M, Tan WH. Chem-Eur J. 2009;15:5327–5336. doi: 10.1002/chem.200802305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Firaguay G, Nunes JA. Sci Signal. 2009;2:1–19. doi: 10.1126/scisignal.286pl3. [DOI] [PubMed] [Google Scholar]
- 75.Li YM, Shi J, Wu X, Luo ZF, Wang FL, Guo QX. Cell Biochem Funct. 2009;27:417–423. doi: 10.1002/cbf.1598. [DOI] [PubMed] [Google Scholar]
- 76.Paix O, Rodrigues LL, Couto I, Martins M, Fernandes P, De Carvalho CCCR, Monteiro GA, Sansonetty F, Amaral L, Viveiros M. J Biol Eng. 2009;3:1–1. doi: 10.1186/1754-1611-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weekley CM, Aitken JB, Vogt S, Finney LA, Paterson DJ, de Jonge MD, Howard DL, Musgrave IF, Harris HH. Biochemistry. 2011;50:1641–50. doi: 10.1021/bi101678a. [DOI] [PubMed] [Google Scholar]
- 78.Han SW, Nakamura C, Imai Y, Nakamura N, Miyake J. Biosens Bioelectron. 2009;24:1219–1222. doi: 10.1016/j.bios.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, Fu Y, Liang D, Zhao RY, Lakowicz JR. Anal Chem. 2009;81:883–889. doi: 10.1021/ac801932m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, Lee HG, Kang SH. Anal Chem. 2009;81:538–542. doi: 10.1021/ac8013324. [DOI] [PubMed] [Google Scholar]
- 81.Sasuga Y, Iwasawa T, Terada K, Oe Y, Sorimachi H, Ohara O, Harada Y. Anal Chem. 2008;80:9141–9149. doi: 10.1021/ac8016423. [DOI] [PubMed] [Google Scholar]
- 82.Zheng XT, Li CM. Biosens Bioelectron. 2010;25:1548–1552. doi: 10.1016/j.bios.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 83.Duhamel S, Gregori G, Van Wambeke F, Mauriac R, Nedoma J. J Microbiol Meth. 2008;75:269–278. doi: 10.1016/j.mimet.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 84.Picazo F, Domingo B, Perez-Ortiz JM, Llopis J. Biosens Bioelectron. 2011;26:2147–2153. doi: 10.1016/j.bios.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 85.Gaggia F, Nielsen DS, Biavati B, Siegumfeldt H. Int J Food Microbiol. 2010;141:S188–S192. doi: 10.1016/j.ijfoodmicro.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 86.Geissbuehler M, Spielmann T, Formey A, Marki I, Leutenegger M, Hinz B, Johnsson K, Van De Ville D, Lasser T. Biophys J. 2010;98:339–349. doi: 10.1016/j.bpj.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu QR, Heikal AA. J Photoch Photobio B. 2009;95:46–57. doi: 10.1016/j.jphotobiol.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blinks L, Skow R. Proc Natl Acad Sci USA. 1938;24:420–427. doi: 10.1073/pnas.24.10.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amatore C, Arbault S, Guille M, Lemaître F. Chem Rev. 2008;108:2585–2621. doi: 10.1021/cr068062g. [DOI] [PubMed] [Google Scholar]
- 90.Adams KL, Puchades M, Ewing AG. Annu Rev Anal Chem. 2008;1:329–355. doi: 10.1146/annurev.anchem.1.031207.113038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang W, Zhang S-H, Li L-M, Wang Z-L, Cheng J-K, Huang W-H. Anal Bioanal Chem. 2009;394:17–32. doi: 10.1007/s00216-009-2703-2. [DOI] [PubMed] [Google Scholar]
- 92.Gonon F, Cespuglio R, Ponchon J, Buda M, Jouvet M, Adams R, Pujol J. C R Acad Sci Hebd Seances Acad Sci. 1978;286:1203–1206. [PubMed] [Google Scholar]
- 93.Dayton MA, Ewing AG, Wightman RM. Eur J Pharmacol. 1981;75:141–144. doi: 10.1016/0014-2999(81)90074-1. [DOI] [PubMed] [Google Scholar]
- 94.Wightman RM, Jankowski JA, Kennedy RT, Kawagoe KT, Schroeder TJ, Leszczyszyn DJ, Near JA, Diliberto EJ, Jr, Viveros OH. Proc Natl Acad Sci USA. 1991;88:10754–10758. doi: 10.1073/pnas.88.23.10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Westerink RHS, Ewing AG. Acta Pysiol. 2008;192:273–285. doi: 10.1111/j.1748-1716.2007.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amatore C, Arbault S, Bouret Y, Guille M, Lemaître F, Verchier Y. Anal Chem. 2009;81:3087–3093. doi: 10.1021/ac900059s. [DOI] [PubMed] [Google Scholar]
- 97.Fernández-Morales JC, Yáñez M, Orallo F, Cortés L, González JC, Hernández-Guijo JM, García AG, de Diego AMG. Mol Pharmacol. 2010;78:34–744. doi: 10.1124/mol.110.066423. [DOI] [PubMed] [Google Scholar]
- 98.Doreian BW, Fulop TG, Meklemburg RL, Smith CB. Mol Biol Cell. 2009;20:3142–3154. doi: 10.1091/mbc.E09-03-0197. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Fulop T, Doreian B, Smith C. Arch Biochem Biophys. 2008;477:146–154. doi: 10.1016/j.abb.2008.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adams KL, Maxson MM, Mellander L, Westerink RHS, Ewing AG. Cell Mol Neurobiol. 2010;30:1235–1242. doi: 10.1007/s10571-010-9570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Berberian K, Torres AJ, Fang Q, Kisler K, Lindau M. J Neurosci. 2009;29:863–870. doi: 10.1523/JNEUROSCI.2818-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xue R, Zhao Y, Su L, Ye F, Chen P. Pflug Arch Eur J Phy. 2009;458:1137–1149. doi: 10.1007/s00424-009-0697-4. [DOI] [PubMed] [Google Scholar]
- 103.Amatore C, Arbault S, Bonifas I, Guille M. Biophys Chem. 2009;143:124–131. doi: 10.1016/j.bpc.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 104.Markov D, Mosharov EV, Setlik W, Gershon MD, Sulzer D. J Neurochem. 2008;107:1709–1721. doi: 10.1111/j.1471-4159.2008.05737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Montesinos MS, Camacho M, Machado JD, Viveros OH, Beltran B, Borges R. Brit J Pharmacol. 2010;159:1548–1556. doi: 10.1111/j.1476-5381.2010.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Churchward MA, Coorsen JR. Biochem J. 2009;423:1–14. doi: 10.1042/BJ20090969. [DOI] [PubMed] [Google Scholar]
- 107.Wang N, Kwan C, Gong X, Posse de Chaves E, Tse A, Tse FW. J Neurosci. 2010;30:3904–3911. doi: 10.1523/JNEUROSCI.4000-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang J, Xue R, Ong W-Y, Chen P. Biophys J. 2009;97:1371–1380. doi: 10.1016/j.bpj.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma M-T, Yeo J-F, Farooqui AA, Zhang J, Chen P, Ong W-Y. J Neural Transm. 2010;117:301–308. doi: 10.1007/s00702-009-0355-1. [DOI] [PubMed] [Google Scholar]
- 110.Ge S, White JG, Haynes CL. Anal Chem. 2009;81:2935–2943. doi: 10.1021/ac8024202. [DOI] [PubMed] [Google Scholar]
- 111.Ge S, White JG, Haynes CL. ACS Chem Biol. 2010;5:819–828. doi: 10.1021/cb100130b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marquis BJ, McFarland AD, Braun KL, Haynes CL. Anal Chem. 2008;80:3431–3437. doi: 10.1021/ac800006y. [DOI] [PubMed] [Google Scholar]
- 113.Marquis BJ, Maurer-Jones MA, Braun KL, Haynes CL. Analyst. 2009;134:2293–2300. doi: 10.1039/b913967b. [DOI] [PubMed] [Google Scholar]
- 114.Marquis BJ, Haynes CL. Anal Bioanal Chem. 2010;398:2979–2985. doi: 10.1007/s00216-010-4263-x. [DOI] [PubMed] [Google Scholar]
- 115.Dong Y, Heien ML, Maxson MM, Ewing AG. J Neurochem. 2008;107:1589–1595. doi: 10.1111/j.1471-4159.2008.05721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Z-H, Zhou W, Wu Z-X, Zhang R-Y, Xu T. Biosens Bioelectron. 2009;24:1358–1364. doi: 10.1016/j.bios.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 117.Amatore C, Arbault S, Erard M. Anal Chem. 2008;80:9635–9641. doi: 10.1021/ac801269e. [DOI] [PubMed] [Google Scholar]
- 118.Amatore C, Arbault S, Bouton C, Drapier J-C, Ghandour H, Koh ACW. ChemBioChem. 2008;9:1472–1480. doi: 10.1002/cbic.200700746. [DOI] [PubMed] [Google Scholar]
- 119.Hu R, Guille M, Arbault S, Lin CJ, Amatore C. Phys Chem Chem Phys. 2010;12:10048–10054. doi: 10.1039/c0cp00398k. [DOI] [PubMed] [Google Scholar]
- 120.Amatore C, Arbault S, Koh ACW. Anal Chem. 2010;82:1411–1419. doi: 10.1021/ac902486x. [DOI] [PubMed] [Google Scholar]
- 121.Ai F, Chen H, Zhang S-H, Liu S-Y, Wei F, Dong X-Y, Huang W-H. Anal Chem. 2009;81:8453–8458. doi: 10.1021/ac901300b. [DOI] [PubMed] [Google Scholar]
- 122.Amatore C, Arbault S, Ferreira DCM, Tapsoba I, Verchier Y. J Electroanal Chem. 2008;615:34–44. [Google Scholar]
- 123.Ferreira DCM, Tapsoba I, Arbault S, Bouret Y, Moreira SAM, Pinto AV, Goulart MOF, Amatore C. ChemBioChem. 2009;10:528–538. doi: 10.1002/cbic.200800517. [DOI] [PubMed] [Google Scholar]
- 124.Tapsoba I, Arbault S, Walter P, Amatore C. Anal Chem. 2010;82:457–460. doi: 10.1021/ac902348g. [DOI] [PubMed] [Google Scholar]
- 125.Amatore C, Arbault S, Jaouen G, Koh ACW, Leong WK, Top S, Valleron M-A, Woo CH. ChemMedChem. 2010;5:296–301. doi: 10.1002/cmdc.200900464. [DOI] [PubMed] [Google Scholar]
- 126.Filipović MR, Koh ACW, Arbault S, Niketić V, Debus A, Schleicher U, Bogdan C, Guille M, Lemaître F, Amatore C, Ivanović-Burmazović I. Angew Chem Int Ed. 2010;49:4228–4232. doi: 10.1002/anie.200905936. [DOI] [PubMed] [Google Scholar]
- 127.Du F, Huang W, Shi Y, Wang Z, Cheng J. Biosens Bioelectron. 2008;24:415–42. doi: 10.1016/j.bios.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 128.Patel BA, Arundell M, Parker KH, Yeoman MS, O’Hare D. Phys Chem Chem Phys. 2010;12:10065–10072. doi: 10.1039/c0cp00310g. [DOI] [PubMed] [Google Scholar]
- 129.Zhang B, Adams KL, Luber SJ, Eves DJ, Heien ML, Ewing AE. Anal Chem. 2008;80:1394–1400. doi: 10.1021/ac702409s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang B, Heien MLAV, Santillo MF, Mellander L, Ewing AG. Anal Chem. 2011;83:571–577. doi: 10.1021/ac102502g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mosharov EV, Sulzer D. Nat Methods. 2005;2:651–658. doi: 10.1038/nmeth782. [DOI] [PubMed] [Google Scholar]
- 132.Amatore C, Arbault s, Bouret Y, Guille M, Lemaître F. ChemPhysChem. 2010;11:2931–2941. doi: 10.1002/cphc.201000102. [DOI] [PubMed] [Google Scholar]
- 133.Amatore C, Oleinick AI, Svir I. ChemPhyschem. 2010;11:159–174. doi: 10.1002/cphc.200900647. [DOI] [PubMed] [Google Scholar]
- 134.Pemberton RM, Rawson FJ, Xu J, Pittson R, Drago GA, Griffiths J, Jackson SK, Hart JP. Microchim Acta. 2010;170:321–330. [Google Scholar]
- 135.Douglas ES, Hsiao SC, Onoe H, Bertozzi CR, Francis MB, Mathies RA. Lab Chip. 2009;9:2010–2015. doi: 10.1039/b821690h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lin Z, Takahashi Y, Murata T, Takeda M, Ino K, Shiku H, Matsue T. Angew Chem Int Ed. 2009;48:2044–2046. doi: 10.1002/anie.200805743. [DOI] [PubMed] [Google Scholar]
- 137.Berberian K, Kisler K, Fang Q, Lindau M. Anal Chem. 2009;81:8734–8740. doi: 10.1021/ac900674g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carabelli V, Gosso S, Marcantoni A, Xu Y, Colombo E, Gao Z, Vittone E, Kohn E, Pasquarelli A, Carbone E. Biosens Bioelectron. 2010;26:92–98. doi: 10.1016/j.bios.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 139.Spégel C, Heiskanen A, Pedersen S, Emnéus J, Ruzgas T, Taboryski R. Lab Chip. 2008;8:323–329. doi: 10.1039/b715107a. [DOI] [PubMed] [Google Scholar]
- 140.Chen X, Gao Y, Hossain M, Gangopadhyay S, Gillis KD. Lab Chip. 2008;8:161–169. doi: 10.1039/b715308m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu H, Newton GJ, Nakamura R, Hashimoto K, Nakanishi S. Angew Chem Int Ed. 2010;49:6596–6599. doi: 10.1002/anie.201000315. [DOI] [PubMed] [Google Scholar]
- 142.Ayers S, Berberian K, Gillis KD, Lindau M, Minch BA. IEEE Trans Biomed Circuits Syst. 2010;4:86–92. doi: 10.1109/TBCAS.2009.2033706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsai C-C, Yang C-C, Shih P-Y, Wu C-S, Chen C-D, Pan C-Y, Chen Y-T. J Phys Chem B. 2008;112:9165–9173. doi: 10.1021/jp803000a. [DOI] [PubMed] [Google Scholar]
- 144.Shao N, Wickstrom E, Panchapakesan B. Nanotechnology. 2008;19:465101.1–465101.11. doi: 10.1088/0957-4484/19/46/465101. [DOI] [PubMed] [Google Scholar]
- 145.Sommerhage F, Baumann A, Wrobel G, Ingebrandt S, Offenhäuser A. Biosens Bioelectron. 2010;26:155–161. doi: 10.1016/j.bios.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 146.Malleo D, Nevill JT, Lee LP, Morgan H. Microfluid Nanofluid. 2010;9:191–198. doi: 10.1007/s10404-009-0534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Senez V, Lennon E, Ostrovidov S, Yamamoto T, Fujita H, Sakai Y, Fujii T. IEEE Sens J. 2008;8:548–557. [Google Scholar]
- 148.Jiang X, Spencer MG. Biosens Bioelectron. 2010;25:1622–1628. doi: 10.1016/j.bios.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 149.Asphahani F, Thein M, Veiseh O, Edmondson D, Kosai R, Veiseh M, Xu J, Zhang M. Biosens Bioelectron. 2008;23:1307–1313. doi: 10.1016/j.bios.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thein M, Asphahani F, Cheng A, Buckmaster R, Zhang M, Xu J. Biosens Bioelectron. 2010;25:1963–1969. doi: 10.1016/j.bios.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Amemiya S, Bard AJ, Fan F-RF, Mirkin MV, Unwin PR. Annu Rev Anal Chem. 2008;1:95–131. doi: 10.1146/annurev.anchem.1.031207.112938. [DOI] [PubMed] [Google Scholar]
- 152.Bergner S, Palatzky P, Wegener J, Matysik F-M. Electroanal. 2011;23:196–200. [Google Scholar]
- 153.Bergner S, Wegener J, Matysik F-M. Anal Chem. 2011;83:169–174. doi: 10.1021/ac1021375. [DOI] [PubMed] [Google Scholar]
- 154.Takahashi Y, Miyamoto T, Shiku H, Asano R, Yasukawa T, Kumagai I, Matsue T. Anal Chem. 2009;81:2785–2790. doi: 10.1021/ac900195m. [DOI] [PubMed] [Google Scholar]
- 155.Xue Y, Ding L, Lei J, Yan F, Ju H. Anal Chem. 2010;82:7112–7118. doi: 10.1021/ac101688p. [DOI] [PubMed] [Google Scholar]
- 156.Koley D, Bard AJ. Proc Natl Acad Sci USA. 2010;107:16783–16787. doi: 10.1073/pnas.1011614107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hirano Y, Nishimaya Y, Kowata K, Mizutani F, Tsuda S, Komatsu Y. Anal Chem. 2008;80:9349–9354. doi: 10.1021/ac8018334. [DOI] [PubMed] [Google Scholar]
- 158.Shiku H, Suzuki J, Murata T, Ino K, Matsue T. Electrochim Acta. 2010;55:8263–8267. [Google Scholar]
- 159.Murata T, Yasukawa T, Shiku H, Matsue T. Biosens Bioelectron. 2009;25:913–919. doi: 10.1016/j.bios.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 160.Takahashi Y, Shiku H, Murata T, Yasukawa T, Matsue T. Anal Chem. 2009;81:9674–9681. doi: 10.1021/ac901796r. [DOI] [PubMed] [Google Scholar]
- 161.Sun P, Laforge FO, Abeyweera TP, Rotenberg SA, Carpino J, Mirkin MV. Proc Natl Acad Sci USA. 2008;105:443–448. doi: 10.1073/pnas.0711075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhao X, Diakowski PM, Ding Z. Anal Chem. 2010;82:8371–8373. doi: 10.1021/ac101524v. [DOI] [PubMed] [Google Scholar]
- 163.Zhao X, Lam S, Jass J, Ding Z. Electrochem Commun. 2010;12:773–776. [Google Scholar]
- 164.Zhu L, Gao N, Zhang X, Jin W. Talanta. 2008;77:804–808. [Google Scholar]
- 165.Ciobanu M, Taylor DE, Jr, Wilburn JP, Cliffel DE. Anal Chem. 2008;80:2717–2727. doi: 10.1021/ac7021184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen Z, Xie S, Shen L, Du Y, He S, Li Q, Liang Z, Meng X, Li B, Xu X, Ma H, Huang Y, Shao Y. Analyst. 2008;133:1221–1228. doi: 10.1039/b807057a. [DOI] [PubMed] [Google Scholar]
- 167.Messerli MA, Collis LP, Smith PJS. Electroanal. 2009;21:1906–1913. [Google Scholar]
- 168.Lew RR. Plant Cell Physiol. 2010;51:1889–1899. doi: 10.1093/pcp/pcq149. [DOI] [PubMed] [Google Scholar]
- 169.Messerli MA, Collis LP, Smith PJS. Biophys J. 2009;96:1597–1605. doi: 10.1016/j.bpj.2008.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Heinemann M, Zenobi R. Curr Opin Biotech. 2011;22:26–31. doi: 10.1016/j.copbio.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 171.Wang D, Bodovitz S. Trends Biotechnol. 2010;28:81–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Han X, Aslanian A, Yates JR. Current Opinion in Chem Biol. 2008;12:83–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Suzuki T, Miyazono A, Baba K, Sugawara R, Kamiyama T. Harmful Algae. 2009;8:233–238. [Google Scholar]
- 174.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Manna M. Proc Natl Acad Sci USA. 2009;106:18903–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Nature Meth. 2010;7:383–387. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 176.Williamson AJK, Smith DL, Blinco D, Unwin RD, Pearson S, Wilson C, Miller C, Lancashire L, Lacaud G, Kouskoff V, Whetton AD. Mol Cell Proteom. 2008;3:459–472. doi: 10.1074/mcp.M700370-MCP200. [DOI] [PubMed] [Google Scholar]
- 177.Greco TM, Seeholzer SH, Mak A, Spruce L, Ischiropoulos H. J Proteome Res. 2010;9:2764–2774. doi: 10.1021/pr100134n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liu Y, Zhuang D, Hou R, Li J, Xu G, Song T, Chen L, Yan G, Pang Q, Zhu J. Anal Chim Acta. 2011;688:183–190. doi: 10.1016/j.aca.2010.12.032. [DOI] [PubMed] [Google Scholar]
- 179.Liu Y, Wu J, Yan G, Hou R, Zhuang D, Chen L, Pang Q, Zhu J. Proteome Sci. 2010;8:2–12. doi: 10.1186/1477-5956-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hemmen K, Reinl T, Buttler K, Behler F, Dieken H, Jänsch L, Wilting J, Weich HA. Angiogenesis. 2011 doi: 10.1007/s10456-011-9200-x. [DOI] [PubMed] [Google Scholar]
- 181.Wang N, Xu M, Wang P, Li L. Anal Chem. 2010;82:2262–2271. doi: 10.1021/ac9023022. [DOI] [PubMed] [Google Scholar]
- 182.Johann DJ, Rodriguez-Canales J, Mukherjee S, Prieto DA, Hanson JC, Emmert-Buck M, Blonder J. J Proteome Res. 2009;8:310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Katoa M, Onda Y, Sekimoto M, Degawa M, Toyooka TJ. Chromatogr A. 2009;1216:8277–8282. doi: 10.1016/j.chroma.2009.06.074. [DOI] [PubMed] [Google Scholar]
- 184.Lapainis T, Rubakhin SS, Sweedler JV. Anal Chem. 2009;81:5858–5864. doi: 10.1021/ac900936g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mayboroda OA, Neususs C, Pelzing M, Zurek G, Derks R, Meulenbelt I, Kloppenburg M, Slagboom EP, Deelder AMJ. Chromatogr A. 2007;1159:49–153. doi: 10.1016/j.chroma.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 186.Soga T, Heiger DN. Anal Chem. 2000;72:236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 187.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Anal Chem. 2010;82:67–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nordman N, Sikanen T, Aura S, Tuomikoski S, Vuorensola K, Kotiaho T, Franssila S, Kostiainen R. Electrophoresis. 2010;31:745–3753. doi: 10.1002/elps.201000373. [DOI] [PubMed] [Google Scholar]
- 189.Tejedor ML, Mizuno H, Tsuyama N, Harada T, Masujima T. Anal Sci. 2009;25:1053–1055. doi: 10.2116/analsci.25.1053. [DOI] [PubMed] [Google Scholar]
- 190.Tsuyama N, Mizuno H, Tokunaga E, Masujima T. Anal Sci. 2008;24:59–561. doi: 10.2116/analsci.24.559. [DOI] [PubMed] [Google Scholar]
- 191.Mizuno H, Tsuyama N, Harada T, Masujima T. J Mass Spectrom. 2008;43:1692–1700. doi: 10.1002/jms.1460. [DOI] [PubMed] [Google Scholar]
- 192.Nemes P, Vertes A. J Vis Exp. 2010;43:1–4. doi: 10.3791/2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shrestha B, Vertes A. J Vis Exp. 2010;4:2144–2148. doi: 10.3791/2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shrestha B, Nemes P, Vertes A. Appl Phys A. 2010;101:21–126. [Google Scholar]
- 195.Shrestha B, Vertes A. Anal Chem. 2009;81:265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- 196.Nemes P, Barton AA, Li Y, Vertes A. Anal Chem. 2008;80:575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- 197.Lewis JK, Wei J, Siuzdak G. In: Encyclopedia of Analytical Chemistry. Meyers RA, editor. John Wiley & Sons Ltd; Chichester: 2000. pp. 5880–5894. [Google Scholar]
- 198.Amantonico A, Oh JY, Sobek J, Heinemann M, Zenobi R. Angew Chem Int Ed. 2008;47:5382–5385. doi: 10.1002/anie.200705923. [DOI] [PubMed] [Google Scholar]
- 199.Amantonico A, Urban PL, Oh JY, Zenobi R. Chimia. 2009;63:185–188. [Google Scholar]
- 200.Miura D, Fujimura Y, Tachibana H, Wariishi H. Anal Chem. 2010;82:498–504. doi: 10.1021/ac901083a. [DOI] [PubMed] [Google Scholar]
- 201.Rubakhin SS, Sweedler JV. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dieckmann R, Helmuth R, Erhard M, Malorny B. Appl Environ Microbiol. 2008;74:7767–7778. doi: 10.1128/AEM.01402-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. ACS Chem Neurosci. 2010;1:05–519. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Yew JY, Wang Y, Barteneva N, Dikler S, Kutz-Naber KK, Li L, Kravitz EA. J Proteome Res. 2009;8:1271–1284. doi: 10.1021/pr800601x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Karger A, Bettin B, Lenk M, Mettenleiter TC. J Virolog Methods. 2010;164:116–121. doi: 10.1016/j.jviromet.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 206.Urban PL, Jefimovs K, Amantonico A, Fagerer SR, Schmid T, Mädler S, Puigmarti-Luis J, Goedecke N, Zenobi R. Lab Chip. 2010;10:3206–3209. doi: 10.1039/c0lc00211a. [DOI] [PubMed] [Google Scholar]
- 207.Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Aerni H-R, Cornett DS, Caprioli RM. Anal Chem. 2009;81:490–7495. doi: 10.1021/ac900974j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, Vinatier D, Wisztorski M, Day R, Fournier I, Salzet M. Mol Cell Proteom. 2009;8:2023–2033. doi: 10.1074/mcp.R800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schwamborn K, Caprioli RM. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 211.Reyzer ML, Caprioli RM. In: Detection of Biological Agents for the Prevention of Bioterrorism NATO Science for Peace and Security Series A: Chemistry and Biology. Banoub J, editor. Springer; Netherlands: 2011. pp. 267–283. [Google Scholar]
- 212.Chaurand P, Cornett DS, Angel PM, Caprioli RM. Mol Cell Proteom. 2011 doi: 10.1074/mcp.O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sugiura Y, Setou M. J Neuroimmune Pharm. 2010;5:31–43. doi: 10.1007/s11481-009-9162-6. [DOI] [PubMed] [Google Scholar]
- 214.Yang H-J, Sugiura Y, Ishizaki I, Sanada N, Ikegami K, Zaima N, Shrivas K, Setou M. Surf Interface Anal. 2010;42:1606–1611. [Google Scholar]
- 215.Perdian DC, Cha S, Oh J, Sakaguchi DS, Yeung ES, Lee YJ. Rapid Commun Mass Spectrom. 2010;24:1147–1154. doi: 10.1002/rcm.4491. [DOI] [PubMed] [Google Scholar]
- 216.Urban PL, Schmid T, Amantonico A, Zenobi R. Anal Chem. 2011;83:1843–1849. doi: 10.1021/ac102702m. [DOI] [PubMed] [Google Scholar]
- 217.Hölscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B, Heckel DG, Schubert US, Svatos A. PlantJ. 2009;60:907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 218.Russel SC. Mass Spec Rev. 2009;28:376–387. doi: 10.1002/mas.20198. [DOI] [PubMed] [Google Scholar]
- 219.Greving MP, Patti GJ, Siuzdak G. Anal Chem. 2011;83:2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Amantonico A, Flamigni L, Glaus R, Zenobi R. Metabolomics. 2009;5:346–353. [Google Scholar]
- 221.Lin H-C, Lin H-H, Kao C-Y, Yu AL, Peng W-P, Chen C-H. Angew Chem Int Ed. 2010;49:3460–3464. doi: 10.1002/anie.201000891. [DOI] [PubMed] [Google Scholar]
- 222.Brunelle A, Laprévote O. Anal Bioanal Chem. 2009;393:31–35. doi: 10.1007/s00216-008-2367-3. [DOI] [PubMed] [Google Scholar]
- 223.Orphan VJ, House CH. Geobiology. 2009;7:360–372. doi: 10.1111/j.1472-4669.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- 224.Kurczy ME, Kozole J, Parry SA, Piehowski PD, Winograd N, Ewing AG. Appl Surf Sci. 2008;255:1158–1161. doi: 10.1016/j.apsusc.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kurczy ME, Piehowski PD, Van Bell CT, Heien M, Winograd N, Ewing AG. Proc Natl Acad Sci USA. 2010;107:2751–2756. doi: 10.1073/pnas.0908101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Piehowski PD, Kurczy ME, Willingham D, Parry SA, Heien M, Winograd N, Ewing AG. Langmuir. 2008;24:906–7911. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dague E, Delcorte A, Latge J-P, Dufrene YF. Langmuir. 2008;24:2955–2959. doi: 10.1021/la703741y. [DOI] [PubMed] [Google Scholar]
- 228.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, Vickerman JC. Anal Chem. 2008;80:9058–9064. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 229.Malm J, Giannaras D, O’Riehle M, Gadegaard N, Sjövall P. Anal Chem. 2009;81:197–7205. doi: 10.1021/ac900636v. [DOI] [PubMed] [Google Scholar]
- 230.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjövall P, Ewing AG. Anal Chem. 2010;82:6652–6659. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Szakal C, Narayan K, Fu J, Lefman J, Subramaniam S. Anal Chem. 2011 doi: 10.1021/ac1030607. dx.doi.org/10.1021/ac1030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, Hutcheon ID, Nealson KH, Capone DG. Proc Natl Acad Sci USA. 2009;106:6345–6350. doi: 10.1073/pnas.0810547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Li T, Wu T-D, Mazéas L, Toffin L, Guerquin-Kern J-L, Leblon G, Bouchez T. Environ Microbiol. 2008;10:580–58. doi: 10.1111/j.1462-2920.2007.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. Appl Environm Microbiol. 2008;74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Heeren RMA, Smith DF, Stauber J, Kükrer-Kaletas B, MacAleese L. J Am Soc Mass Spec. 2009;20:1006–1014. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 236.Schwartz SA, Caprioli RM. Meth Mol Biol. 2010;656:3–19. doi: 10.1007/978-1-60761-746-4_1. [DOI] [PubMed] [Google Scholar]
- 237.Touboul D, Brunelle A, Laprévote O. Biochimie. 2011;93:113–119. doi: 10.1016/j.biochi.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 238.Tsuyama N, Mizuno H, Masujima T. Anal Sci. 2011;27:163–170. doi: 10.2116/analsci.27.163. [DOI] [PubMed] [Google Scholar]
- 239.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 240.Tanner SD, Bandura DR, Ornatsky O, Baranov VI, Nitz M, Winnik MA. Pure Appl Chem. 2008;80:2627–2641. [Google Scholar]
- 241.Ho K-S, Chan W-T. J Anal At Spectrom. 2010;25:1114–1122. [Google Scholar]
- 242.Chtcheglova LA, Wildling L, Waschke J, Drenchkahn D, Hinterdorfer P. J Mol Recognit. 2010;23:589–596. doi: 10.1002/jmr.1052. [DOI] [PubMed] [Google Scholar]
- 243.Ounkomol C, Yamada S, Heinrich V. Biophys J. 2010;99:L100–L102. doi: 10.1016/j.bpj.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bai S-J, Fabian T, Prinz FB, Fasching RJ. Sensor Actuat B-Chem. 2008;130:249–257. [Google Scholar]
- 245.Lulevich V, Yang H-Y, Isseroff RR, Liu G-Y. Ultramicroscopy. 2010;110:1435–1442. doi: 10.1016/j.ultramic.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 246.Arfsten J, Leupold S, Bradtmöller C, Kampen I, Kwade A. Colloid Surface B. 2010;79:284–290. doi: 10.1016/j.colsurfb.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 247.De Jonge N, Peckys DB, Kremers JG, Piston DW. Proc Natl Acad Sci USA. 2009;106:2159–2164. doi: 10.1073/pnas.0809567106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Greif D, Wesner D, Regtmeier J, Anselmetti D. Ultramicroscopy. 2010;110:1290–1296. doi: 10.1016/j.ultramic.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 249.Nägerl UV, Bonhoeffer T. J Neurosci. 2010;30:9341–9346. doi: 10.1523/JNEUROSCI.0990-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Blom H, Römlund D, Scott L, Spicarova Z, Widegren J, Bondar A, Aperia A, Brismar H. BMC Neurosci. 2011 doi: 10.1186/1471–2202–12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 252.Evans CL, Xie XS. Annu Rev Anal Chem. 2008;1:883–909. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]
- 253.Åkeson M, Brackmann C, Gustafsson L, Enejder A. J Raman Spectrosc. 2010;41:1638–1644. [Google Scholar]
- 254.Freudiger CW, et al. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wang Y, Chen Z-Z, Li Q-L. Microchim Acta. 2010;168:177–195. [Google Scholar]
- 256.Chao T-C, Ros A. J R Soc Interface. 2008;5:S139–S150. doi: 10.1098/rsif.2008.0233.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Frimat J-P, Becker M, Chiang Y-Y, Marggraf U, Janasek D, Hengstler JG, Franzke J, West J. Lab Chip. 2011;11:231–237. doi: 10.1039/c0lc00172d. [DOI] [PubMed] [Google Scholar]
- 258.Honarmandi P, Lee H, Lang MJ, Kamm RD. Lab Chip. 2011;11:684–694. doi: 10.1039/c0lc00487a. [DOI] [PubMed] [Google Scholar]
- 259.Christ KV, Williamson KB, Masters KS, Turner KT. Biomed Microdevices. 2010;12:443–455. doi: 10.1007/s10544-010-9401-x. [DOI] [PubMed] [Google Scholar]
- 260.Yan H, Zhang B, Wu H. Electrophoresis. 2008;29:1775–1786. doi: 10.1002/elps.200700561. [DOI] [PubMed] [Google Scholar]
- 261.Caselli F, Bisegna P, Maceri F. J Microelectromech S. 2010;19:1029–1040. [Google Scholar]
- 262.Chabert M, Viovy J-L. Proc Natl Acad Sci USA. 2008;105:3191–3196. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wlodkowic D, Faley S, Zagnoni M, Wikswo JP, Cooper JM. Anal Chem. 2009;81:5517–5523. doi: 10.1021/ac9008463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Taylor RJ, Falconnet D, Niemistö A, Ramsey SA, Prinz S, Shmulevich I, Galiktski T, Hansen CL. Proc Natl Acad Sci USA. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Singhal A, Haynes CA, Hansen CL. Anal Chem. 2010;82:8671–8679. doi: 10.1021/ac101956e. [DOI] [PubMed] [Google Scholar]
- 266.Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, Diaz-Flores E, Coram M, Shannon KM, Nolan GP, Loh ML. Cancer Cell. 2008;14:335–343. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Santillo MF, Heien ML, Ewing AG. Lab Chip. 2009;9:2796–2802. doi: 10.1039/b907942d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Choi JH, Ogunniyi AO, Du M, Du M, Kretschmann M, Eberhardt J, Love JC. Biotechnol Prog. 2010;26:888–895. doi: 10.1002/btpr.374. [DOI] [PubMed] [Google Scholar]
- 269.Guldevall K, Vanherberghen B, Frisk T, Hurtig J, Christakou AE, Manneberg O, Lindström S, Andersson-Svahn H, Wiklund M, Önfelt B. PLoS ONE. 2010;5:e15453. doi: 10.1371/journal.pone.0015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Wang Y, Phillips C, Xu W, Pai J-H, Dhopeshwarkar R, Sims CE, Allbritton N. Lab Chip. 2010;10:2917–2924. doi: 10.1039/c0lc00186d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mauritz JMA, Tiffert T, Seear R, Lautenschläger F, Esposito A, Lew VL, Guck J, Kaminski CF. J Biomed Opt. 2010;15:030517-1–030517-3. doi: 10.1117/1.3458919. [DOI] [PubMed] [Google Scholar]
- 272.Tanaka T, Sunaga Y, Hatakeyama K, Matsunaga T. Lab Chip. 2010;10:3348–3354. doi: 10.1039/c0lc00039f. [DOI] [PubMed] [Google Scholar]
- 273.Takahashi S, Iida M, Furusawa H, Shimizu Y, Ueda T, Okahata Y. J Am Chem Soc. 2009;131:9326–9332. doi: 10.1021/ja9019947. [DOI] [PubMed] [Google Scholar]
- 274.Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sánchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, Myers EW, Kim SK. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 276.Eid J, Fehr A, et al. Science. 2009;23:133–138. [Google Scholar]
- 277.Harris TD, Buzby PR, et al. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 278.Fan HC, Wang J, Potanina A, Quake SR. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Petriv OI, Kuchenbauer F, et al. Proc Natl Acad Sci USA. 2010;107:15443–15448. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 281.Salehi-Reyhani A, Kaplinsky J, Burgin E, Novakova M, de Mello AJ, Templer RH, Parker P, Neil MAA, Ce O, French P, Willison KR, Klug D. Lab Chip. 2011 doi: 10.1039/c0lc00613k. [DOI] [PubMed] [Google Scholar]
- 282.Kortmann H, Kurth F, Blank LM, Dittrich PS, Schmid A. Lab Chip. 2009;9:3047–3049. doi: 10.1039/b908679j. [DOI] [PubMed] [Google Scholar]
- 283.Ogilby PR. Chem Soc Rev. 2010;39:3181–3209. doi: 10.1039/b926014p. [DOI] [PubMed] [Google Scholar]
- 284.Breitenbach T, Kuimova MK, Gbur P, Hatz S, Schack NB, Pedersen BW, Lambert JDC, Poulsen L, Ogilby PR. Photochem Photobiol Sci. 2009;8:442–452. doi: 10.1039/b809049a. [DOI] [PubMed] [Google Scholar]
- 285.Kuimova1 MK, Botchway SW, Parker AW, Balaz M, Collins HA, Anderson HL, Suhling K, Ogilby PR. Nature Chemistry. 2009;1:69–73. doi: 10.1038/nchem.120. [DOI] [PubMed] [Google Scholar]