Abstract
PLP is well-regarded for its role as a coenzyme in a number of diverse enzymatic reactions. Transamination, deoxygenation, and aldol reactions mediated by PLP-dependent enzymes enliven and enrich deoxy sugar biosynthesis, endowing these compounds with unique structures and contributing to their roles as determinants of biological activity in many natural products. The importance of deoxy amino sugars in natural product biosynthesis has spurred several recent structural investigations of sugar aminotransferases. The structure of a PMP-dependent enzyme catalyzing the C-3 deoxygenation reaction in the biosynthesis of ascarylose was also determined. These studies, and the crystal structures they have provided, offer a wealth of new insights regarding the enzymology of PLP/PMP-dependent enzymes in deoxy sugar biosynthesis. In this review, we consider these recent achievements in the structural biology of deoxy sugar biosynthetic enzymes and the important implications they hold for understanding enzyme catalysis and natural product biosynthesis in general.
Keywords: vitamin B6 coenzyme, deoxy sugars, amino sugars, biosynthesis, deoxygenation, enzyme mechanisms
1. Introduction
The roles played by deoxy sugars as bacterial surface antigenic determinants, ligands for cell-cell interactions, as targets for toxins, antibodies, and microorganisms, and even in controlling the half-life of proteins in serum, have long been recognized [1–5]. Their contributions to the activities of numerous secondary metabolites have also been appreciated through herbal healing practices since prehistoric times. Studies of their formation, fundamental properties, and biological activities are major research foci in the fields of glycobiology, glycobiochemistry, natural product chemistry, and drug discovery [6–11]. A recent emphasis in natural product research is on glycodiversification [10–16], a combinatorial biosynthetic effort towards the development of novel glycoconjugates carrying unusual deoxy sugars, with the ultimate aim of enhancing the bioactivity of natural products or endowing these compounds with altered target specificity. This approach takes advantage of the structural diversity of bacterial carbohydrates: whereas eukaryote glycoforms are synthesized using only nine different monosaccharides, biosynthesis of prokaryotic glycoforms utilizes over 100 different sugars as building blocks [11].
Since most of the prokaryotic deoxy sugar biosynthetic pathways share some common intermediates, enzymes from different sugar biosynthetic pathways can be combined to create novel deoxy sugar pathways [10–16]. However, to maximize the feasibility of this technique, the types of reactions involved in generating these novel sugars and the substrate and product preferences of the enzymes catalyzing these reactions must be well understood. Besides diversifying our antimicrobial arsenal, an understanding of deoxy sugar biosynthetic pathways may elucidate novel drug targets since many pathogenic bacteria rely on sugars unique to prokaryotes for virulence and pathogenesis.
PLP-dependent enzymes are widely regarded for their ability to catalyze a wide repertoire of reactions on a variety of substrates [17–21]. The broad utility of these enzymes also extends to deoxy sugar biosynthesis, where they can catalyze aminotransfer reactions, deoxygenation reactions, and aldolase reactions [22–33]. Thus, PLP-dependent enzymes involved in deoxy sugar biosynthesis offer tremendous potential for the development of novel deoxy sugars and will be useful in furthering combinatorial biosynthetic efforts towards the development of glycoconjugates with new and/or improved biological activities.
Clearly, the development of new pharmaceutical agents is an ongoing effort on account of our ever expanding knowledge of the molecular basis of cancer and other chronic diseases as well as the continual evolution of highly antibiotic-resistant bacterial pathogens [33]. Therefore, a fundamental understanding of PLP-dependent enzymes in deoxy sugar biosynthesis has considerable biomedical potential due to the wide breath of reactions catalyzed by these enzymes coupled with the versatility the resulting compounds demonstrate in modulating the pharmacological activity of a number of natural products. In this review, a survey of the PLP-dependent enzymes in prokaryotic deoxy sugar biosynthesis is undertaken with an emphasis on recent structural studies of sugar aminotransferases and dehydratases.
2. Transamination
2.1. Natural Occurrence and Biological Significance of Deoxy Aminosugars
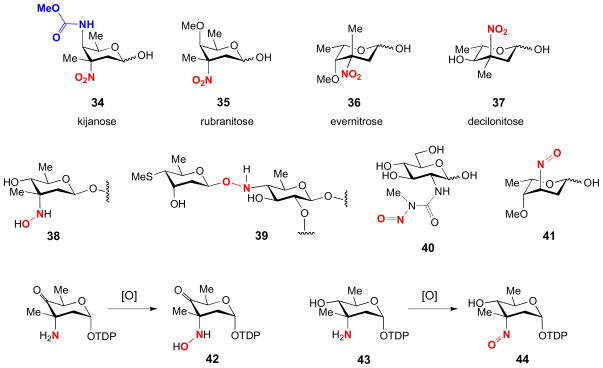
Aminotransferases introduce an amino group into a ketosugar substrate and are probably the most obvious manifestation of PLP-dependent enzymes in deoxy sugar biosynthesis. The resultant deoxy aminosugars are found ubiquitously in nature as components of bacterial cell membranes, flagellar glycolipids, bacterial toxins, and as components of diverse secondary metabolites, including many clinically useful bioactive pharmaceutics [10,11,22]. Shown in Fig. 1 are some examples of naturally occurring deoxy aminosugars (1–33). In addition, the amino group serves as the precursor for the biosynthesis of more exotic functional groups through modifications like acetylation, alkylation, oxidation, and others to generate N-acyl, N-alkyl, hydroxyamino, nitroso, nitro, and carbamate moieties. Shown in Fig. 2 are some examples of nitro-, nitroso-, hydroxyamino-, and carbamate-containing sugars (34–41), and the isolated hydroxylamino and nitroso intermediates (42, 44) from two nitro sugar biosynthetic pathways [34–37]. Thus, many of the existing nitrogen-containing unusual sugars are likely evolved from the corresponding deoxy aminosugars. This logic can be exploited in the development of diverse novel deoxy sugars through combinatorial biosyntheses employing deoxy aminosugars as design templates for further modification [11,38,39].
Figure 1.
A collection of commonly encountered deoxy aminosugars found in natural products.
Figure 2.
Examples of nitro-, nitroso-, hydroxyamino-, and carbamate-containing sugars (34–41), and the isolated hydroxylamino and nitroso intermediates (42, 44) from two nitro sugar biosynthetic pathways.
The ubiquity of aminosugars in a wide variety of natural products underscores their importance, as these sugars are often found to mediate interactions between the natural products they decorate and the targets of these molecules. Examples that highlight the significance of the amino substituent to the bioactivity of natural products include macrolides (e.g., desosamine (16) in clarithromycin), anthracyclines (e.g., daunosamine (23) in daunomycin), glycopeptides (e.g., vancosamine (20) in vancomycin), polyene antifungals (e.g., mycosamine (13) in amphotericin B), the aminoglycosides (e.g., purpurosamines (27–29) in gentamycins), and many others. In particular, the importance of aminosugars to the activity of the macrolides has been extensively studied. Macrolides are composed of a polyketide-derived macrolactone that is often glycosylated, including at least one deoxy aminosugar. The mechanism of many macrolides as antimicrobial agents is mainly due to interference with protein synthesis by blocking the peptide exit tunnel of the bacterial 50S ribosomal subunit [40,41]. High-resolution structures of bacterial ribosomes co-crystallized with several macrolides have demonstrated that these compounds make many contacts with the bacterial rRNA [42]. While the macrolactone appears to contribute the majority of the binding interactions with the ribosome, macrolides devoid of their deoxysugar moieties lack antibacterial activity. Further, the aminosugars appear indispensable for the proper function of the macrolides, as compounds bearing only their aminosugars remain biologically active. Besides antibacterial compounds, the macrolide scaffold is also found in diverse compounds featuring antihelmithic or insecticidal activity. One such group of compounds is the spinosyns, which are polyketide insecticides containing forosamine (17) and a permethylated rhamnose [43]. Interestingly, the biological targets of spinosyns are insect nicotinic acetylcholine receptors and γ-aminobutyric acid receptors; and, like antibacterial macrolides, while both sugar components are required for binding to these receptors, the full activity is tolerable of modifications to the rhamnose moiety but not to the forosamine residue.
2.2. General Mechanism of Transamination Catalyzed by Aminotransferases
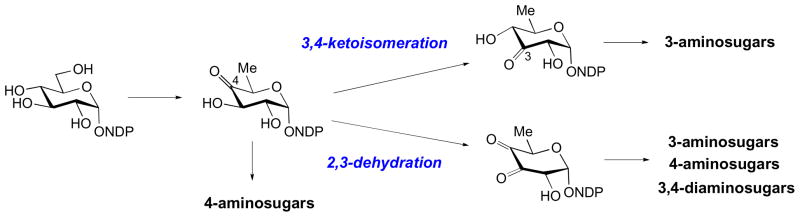
As in all transamination reactions, sugar aminotransferases (SATs) transfer an amino group from an amino acid donor (46) to a ketone-bearing acceptor, in this case an NDP-activated ketosugar (47). For most SAT, the amino donor is either L-glutamate or L-glutamine, but L-aspartate may also be used in some cases [17–21]. The substitution position of the aminotransfer is dictated by the aminotransferase involved and the location of the substrate’s keto group, which is typically generated in a preceding dehydrogenation or isomerization reaction in the biosynthetic pathway. Since the biosyntheses of most deoxy sugars share similar precursors, many of which are 3- or 4-ketosugars (e.g., 47 is a NDP-4-keto-6-deoxyglucose), the commonly encountered deoxy aminosugars are, thus, aminated at the C-3 and C-4 positions (Fig. 3) [10,11].
Figure 3.
Conserved biosynthetic intermediates dictate frequently observed positions of amination of deoxy sugar hexose ring.
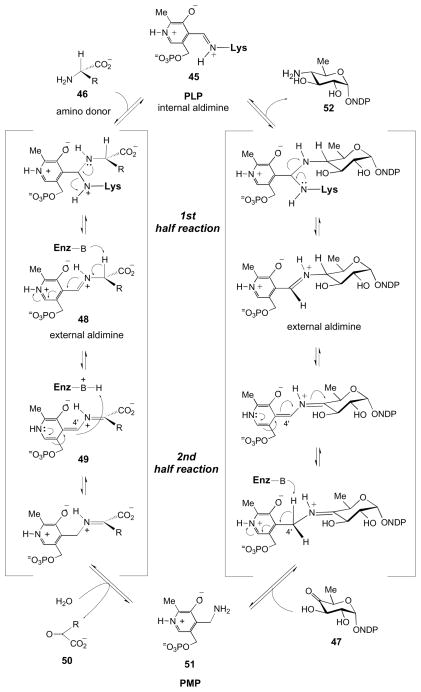
The reaction mechanism of SATs follows the well-established paradigm for other aminotransferases [17–21]. The PLP cofactor serves as an electron sink throughout the reaction, and in the aspartate aminotransferases, an invariant aspartate residue in their active sites helps to maintain this role by stabilizing the positively charged pyridinium ring of the cofactor. Transamination is mechanistically composed of two stages, often referred to as the first-half and the second-half reactions (Fig. 4) [19]. Prior to reaction, the PLP cofactor is covalently bound to the enzyme via a conserved lysine residue; this adduct is known as the internal aldimine (45). In the first-half reaction, the lysine is exchanged for the amino acid (46) serving as an amino donor, resulting in the formation of the external aldimine (48). At this point, the PLP is no longer covalently bound to the enzyme, but, rather, is anchored in place through an extensive hydrogen-bonding network centered on the phosphate of the cofactor. In the second-half reaction, a conserved base abstracts a proton from Cα of the PLP-amino acid complex to form a Cα anion or a quininoid species (49). The abstracted proton is then categorically added back on the si-face of the quininoid complex at C4′ of the cofactor [17,18]. Finally, the PLP-amino acid complex is hydrolyzed resulting in the expulsion of the α-keto acid (50) and the formation of PMP (51). The remainder of the reaction, commencing with PMP and the keto sugar substrate (47) is mechanistically the reverse of the initial sequence and results in the formation of the aminosugar product (52) and regeneration of the internal aldimine (45).
Figure 4.
Prototypical transamination mechanism catalyzed by vitamin B6-dependent enzymes.
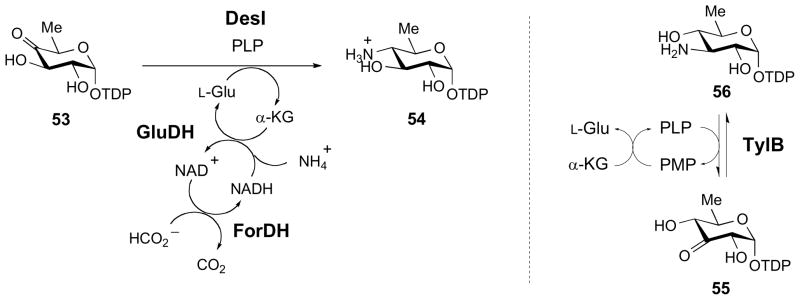
In many cases, the in vitro conversion of the keto sugar to the corresponding amino sugar requires the addition of excess amino donor to help drive the reaction to completion. Alternatively, the amino acid donor can be regenerated in the presence of ammonia and the appropriate enzyme, such as glutamate dehydrogenase (GluDH) when L-glutamate is the amino donor. For example, in the DesI catalyzed C4 transamination of TDP-4-keto-6-deoxy-D-glucose (53) [44], a nearly quantitative conversion to 54 can be achieved by coupling the transamination reaction with the GluDH reaction and the oxidation of formate by formate dehydrogenase (ForDH), as shown in Fig. 5 [45]. This coupled reaction drives the transamination equilibrium to completion and, thus, improves the conversion yield from approximately 20% to nearly quantitative. Since the enzyme-catalyzed transamination is a reversible process, the activity of the SATs can also be assayed in the reverse direction. Considering the fact that the amino sugar product is generally more stable than the keto sugar substrate and its chemical synthesis is comparatively much less demanding, assaying the transamination activity in the reverse direction is more convenient in determining the activities of aminotransferases involved in amino sugar biosynthesis. Some applications can be found in the study of TylB (Fig. 4, 56 → 55) and SpnR in the biosynthesis of mycaminose (14) in tylosin [26,46–48] and forosamine (17) in spinosyn A [29,49], respectively.
Figure 5.
Methods for improving the yield of in vitro enzymatic transamination reactions.
2.3. Known Structures of Sugar Aminotransferases (SATs)
PLP-utilizing enzymes are classified into five fold types based on a combination of structural considerations and reaction mechanism details [17,24]. The vast majority of PLP-utilizing enzymes (including SATs) are of Fold Type I, the so-called aspartate aminotransferases [17,24]. Furthermore, aminotransferases can also be classified into subgroups based on multiple sequence alignments following the “pfam” classification. Based on this classification, SATs belong to Subgroup VI, known as “DegT_DnrJ_EryCII_StrC.” While only encompassing more commonly observed substrate preferences, it has also been suggested that Subgroup VI can be further divided into Subgroups V1a, VIb, and VIg, where SATs in Subgroup VIa utilize NDP-3-keto-sugar substrates, SATs in Subgroup VIb utilize NDP-4-keto-sugar substrates, and SATs in subgroup VIg utilize scyllo-inosose as substrates [24].
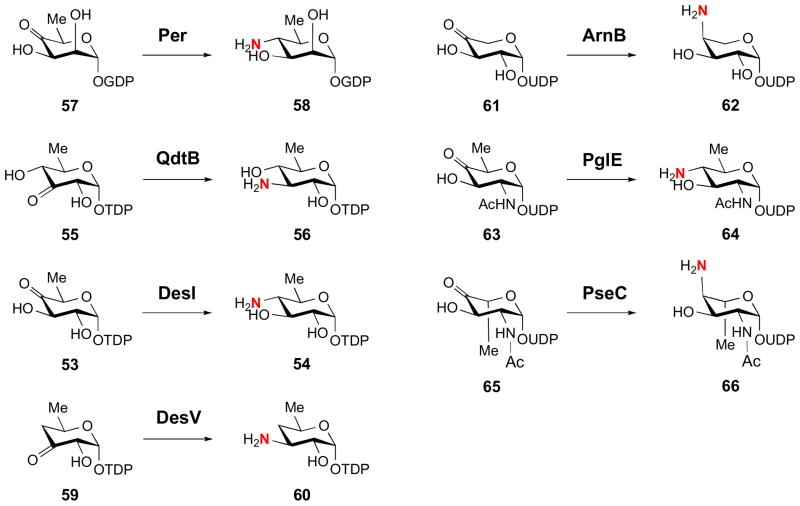
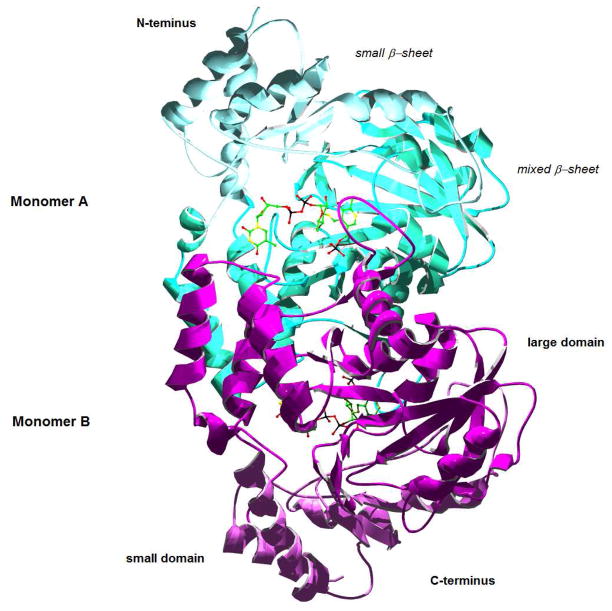
During the last eight years, crystal structures of seven bacterial SATs have been solved: Per from the perosamine (12) pathway [50], QdtB from the mycaminose (14) pathway [51], DesI and DesV from the desosamine (16) pathway [52,53], ArnB from the 4-amino-L-arabinose (24) pathway [27], PglE from the bacillosamine (10) pathway [54], and PseC from the pseudaminic acid (31) pathway [31]. The substrates and products of these enzymes are shown in Fig. 6. In general, the enzymes share many structural characteristics [55]. Each of the enzymes was found to be dimeric with extensive dimer interfaces, featuring a large, mixed β-sheet surrounded by α-helices (see Fig. 7). The active sites lie at the interface of the dimer, and while each monomer contributes critical residues to each active site, the active sites are generally independent. Based on these structural details and on the presence of a conserved active site aspartate, these SATs clearly belong to Fold Type I. The invariant aspartate residue in the active site facilitates formation of the protonated state of the PLP pyridinium ring, rendering the coenzyme electrophilic, which is important for the transamination reaction catalyzed by SATs.
Figure 6.
Substrates and products of the sugar aminotransferases crystallized to date.
Figure 7.
QdtB (3FRK) as a model sugar aminotransferase, highlighting dominant structural features, including the large and small domains. The bound ligand is the PLP-substrate complex (see Figure 6).
These Fold Type I SATs are further characterized by an architecture composed of a “large domain” and a “small domain.” The large domain features a seven- or eight-stranded β-sheet, and the small domain is formed by the C-terminal portion of a monomer, which folds into a four-stranded β-sheet surrounded by helices. The N-terminal portion often contributes to the small domain as well. In some enzymes these domains have been observed, or, in some cases, predicted, to move considerably upon substrate binding to create a “closed” conformation, which may help to confer substrate and reaction specificity [17, 50–55].
2.4. Structural and Mechanistic Correlations of SATs
While the mechanistic details of the aminotransfer reaction are well-established, and a good number of crystal structures of SAT have been characterized [55], comparatively little is known about the structural factors determining the regiospecificity or stereospecificity of the reaction. The SATs are paradoxical in that their structures, including active sites, share a high degree of similarity to each other, yet these enzymes are often capable of differentiating between similar substrates to produce a variety of products with distinct stereochemistry and regiochemistry. While the substrate and product specificity is more likely the result of many subtle contributing factors, the structures did provide some tantalizing examples of clear substrate specification.
The first structure of a SAT co-crystallized with its native substrate was PseC [31]. It was noted that the binary complex lacks specific contacts between the protein and the pyranose portion of the sugar substrate (65). Instead, the structure features a hydrogen-bonding network centered on the nucleotide diphosphate portion of the substrate. In fact, this appears to be a recurring theme seen in the structures of many SAT enzymes co-crystallized with their substrates [31,51,52]. Thus, it has been suggested that minute changes in the orientation of select residues distal to the active site might be responsible for discriminating between different NDP-activated substrates [51].
In support of the idea that recognition of the NDP group is important in binding of the substrate, the flexibility of the aminotransferases WecE and Per towards sugar substrates substituted with different NDP groups was explored [50,56,57]. WecE is a 4-aminotransferase which uses TDP-4-keto-6-deoxy-D-glucose (53) as its substrate. While this enzyme could turn over TDP-4-keto-6-deoxy-D-mannose, the 2-epimer of its substrate, GDP-4-keto-6-deoxy-D-mannose (57) could not be utilized [56]. Similarly, Per, whose substrate is GDP-4-keto-6-deoxy-D-mannose (57), could not accept the TDP-form of the native substrate [50]. These results indicated that the identity of the NDP group is an important factor in discriminating between substrates.
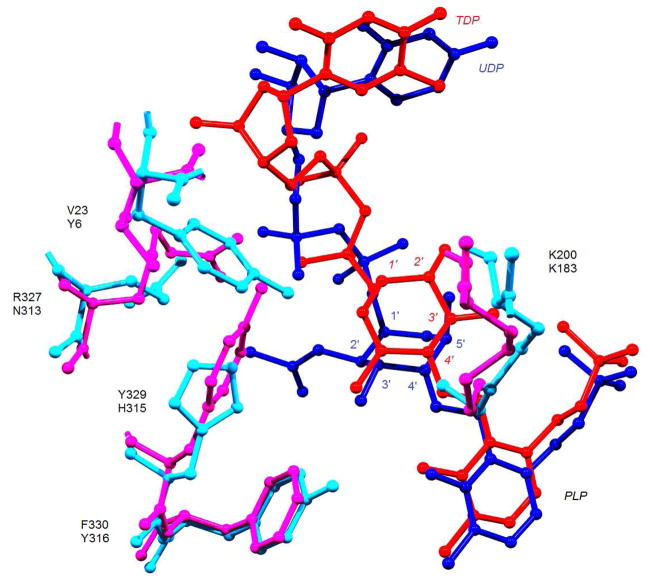
In a separate example, comparison of the structures of PseC and DesI, both of which were co-crystallized with their substrates, provided some basis for controlling the stereochemistry of their products (Fig. 8) [50,52]. These two SATs catalyze aminotransfer reactions using a 4-ketohexose substrate to generate a 4-aminohexose, but, in the former case, the amino group is installed with axial stereochemistry (see 66), whereas in the latter case the amino group is installed with equatorial stereochemistry (54). The crystal structures of PseC and DesI revealed that the binding modes of the pyranose portions of their respective sugar substrates differ by a 180° rotation in these two enzymes (Fig. 8) [52]. These opposite binding modes are likely responsible for the observed stereochemical outcomes of these enzymes, but while it is tempting to attribute the stereoselectivity of the reaction solely to the observed differences in the binding mode of their substrates, other structural factors may also be at play. For instance, the PseC substrate features an acetamido group at the C2 position and the C5 methyl group has an axial orientation (see 65). In contrast, the DesI substrate features a hydroxyl group at C2 and its C5 methyl group is oriented in an equatorial configuration (see 53). Unfortunately, this is the only pair of SATs that catalyze aminotransfer reactions at the same position with opposite stereochemistry and whose structures are known. Whether this is a common theme conserved among all such pairs of enzymes remains to be seen.
Figure 8.
Composite of DesI and PseC active sites (2PO3 and 2FNU, respectively). PseC residues are shown in light blue, and its substrate-PLP complex is in dark blue. DesI residues are shown in magenta, and its substrate-PLP complex is in red. DesI residue labels are on top of PseC residue labels, and their hexose positions are italicized and non-italicized, respectively.
Interestingly, the apparent lack of specific protein contacts to the pyranose portion of the substrate could enable some SATs to exhibit relaxed substrate specificity. For example, the structure of QdtB was shown to lack contacts with the C4 position of the pyranose substrate (55). This feature was exploited in QdtB activity assays which showed that the enzyme could also turn over the C4 epimer of its natural substrate, 3-amino-3,6-dideoxy-D-galactose [51]. Further, it was shown that DesV, a 3-aminotransferase whose natural substrate is deoxygenated at the C4 position (see 59) [58], could accept a substrate analog with an equatorially positioned hydroxyl group at C4 (see 55) [53].
3. Deoxygenation
3.1. Natural Occurrence of Deoxy Sugars and Their Formation
The simple deoxy sugars are derived from common sugars (e.g., glucose and mannose) by the replacement of at least one hydroxyl group with a hydrogen atom. Most naturally occurring deoxyhexoses are deoxygenated at the C6 position; however, further deoxygention at C2, C3 and C4 is also observed [3,6,59]. Although the 2,6-dideoxyhexoses are by far the most abundant deoxy sugars found in natural products, deoxy sugars featuring different patterns of deoxygenation, e.g., 3,6-dideoxy hexoses, are commonly found as components of Gram negative lipopolysaccharide (LPS) O-antigens, where their structural variety contributes to their complex roles in cellular recognition and virulence [60,61]. Other alternatively deoxygenated sugars such as 4,6-dideoxy, 2,3,6-trideoxy, 2,4,6-trideoxy, 3,4,6-trideoxy, and 2,3,4,6-tetradeoxy hexoses are much less abundant and are found mainly as components of bioactive natural products, where they are often found to be essential to the biological efficacy or target specificity of the natural products they decorate [1–5,10,11].
Since common monosaccharides possess a carbon skeleton that is fully substituted with hydroxyl groups, a variety of mechanisms have evolved for the deoxygenation of these precursors to generate modified sugar structures. Mechanisms for deoxygenation at the C2 or C6 position of a hexose substrate have been well studied and understood to proceed through an α-anion-induced β-dehydration mechanism [8–11,59,62]. In contrast, the removal of the 4-hydroxyl group to form 4-deoxyhexoses is catalyzed by a radical SAM-dependent enzyme [45,63–65]. While some aspects of C4-deoxygenation have been established, the detailed mechanism remains to be elucidated.
Interestingly, C3-deoxygenation in the biosynthesis of 3-deoxyhexoses commences with a 4-ketohexose substrate, requires the participation of coenzyme B6, and involves the formation of radical intermediates [8,9,59,66,67]. The enzymes carrying out this reaction are all aspartate aminotransferases based on sequence alignments and the available structural information, however, unlike other B6-dependent enzymes, they employ PMP instead of PLP in catalysis. Moreover, three related but distinct C3-deoxygenation mechanisms have been identified and are exemplified in the biosyntheses of CDP-D-ascarylose (67) and GDP-D-colitose (73), which are 3,6-dideoxy sugars, and TDP-D-forosamine (see 17), a 2,3,4,6-tetradeoxy sugar. Since the 3,6-dideoxyhexoses are major antigenic determinants of many pathogenic bacteria, including Eschericia coli, Vibrio cholerae, and species of Shigella and Yersinia, the enzymes carrying out the C-3 deoxygenation reaction in each pathway are potential antibiotic targets [61,68].
3.2. C-3 Deoxygenation in the Biosynthesis of Ascarylose
3.2.1. Biochemical Properties of E1 and E3 Involved in C-3 Deoxygenation
CDP-D-ascarylose (67) is found widely in Gram-negative bacterial cell walls. Studies of its biosynthesis led to the discovery of CDP-6-deoxy-L-threo-D-glycero-4-hexulose 3-dehydratase, E1, which catalyzes C-O bond cleavage at C3 of the CDP-4-keto-6-deoxysugar substrate (68) [25,69]. Interestingly, this enzyme was found to have little activity after purification by ion-exchange chromatography [69,70]. PMP was subsequently discovered in the early column fractions, and it was found that its addition could restore enzyme function [70]. This finding was significant because it provided the first hint about the PMP-dependence of E1 whose catalysis is not associated with transamination. Later, it was found that the “as-purified” E1 is a mixture of holo- and apoenzyme, which requires reconstitution with not only PMP, but also iron and sulfur [71,72]. The fully reconstituted E1 is a dark red-brown protein that exists as a homodimer in its native state containing 1 equivalent of PMP and one [2Fe-2S] cluster per subunit with a subunit molecular mass of approximate 49 kDa. Despite being classified as a member of the aspartate aminotransferase family, E1 has several unique features distinguishing it from typical aminotransferases, namely: 1) the enzyme lacks the highly conserved Schiff-base-forming lysine found in PLP-dependent enzymes, featuring instead a histidine residue in its place [73]; and 2) E1 bears a [2Fe-2S] cluster bound by a unique motif, C-X57-C-X1-C-X7-C, predicted based on sequence alignment [74]. These characteristics of E1 reflect a distinct evolutionary path for E1, and distinguish it from other B6-dependent enzymes. It should be noted that GABA-aminotransferase is another B6-dependent enzyme carrying a [2Fe-2S] cluster [75], however, the catalytic role of the iron-sulfur cluster in this case remains elusive. E1 is, thus, the only example of a PMP-dependent enzyme that contains a mechanistically defined and relevant [2Fe-2S] cluster.
3.2.2. Proposed Mechanism for E1 and E3 Catalyzed Reaction
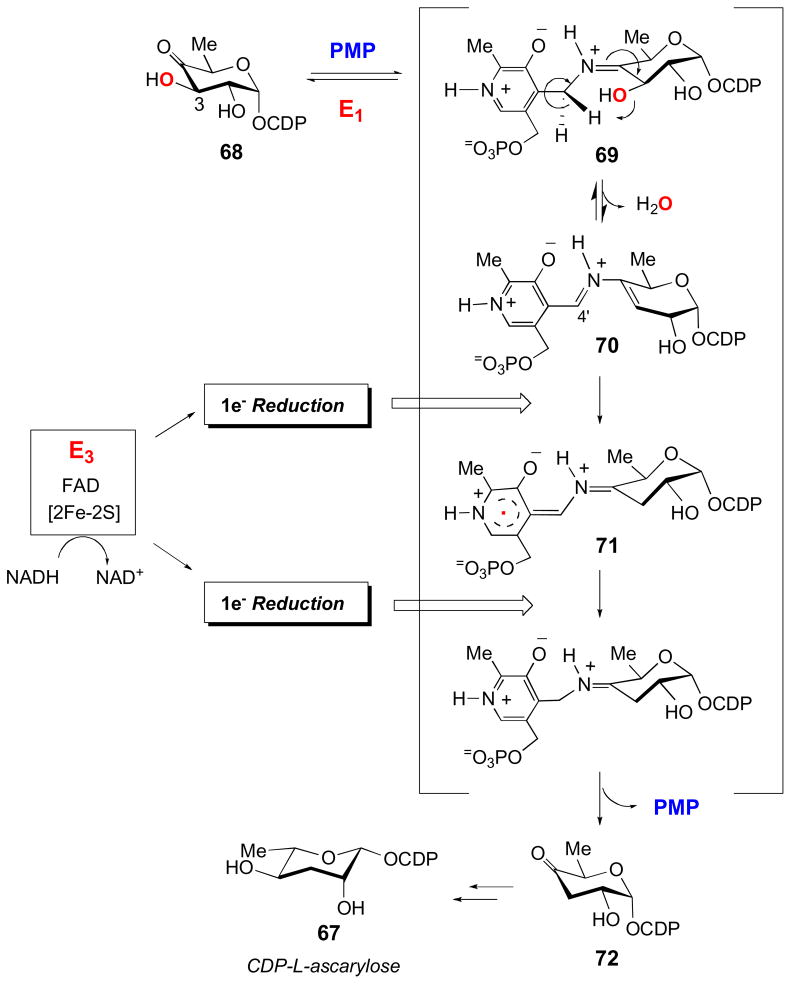
The following mechanism of deoxygenation by E1 has been characterized in detail (Fig. 9) [6–8]. First, the enzyme forms a Schiff base (69) between its PMP cofactor and the 4-keto group of its substrate, CDP-4-keto-6-deoxy-D-glucose (68). Following C4′-proton abstraction from PMP by His220 of E1, the 3-hydroxyl group of the substrate is expelled in a 1,4-dehydration reaction, which results in the formation of a Δ3,4-glucoseen intermediate (70). The C-O bond cleavage is reversible with an equilibrium favoring the reverse direction (70 → 69) [76], so this intermediate must be reduced by two electrons to drive the reaction to completion. The reduction, in which NADH serves as the ultimate electron donor, is mediated by E3, an iron-sulfur containing flavodoxin-NADH reductase. E3 is a red-brown monomeric protein containing 1 mol of FAD and one plant-type ferredoxin [2Fe-2S] center per molecule with a molecular mass of 36 kDa [77–80]. The reduction is initiated by a hydride transfer from NADH to the FAD of E3 to generate the reduced hydroquinone form of this cofactor. This is followed by two successive one-electron transfers from the reduced FAD in E3 to E1 via these enzymes’ [2Fe-2S] clusters to reduce the E1-bound glucoseen intermediate (70) [81,82]. Subsequent hydrolysis releases the product, CDP-4-keto-3,6-dideoxy-D-glucose (72), from the active site of E1 and regenerates the PMP cofactor. Interestingly, E1 can also utilize other reduction systems, albeit with greatly reduced efficiency.
Figure 9.
The mechanism of C-3 deoxygenation reaction catalyzed by E1, CDP-6-deoxy-L-threo-D-glycero-4-hexulose 3-dehydratase, and its reductase partner E3 in the biosynthesis of CDP-L-ascarylose. See section 3.2.2 for details.
The involvement of iron-sulfur clusters, which are obligatory one-electron transfer cofactors, implies that radical intermediates must be formed in the reduction of the glucoseen intermediate (70) [71,72]. Evidence of the existence of radical intermediates in E1 was demonstrated experimentally using both EPR and ENDOR spectroscopic techniques [83,84]. This set of experiments revealed the presence of two radical species during turnover, one of which could be assigned to the flavin semiquinone radical. The other radical was demonstrated to be associated with the PMP cofactor-glucoseen complex (see 71). The involvement of glucoseen in carrying the unpaired electron spin was dismissed as the EPR shows a featureless singlet with no obvious coupling. Given that every carbon of the glucoseen intermediate (70) bears at least one hydrogen atom, such a featureless singlet would not be expected for a glucoseen radical. Thus, the radical is more likely carried by the PMP cofactor in the complex [83]. While it was initially thought that the radical existed as a phenoxy radical, mechanistic probes failed to substantiate this. On the other hand, using deuterated PMP analogs, pulsed ENDOR studies were able to detect hyperfine couplings between the unpaired electron spin and the deuterium labels of the PMP cofactor. These studies clearly indicated that the radical is delocalized primarily within the PMP pyridinium ring (see 71) [84]. The participation of PMP in deoxygenation is unusual, as is the direct involvement of PMP in the stabilization of an unpaired electron spin in an electron-transfer reduction. It should be pointed out that the 1,2-shift of the amino group catalyzed by lysine aminomutases also involves PLP stabilizing radical intermediates during turnover [85–88], which is covered in another review by Frey and Reed in this issue. Despite this departure from more typical B6-dependent catalysis, E1 was found to share the stereospecificity of proton abstraction of the PMP cofactor displayed by other PLP-utilizing enzymes. Namely, the pro-S proton is abstracted from C4′ of both PMP and the cofactor-substrate complex [76].
3.2.3. Crystal Structure of E1
As mentioned above, E1 is considered an aspartate aminotransferase, but given the unique reaction this enzyme catalyzes, structural studies were performed to elucidate the determinants of an NDP-hexose C3-dehydrase. The crystal structures of the wild type E1 from Yersinia pseudotuberculosis and its H220K mutant display the hallmark structural details observed in aspartate aminotransferases except for a loop which coordinates the [2Fe-2S] cluster (Fig. 10A) [89]. Both structures were dimeric and dominated by a large seven-stranded mixed β-sheet clothed in α-helices. Also, each features a single helix which “points” at the PLP cofactor present in the H220K mutant, and the dipole of this helix is aligned with its positive end towards the cofactor phosphate group providing stabilization. This helix is a common structural theme also observed in the SATs crystallized to date [50,52,53].
Figure 10.
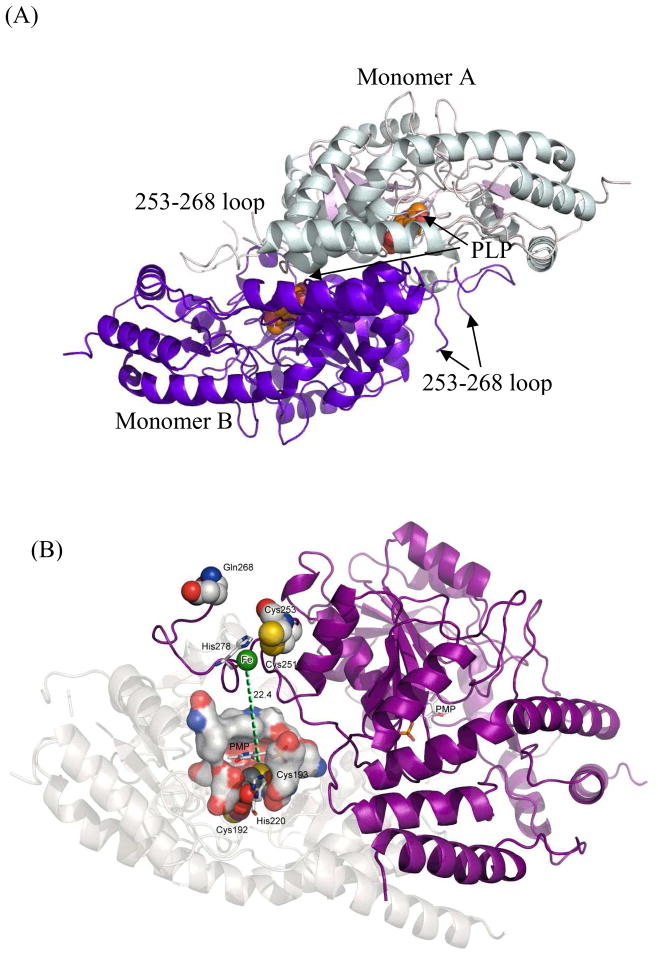
(A) The overall topology of dimeric H220K-E1, including the cofactor binding site and the putative [2Fe-2S]-binding loop. The cofactor PLP is shown in spheres since H220K-E1 has PLP in the active site. (B) The anomalous map (sigma at 5) shows strong Fe peak near the 253–268 loop region. The distance of C192 and C193 is 22–30 Å away from the main [Fe-S] coordinating loop. Hence, the fourth ligand is more likely H278. See 3.2.3 for details.
E1 is also set apart from other AAT structures by the presence of a [2Fe-2S] cluster, although this cluster was not seen in the crystal structures [89]. Mutagenesis studies showed this cluster is coordinated by three cysteine residues found on a flexible loop composed of residues 253–268: C251, C253, and C261 [74]. This arrangement would hold the cluster on the surface of the protein. The fourth coordinating residue has been a point of contention, as mutagenesis of C192 and C193 resulted in the inability of E1 to bind its [2Fe-2S] cluster [74]. However, the crystal structures of E1 show that C192 and C193 lie quite far from the other three cysteine residues in a buried region that is inaccessible to both solvent and the flexible loop (Fig. 10B). As a dramatic change in protein conformation to bring these residues close to the other three is unlikely, it seems more reasonable that these residues play a structural role. On the other hand, the crystal structures of E1 suggest that H278 is more likely the fourth [2Fe-2S] cluster-coordinating residue. This was substantiated by mutagenesis studies which showed that H278A results in an enzyme that cannot hold the cluster, whereas H278C leads to half of the wild type iron-binding capability [89]. Thus, E1 binds its [2Fe-2S] cluster using an unprecedented structural motif.
Another unusual structural feature of E1 is the disorder in the 253–268 loop region [89]. This loop region is the least conserved region between E1 and other PLP-dependent enzymes. When the structure of E1 is superimposed on the homologous structures, it becomes obvious that the 253–268 loop region of one monomer is likely adjacent to the active site of the other monomer. A possible explanation for the loop flexibility observed in E1 is that it allows “open” and “closed” conformational changes of the substrate binding pocket as a gating mechanism. However, in the homologous ColD [90–92], BtrR [93], and especially PseC [31], whose apo- and substrate/inhibitor-bound structures are available for comparisons, this loop region remains unchanged with and without substrate binding [89]. These observations suggest that a second possible reason for the E1 loop flexibility is to allow close contact with the reductase E3 to form a reasonably tight binary complex. Such a complex might facilitate the electron transfer from the reduced iron-sulfur center of E3 via the iron-sulfur cluster of E1 to reduce the dehydration product 70 in the E1 active site to complete the deoxygenation reaction [82,83].
Because co-crystals with an E1 reaction analog, CDP-4-amino-4,6-dideoxy-D-glucose, consistently revealed incomplete occupancy of the sugar in electron density maps, the active site was studied using more indirect methods [89]. Specifically, while the sugar displayed incomplete occupancy, omit maps of the co-crystals showed clear electron density for the cofactor and partial electron density of the CDP-sugar. This permitted docking studies which revealed, at best, semiconserved hydrophobic residues mediating substrate interactions. However, more definitive characterization of the substrate binding pocket must await further study.
3.2.4. Mutations of E1 Enable Aminotransferase Activity
Demonstration that E1 shares the stereospecificity of proton transfer observed in the vast majority of PLP-dependent enzymes [76] lent credence to the idea that all PLP-dependent enzymes developed from shared evolutionary precursors [17]. The most obvious residue in E1 that is incongruent with common PLP-dependent aminotransferases is the histidine found in place of the strictly conserved lysine present in all PLP-containing enzymes. The conserved histidine found in place of this residue was mutated to lysine (H220K mutant) which immediately endowed E1 with aminotransferase activity, allowing production of CDP-4-amino-4,6-dideoxy-D-glucose using the same substrate of the wild type E1 reaction, CDP-4-keto-6-deoxy-D-glucose (68) [94]. However, this reaction was non-catalytic, as only one round of reaction was possible because the PLP cofactor could not be regenerated. Analysis of the H220K mutant showed that E1 was capable of forming the internal aldimine species with PLP, as it was seen in the crystal structure. Since H220K was only sufficient to create a non-catalytic aminotransferase, the structure of H220K was further scrutinized to uncover more residues important for aminotransferase activity. This process was facilitated by comparison of the E1 active site to the active site of ArnB, a bona fide aminotransferase [27]. Since both E1 and ArnB accept similar substrates (ArnB uses a UDP version of the E1 substrate lacking the 5-methyl group, see 61 in Fig. 6), the comparison seemed valid. This analysis revealed four active site residues that differed between the two enzymes: D194, Y217, H220, and F345 in E1 and H163, H185, K188, and H297 in ArnB. Mutation of all of the E1 active site residues to those found in ArnB converted E1 into a catalytically active aminotransferase [89]. However, kinetic analysis of the E1 quadruple mutant showed it to be much less efficient than ArnB: Whereas both enzymes showed comparable rates for the first half reaction (conversion of PLP to PMP, formation of the external aldimine), the rate for the second half reaction of the E1 mutant is ten times slower than its first half reaction. The overall rate of reaction for ArnB is 1.3 × 103 nmol·min−1·mg−1, while for the mutated E1 the overall rate is 9.6 nmol·min−1·mg−1, suggesting that enzyme characteristics differentiating aminotransferase and deoygenation functions are likely due to additional subtle features.
3.3. C-3 Deoxygenation in the Biosynthesis of Colitose
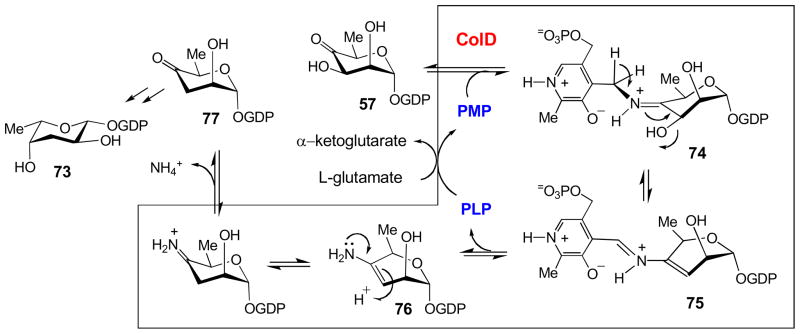
GDP-D-colitose (73) is another unusual deoxy sugar found in Gram-negative cell walls whose biosynthesis, like CDP-D-ascarylose (67), has garnered much attention due to its C3-deoxygenation reaction [28,95]. Unexpectedly, while GDP-D-colitose is also deoxgenated via a PMP-dependent enzyme, the reaction follows a mechanism distinct from E1 in the biosynthesis of CDP-D-ascarylose. Most significantly, deoxygenation of GDP-D-colitose utilizes a one-enzyme system, ColD, which does not require a reductase, NADH, or FAD. Also, similar to E1, ColD has a histidine in place of the conserved lysine residue found in PLP-dependent enzymes, but unlike E1, ColD lacks a [2Fe-2S] cluster [95]. The mechanism of ColD commences similarly to E1, as initially a Schiff base (74) is formed between the substrate, GDP-4-keto-6-deoxy-D-mannose (57), and the PMP cofactor of ColD (Fig. 11) [10,11,59]. Following proton abstraction from C4′ of PMP and the subsequent expulsion of the substrate 3′-hydroxyl group, the Δ3,4-mannoseen intermediate (75) is formed. Then, in contrast to E1, the Δ3,4-mannoseen intermediate-cofactor complex is hydrolyzed, yielding PLP and an enamine sugar (76). The nascent enamine sugar (76) undergoes tautomerization and hydrolysis to yield ammonia and the product of the reaction, GDP-4-keto-3,6-dideoxy-D-mannose (77). Finally, PMP must be regenerated through a transamination reaction with glutamate as an amino donor.
Figure 11.
The mechanism of C-3 deoxygenation reaction catalyzed by ColD in the biosynthesis of CDP-L-colitose. See section 3.3 for details.
Recently, the structure of ColD, has been solved to 1.8 Å resolution [90–92]. Like other aspartate aminotransferases crystallized to date, ColD displays the hallmark structural features consistent with these PLP-dependent enzymes. Namely, ColD forms a dimer with an extensive dimer interface, a mixed eight-stranded β-sheet surrounded by helices dominates the structure, and a helix whose macrodipole positive end “points” at the phosphate of the cofactor. Also, while the active site residues within each monomer are contributed mainly by that monomer, the active site architecture of each monomer is completed by a loop composed of residues F240-E253 from the other monomer. In one structure of ColD crystallized in the presence of α-ketoglutarate and PLP, a hydrated form of the PLP cofactor was obtained, where the aldehyde of the cofactor appeared to exist as a gem-diol [90]. The physiological ramifications of this finding are unknown. In this crystal, the residues anchoring the PLP cofactor via its phosphate group can be seen to be involved in an extensive hydrogen bonding network. In another structure obtained from crystallization of ColD in the presence of PLP and L-glutamate the cofactor is seen as the ketimine species.
Like E1 and CDP-D-ascarylose, attempts to co-crystallize ColD and its substrate consistently failed [90,91]. It was thought that perhaps this was due to the instability of the substrate during the crystallization process, so the enzyme was crystallized in the presence of GDP-D-perosamine (58), which differs from the natural substrate in that it features an amino group instead of a ketone at the C4-position [92]. This analog, besides providing stability, would allow formation of the external aldimine with the PLP cofactor. However, the structures still suffered from poor occupancy of the sugar, so it was reasoned that the enzyme was still active in the crystal lattice and turned over the substrate, which diffused out. In fact, enzymatic assays of ColD with GDP-D-perosamine (58) as a substrate and omitting L-glutamate showed that ColD could turn over the substrate analog, producing the natural ColD product, GDP-4-keto-3,6-dideoxy-D-mannose (77) [90,92]. Apparently GDP-D-perosamine could serve as an amino donor to form the catalytically necessary PMP form of the cofactor. In doing so, the C4-amino sugar becomes a C4-keto sugar, which is a substrate for ColD. Thus, to obtain an enzyme-substrate analog co-crystal, the ColD active-site histidine residue, H188, was mutated to asparagine (H188N mutant). This strategy was successful, and the H188N-GDP-D-perosamine co-crystal was obtained [92]. The cofactor formed an external aldimine with the sugar, but, unexpectedly, the sugar was found to be in a boat configuration. The structure once again revealed a hydrogen-bonding network securing the cofactor in place, and while several residues appeared to be involved in interactions with the GDP-group, no specific interactions between the pyranose and ColD were seen. However, it is likely that the interactions responsible for discriminating between substrates are more subtle.
3.4. C-3 Deoxygenation in the Biosynthesis of Forosamine
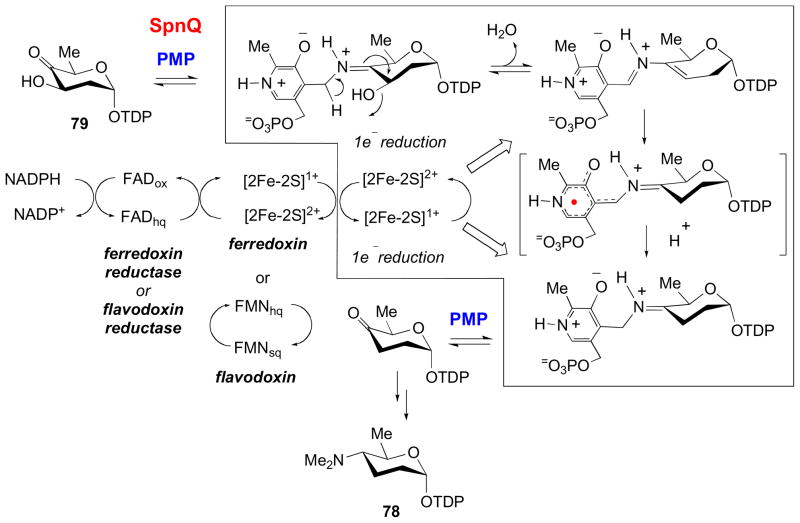
TDP-D-forosamine (78) is unique in its high degree of deoxygenation. This sugar is found as a component of the commercial insecticide Spinosad and a few other natural products [29,43,96]. Isolation and analysis of the spinosyn biosynthetic gene cluster revealed SpnQ, which encodes an enzyme with homology to both E1 and ColD, as well as other putative C3-dehydratases [49,97]. Like E1, work from our laboratory suggests that SpnQ appears to harbor a [2Fe-2S] cluster, but, following the ColD paradigm, the spinosyn biosynthetic gene cluster does not contain an E3 homolog. The implications of the latter finding were borne out by experiments which revealed that SpnQ is capable of deoxgenating its substrate, TDP-4-keto-2,6-dideoxy-D-glucose (79) using either sodium dithionite, flavodoxin/flavodoxin reductase, or ferrodoxin/ferrodoxin reductase systems (Fig. 12). Thus, it seems SpnQ may utilize a general, cellular reductase system [49,97]. These findings suggest that, while carrying out the same reaction as E1 and ColD, SpnQ represents a third, distinct C3 deoxygenation mechanism.
Figure 12.
The mechanism of C-3 deoxygenation reaction catalyzed by SpnQ in the biosynthesis of TDP-D-forosamine. See section 3.4 for details.
Unexpectedly, in the absence of a reductase, SpnQ produces the aminosugar TDP-4-amino-2,4,6-trideoxy-D-glucose [97]. This bears some resemblance to ColD which also catalyzes an aminotransfer reaction. Interestingly, after observing SpnQ catalyzed conversion of its substrate to the C4 amino sugar product using PLP and L-glutamate, we also assayed E1 using its natural substrate 68 in the presence of L-glutamate and a catalytic amount of PLP and observed that it catalyzes C3 deoxygenation rather than C4 aminotransfer under these conditions [97]. Thus, in the absence of reductase and using L-glutamate and their natural substrates, E1 and SpnQ follow different reaction pathways. This is intriguing in that, although E1 and SpnQ seem to catalyze mechanistically identical reactions in the presence of their respective reductase partners, SpnQ, and not E1 or ColD, has retained an ancestral aminotransferase activity, whereas E1 behaves like ColD, catalyzing the reductase-independent C3 deoxygenation, pointing to a unique evolutionary pathway for SpnQ and other closely related TDP-2,6-deoxysugar 3-dehydrases.
4. Glycine Addition
4.1. Aldolase reaction in the biosynthesis of nucleoside antibiotics
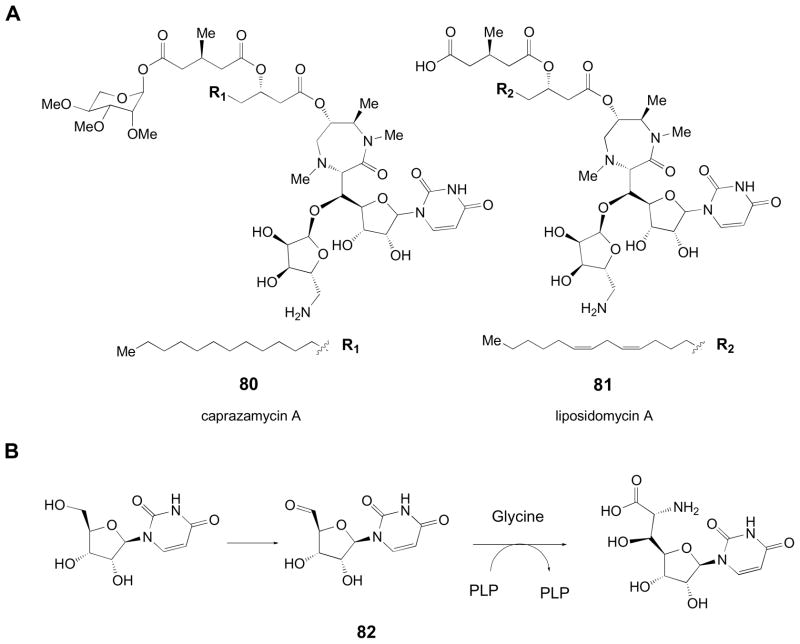
Carbon-carbon bond formation is arguably one of the most important reactions in organic chemistry. While synthetic chemistry employs various strategies to control the stereo- and regiochemistry of the reaction, the ability of enzymes to enable exquisite control over these parameters is unrivaled. Consequently, enzymes catalyzing carbon-carbon bond formation can be valuable, especially in the formation of novel branched deoxysugars [98–101]. Branched-chain sugars [3,11,102] are frequently employed in bacterial cell wall formation, where they serve structural roles or contribute to antigenic determinants [61]. In deoxy sugar combinatorial biosynthesis, enzymes capable of branching could allow access to more complex carbohydrates [103]. The caprazamycins (80), liposidomycins (81), murraymycins, and related compounds are nucleoside antibiotics that feature a unique diazopanone moiety [104,105]. Analysis of the caprazamycin and liposidomycin gene clusters revealed a gene encoding a putative serine-glycine hydroxymethyltransferase (SHMT). The SHMTs from these pathways belong to Fold Type I, the aspartate aminotransferases. Although SHMTs normally participate in the physiological interconversion of serine and glycine with concomitant interconversion of N5,N10-methylene tetrahydrofolate and tetrahydrofolate [106,107], these enzymes have also been shown to be capable of catalyzing the condensation of glycine with acetaldehyde to form threonine [108,109]. Thus, the diazopanone formation is presumed to begin with the condensation of a uracil 5′-aldehyde with glycine in a reaction proposed to be catalyzed by the putative SHMTs found in the biosynthetic gene clusters of caprazamycin and liposidomycin (Fig. 13) [104,105]. While their substrate flexibility remains to be explored, it is expected that the glycine hydroxymethyl transferases may be versatile tools in the development of novel deoxy sugars, especially considering the generic nature of the putative substrates.
Figure 13.
(A) Structures of diazepanone-containing natural products, caprazamycin A and liposidomycin A. (B) Hypothetical reaction sequence for the condensation of glycine and a uridine 5′-ribosyl aldehyde (82) to yield a diazepanone precursor.
5. Conclusions
Owing to the catalytic versatility of the PLP/PMP cofactor, the vitamin B6-dependent enzymes involved in deoxy sugar biosynthesis may carry out transamination, deoxygenation, and even aldolase reactions to generate a staggering number of unique structures. Transaminases alone are responsible for a majority of the diversity of sugar products, since they can aminate nearly every position of the hexose ring. The introduced amino functional groups frequently endow these amino deoxy sugars with their biological activities and provide a platform for the generation of even more diverse functional groups. Thus, vitamin B6-dependent enzymes offer many opportunities for the advancement of glycodiversification as a viable strategy for the development of novel or improved pharmaceuticals.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (GM35906, GM54346) and the Josefina Lesvia Falcon Graduate Fellowship (to A.J.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brandenburg K, Schromm AB, Gutsmann T. Endotoxins: relationship between structure, function, and activity. In: Wang X, Quinn PJ, editors. Endotoxins: Structure, Function, and Recognition. Springer Science+Business Media B.V; New York: 2010. pp. 53–67. [DOI] [PubMed] [Google Scholar]
- 2.Weymouth-Wilson AC. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DA, Liu H-w. Deoxysugars: occurrence, genetics, and mechanism of biosynthesis. In: Barton D, Nakanishi K, Meth-Cohn O, editors. Comprehensive Chemistry of Natural Product Chemistry. Pergamon; New York: 1999. pp. 311–365. [Google Scholar]
- 4.Thorson JS, Hosted TJ, Jiang JQ, Biggins JB, Ahlert J. Nature’s carbohydrate chemists: the enzymatic glycosylation of bioactive bacterial metabolites. Curr Org Chem. 2001;5:139–167. [Google Scholar]
- 5.Kren V, Martinkova L. Glycosides in medicine: the role of glycosidic residue in biological activity. Curr Med Chem. 2001;8:1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]
- 6.Liu H-w, Thorson JS. Pathways and mechanisms in the biosynthesis of novel deoxy sugars by bacteria. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 7.Hallis TM, Liu H-w. Learning nature’s strategies for making deoxy sugars: pathways, mechanisms, and combinatorial applications. Acc Chem Res. 1999;32:579–588. [Google Scholar]
- 8.He X, Agnihotri G, Liu H-w. Novel enzymatic mechanisms in carbohydrate metabolism. Chem Rev. 2000;100:4615–4661. doi: 10.1021/cr9902998. [DOI] [PubMed] [Google Scholar]
- 9.He X, Liu H-w. Formation of unusual sugars: mechanistic studies and biosynthetic applications. Annu Rev Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]
- 10.Thibodeaux CJ, Melancon CE, 3rd, Liu H-w. Unusual sugar biosynthesis and natural product glycodiversification. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeaux CJ, Melancon CE, 3rd, Liu H,-w. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez C, Salas JA. Altering the glycosylation pattern of bioactive compounds. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]
- 13.Pelzer S, Vente A, Bechthold A. Novel natural compounds obtained by genome-based screening and genetic engineering. Curr Opin Drug Discov Devel. 2005;8:228–238. [PubMed] [Google Scholar]
- 14.Griffith BR, Langenhan JM, Thorson JS. Sweetening natural products via glycorandomization. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Thibodeaux CJ, Liu H-w. Manipulating nature’s sugar biosynthetic machineries for glycodiversification of macrolides: recent advances and future prospects. Pure Appl Chem. 2007;79:785–799. [Google Scholar]
- 16.Thibodeaux CJ, Liu Hw, Thorson JS. Complimentary routes to natural product glycodiversification: pathway engineering and glycorandomization. In: Kamerling JP, Boons G-J, Lee Y, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive glycoscience. Vol. 3. Elsevier; Amsterdam: 2007. pp. 373–396. [Google Scholar]
- 17.Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998;8 :759–769. doi: 10.1016/s0959-440x(98)80096-1. [DOI] [PubMed] [Google Scholar]
- 18.Soda K, Yoshimura T, Esaki N. Stereospecificity for the hydrogen transfer of pyridoxal enzyme reactions. Chem Rec. 2001;1:373–384. doi: 10.1002/tcr.1021. [DOI] [PubMed] [Google Scholar]
- 19.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 20.Percudani R, Peracchi A. A genomic overview of pyridoxal-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toney MD. Reaction specificity in pyridoxal phosphate enzymes. Arch Biochem Biophys. 2005;433:279–287. doi: 10.1016/j.abb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 22.Nedal A, Zotchev SB. Biosynthesis of deoxyaminosugars in antibiotic-producing bacteria. Appl Microbiol Biotechnol. 2004;64:7–15. doi: 10.1007/s00253-003-1535-9. [DOI] [PubMed] [Google Scholar]
- 23.Tanner M. Sugar nucleotide-modifying enzymes. Curr Org Chem. 2001;5:169–192. [Google Scholar]
- 24.Hwang B-Y, Chob B-K, Yun H, Koteshwar K, Kim B-G. Revisit of aminotransferase in the genomic era and its application to biocatalysis. J Mol Catal B Enzym. 2005;37:47–55. [Google Scholar]
- 25.Weigel TM, Miller VP, Liu H-w. Mechanistic and stereochemical studies of a unique dehydration catalyzed by CDP-4-keto-6-deoxy-D-glucose-3-dehydrase: a pyridoxamine 5′-phosphate dependent enzyme isolated from Yersinia pseudotuberculosis. Biochemistry. 1992;31:2140–2147. doi: 10.1021/bi00122a035. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Yeung SM, Que NLS, Müller T, Schmidt RR, Liu H-w. Expression, purification, and characterization of TylB, an aminotransferase involved in the biosynthesis of mycaminose. J Am Chem Soc. 1999;121:7166–7167. [Google Scholar]
- 27.Breazeale SD, Ribeiro AA, Raetz CR. Origin of lipid A species modified with 4-amino-4-deoxy-L-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-L-arabinose. J Biol Chem. 2003;278:24731–24739. doi: 10.1074/jbc.M304043200. [DOI] [PubMed] [Google Scholar]
- 28.Beyer N, Alam J, Hallis TM, Guo Z, Liu H-w. The biosynthesis of GDP-L-colitose: C-3 deoxygenation is catalyzed by a unique coenzyme B6-dependent enzyme. J Am Chem Soc. 2003;125:5584–5585. doi: 10.1021/ja030088r. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z, Hong L, Liu H-w. Characterization of protein encoded by spnR from the spinosyn gene cluster of Saccharopolyspora spinosa: mechanistic implications for forosamine biosynthesis. J Am Chem Soc. 2005;21:7692–7693. doi: 10.1021/ja042702k. [DOI] [PubMed] [Google Scholar]
- 30.Schoenhofen IC, McNally DJ, Brisson J-R, Logan SM. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 2006;16:8C–14C. doi: 10.1093/glycob/cwl010. [DOI] [PubMed] [Google Scholar]
- 31.Schoenhofen IC, Lunin VV, Julien J-P, Li Y, Ajamian E, Matte A, Cygler M, Brisson J-R, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 2006;281:8907–8916. doi: 10.1074/jbc.M512987200. [DOI] [PubMed] [Google Scholar]
- 32.Kaysser L, Lutsch L, Siebenberg S, Wemakor E, Kammerer B, Gust B. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem. 2009;284:14987–14996. doi: 10.1074/jbc.M901258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engel LS. The dilemma of multidrug-resistant gram-negative bacteria. Am J Med Sci. 2010;340:232–237. doi: 10.1097/MAJ.0b013e3181e939c3. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, White-Phillip JA, Melançon CE, 3rd, Yu Wl, Kwon H-j, Liu Hw. Elucidation of the kijanimicin gene cluster: insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J Am Chem Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Al-Mestarihi A, Grimes CL, Kahne D, Bachmann BO. A unifying nitrososynthase involved in nitrosugar biosynthesis. J Am Chem Soc. 2008;130:15756–15757. doi: 10.1021/ja8051415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson HD, Thorson JS. Characterization of CalE10, the N-oxidase involved in calicheamicin hydroxyaminosugar formation. J Am Chem Soc. 2008;130:17662–17663. doi: 10.1021/ja807557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruender NA, Thoden JB, Holden HM. X-ray structure of KijD3, a key enzyme involved in the biosynthesis of D-kijanose. Biochemistry. 2010;49:3517–3524. doi: 10.1021/bi100318v. [DOI] [PubMed] [Google Scholar]
- 38.Timmons S, Thorson JS. Increasing carbohydrate diversity via amine oxidation: aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr Opin Chem Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nepal KK, Yoo JC, Sohng JK. Biosynthetic approach for the production of new aminoglycoside derivative. J Biosci Bioeng. 2010;110:109–112. doi: 10.1016/j.jbiosc.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 41.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 42.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 43.Kirst HA. The spinosyn family of insecticides: realizing the potential of natural products research. J Antibiot (Tokyo) 2010;63:101–111. doi: 10.1038/ja.2010.5. [DOI] [PubMed] [Google Scholar]
- 44.Borisova SA, Zhao L, Sherman DH, Liu H-w. Biosynthesis of desosamine: construction of a new macrolide carrying a genetically designed sugar moiety. Org Lett. 1999;1:133–136. doi: 10.1021/ol9906007. [DOI] [PubMed] [Google Scholar]
- 45.Ruszczycky MW, Choi SH, Liu H-w. Stoichiometry of the redox neutral deamination and oxidative dehydrogenation reactions catalyzed by the radical SAM enzyme DesII. J Am Chem Soc. 2010;132:2359–69. doi: 10.1021/ja909451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melançon CE, 3rd, Yu WL, Liu H-w. TDP-mycaminose biosynthetic pathway revised and conversion of desosamine pathway to mycaminose pathway with one gene. J Am Chem Soc. 2005;127:12240–12241. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]
- 47.Melançon CE, 3rd, Hong L, White JA, Takahashi H, Liu Y-n, Liu H-w. Characterization of TDP-4-keto-6-deoxy-D-glucose-3,4-isomerase (Tyl1a) from the TDP- D-mycaminose pathway of Streptomyces fradiae. Biochemistry. 2007;46:577–590. doi: 10.1021/bi061907y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melançon CE, 3rd, Liu H-w. Engineered biosynthesis of macrolide antibiotics bearing the non-natural deoxysugars 4-epi-D-mycaminose and 3-N-monomethylamino-3-deoxy-D-fucose. J Am Chem Soc. 2007;129:4896–4897. doi: 10.1021/ja068254t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong L, Zhao Z, Melançon CE, 3rd, Zhang H, Liu H-w. In vivo characterization of the enzymes involved in TDP-D-forosamine biosynthesis in the spinosyn pathway of Saccharopolyspora spinosa. J Am Chem Soc. 2008;130:4954–4967. doi: 10.1021/ja0771383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook PD, Holden HM. GDP-perosamine synthase: structural analysis and production of a novel trideoxysugar. Biochemistry. 2008;47:2833–2840. doi: 10.1021/bi702430d. [DOI] [PubMed] [Google Scholar]
- 51.Thoden JB, Schffer C, Messner P, Holden HM. Structural analysis of QdtB, an aminotransferase required for the biosynthesis of dTDP-3-acetamido-3,6-dideoxy-α-D-glucose. Biochemistry. 2009;48:1553–1561. doi: 10.1021/bi8022015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burgie ES, Holden HM. Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine. Biochemistry. 2007;46:8999–9006. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgie ES, Thoden JB, Holden HM. Molecular architecture of DesV from Streptomyces venezuelae: a PLP-dependent transaminase involved in the biosynthesis of the unusual sugar desosamine. Protein Sci. 2007;16:887–896. doi: 10.1110/ps.062711007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson J-R, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 55.Holden HM, Cook PD, Thoden JB. Biosynthetic enzymes of unusual microbial sugars. Curr Opin Struct Biol. 2010:543–550. doi: 10.1016/j.sbi.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang B-Y, Lee H-J, Yang Y-H, Joo H-S, Kim B-G. Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem Biol. 2004;11:915–925. doi: 10.1016/j.chembiol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 57.Cook PD, Carney AE, Holden HM. Accommodation of GDP-linked sugars in the active site of GDP-perosamine synthase. Biochemistry. 2008;47:10685–10693. doi: 10.1021/bi801309q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao LNL, Que S, Xue Y, Sherman DH, Liu H-w. Biosynthesis of desosamine: probing the order of C-4 deoxygenation versus C-3 transamination. J Am Chem Soc. 1998;120:12159–12160. [Google Scholar]
- 59.He X, Liu H-w. Mechanisms of enzymatic C-O bond cleavages in deoxyhexose biosynthesis. Cur Opin Chem Biol. 2002;6:590–597. doi: 10.1016/s1367-5931(02)00367-8. [DOI] [PubMed] [Google Scholar]
- 60.Johnson DA, Liu Hw. A mechanistic analysis of C-O bond cleavage events with a comparison to 3,6-dideoxysugar formation. In: Cooper R, Snyder JD, editors. The Biology-Chemistry Interface: A Tribute to Koji Nakanishi. Marcel Dekker; New York: 1999. pp. 351–396. [Google Scholar]
- 61.Samuel G, Reeves P. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res. 2003;338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Chen H, Agnihotri G, Guo ZNL, Que S, Chen XH, Liu H-w. Biosynthesis of mycarose: isolation and characterization of enzymes involved in the C-2 deoxygenation. J Am Chem Soc. 1999;121:8124–8125. [Google Scholar]
- 63.Zhao L, Borisova SA, Yeung S-M, Liu H-w. Study of C-4 deoxygenation in the biosynthesis of desosamine: evidence implicating a novel mechanism. J Am Chem Soc. 2001;123:7909–7910. doi: 10.1021/ja010587x. [DOI] [PubMed] [Google Scholar]
- 64.Szu P-h, He X, Zhao L, Liu H-w. Biosynthesis of TDP-D-desosamine: elucidation of a new strategy for C-4 deoxygenation. Angew Chem Int Ed. 2005;44:6742–6746. doi: 10.1002/anie.200501998. [DOI] [PubMed] [Google Scholar]
- 65.Szu P-h, Ruszczucky M, Choi SH, Yan F, Liu H-w. Characterization and mechanistic studies of DesII: a radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine. J Am Chem Soc. 2009;131:14030–14042. doi: 10.1021/ja903354k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu H-w. Coenzyme B6 dependent novel bond cleavage reactions. Pure Appl Chem. 1998;70:9–16. [Google Scholar]
- 67.Agnihotri G, Liu H-w. Radicals in coenzyme B6 dependent enzyme catalysis. Bioorg Chem. 2001;29:234–257. doi: 10.1006/bioo.2001.1211. [DOI] [PubMed] [Google Scholar]
- 68.Power PM, Jennings MP. The genetics of glycosylation in Gram-negative bacteria. FEMS Microbiol Lett. 2003;218:211–222. doi: 10.1111/j.1574-6968.2003.tb11520.x. [DOI] [PubMed] [Google Scholar]
- 69.Weigel TM, Liu L-d, Liu H-w. Mechanistic studies of the biosynthesis of 3,6-dideoxyhexoses in Yersinia pseudotuberculosis: purification and characterization of CDP- 4-keto-6-deoxy-D-glucose-3-dehydrase. Biochemistry. 1992;31:2129–2139. doi: 10.1021/bi00122a034. [DOI] [PubMed] [Google Scholar]
- 70.Rubenstein PA, Strominger JL. Enzymatic synthesis of cytidine diphosphate 3,6-dideoxyhexoses: VIII. Studies of the properties of E3 and its role in the formation of cytidine diphosphate-4-keto-3,6-dideoxyglucose. J Biol Chem. 1974;249:3782–3788. [PubMed] [Google Scholar]
- 71.Thorson JS, Liu H-w. Characterization of the first PMP-dependent iron-sulfur containing enzyme which is essential for the biosynthesis of 3,6-dideoxyhexoses. J Am Chem Soc. 1993;115:7539–7540. [Google Scholar]
- 72.Thorson JS, Liu H-w. Coenzyme B6 as a redox cofactor — a new role for an old coenzyme? J Am Chem Soc. 1993;115:12177–12178. [Google Scholar]
- 73.Lei Y, Ploux O, Liu H-w. Mechanistic studies of CDP-6-deoxy-D-glycero-L-threo-4-hexulose-3-dehydrase: identification of His220 as the active-site base by chemical modification and site-directed mutagenesis. Biochemistry. 1995;34:4643–4654. doi: 10.1021/bi00014a018. [DOI] [PubMed] [Google Scholar]
- 74.Agnihotri G, Liu Y, Paschal B, Liu H-w. Identification of an unusual [2Fe-2S]-binding motif in the dehydrase responsible for C-3 deoxygenation in the biosynthesis of 3,6-dideoxyhexoses. Biochemistry. 2004;43:14265–14274. doi: 10.1021/bi048841w. [DOI] [PubMed] [Google Scholar]
- 75.Storici P, De Biase D, Bossa F, Bruno S, Mozzarelli A, Peneff C, Silverman RB, Schirmer T. Structures of γ-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5′-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with γ-ethynyl-GABA and with the antiepilepsy drug vigabatrin. J Biol Chem. 2004;279:363–373. doi: 10.1074/jbc.M305884200. [DOI] [PubMed] [Google Scholar]
- 76.Pieper PA, Guo Z, Liu H-w. Mechanistic studies of 3,6-dideoxysugar biosynthesis: stereochemical course of C-3 deoxygenation. J Am Chem Soc. 1995;117:5158–5159. [Google Scholar]
- 77.Han O, Miller VP, Liu H-w. Mechanistic studies of the biosynthesis of 3,6-dideoxy-hexoses in Yersinia pseudotuberculosis: purification and characterization of CDP-6-deoxy-Δ3,4-glucoseen reductase based on its NADH:dichlorophenolindolphenol oxidoreductase activity. J Biol Chem. 1990;265:8033–8041. [PubMed] [Google Scholar]
- 78.Miller VP, Thorson JS, Lo SF, Ploux O, Liu H-w. Cofactor characterization and mechanistic studies of CDP-6-deoxy-Δ3,4-glucoseen reductase: exploration into a novel enzymatic C-O bond cleavage event. Biochemistry. 1993;32:11934–11942. doi: 10.1021/bi00095a025. [DOI] [PubMed] [Google Scholar]
- 79.Lo SF, Miller VP, Lei Y, Thorson JS, Liu H-w, Schottel JL. CDP-6-deoxy-Δ3,4-glucoseen reductase from Yersinia pseudotuberculosis: enzyme purification and characterization of the cloned gene. J Bacteriol. 1994;176:460–468. doi: 10.1128/jb.176.2.460-468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ploux O, Lei Y, Vatanen K, Liu H-w. Mechanistic studies on CDP-6-deoxy-Δ3,4-glucoseen reductase: the role of cysteine residues in catalysis as probed by chemical modification and site directed mutagenesis. Biochemistry. 1995;34:4159–4168. doi: 10.1021/bi00013a003. [DOI] [PubMed] [Google Scholar]
- 81.He X, Ploux O, Liu H-w. Biosynthesis of 3,6-dideoxyhexoses: in vivo and in vitro evidence for protein-protein interaction between CDP-6-deoxy-L-threo-D-glycero-4-hexulose-3-dehydrase (E1) and its reductase (E3) Biochemistry. 1996;35:16412–16420. doi: 10.1021/bi961921i. [DOI] [PubMed] [Google Scholar]
- 82.Burns KD, Pieper PA, Liu H-w, Stankovich MT. Characterization of the spectral and thermodynamic properties of CDP-6-deoxy-L-threo-D-glycero-4-hexulose-3-dehydrase reductase (E3) and CDP-6-deoxy-L-threo-D-glycero-4-hexulose-3-dehydrase (E1) Biochemistry. 1996;35:7879–7889. doi: 10.1021/bi960284t. [DOI] [PubMed] [Google Scholar]
- 83.Johnson DA, Gassner GT, Bandarian V, Ruzicka F, Ballou DP, Reed GH, Liu H-w. Kinetic characterization of an organic radical in the ascarylose biosynthetic pathway. Biochemistry. 1996;35:15846–15857. doi: 10.1021/bi961370w. [DOI] [PubMed] [Google Scholar]
- 84.Chang C-WT, Johnson DA, Bandarian V, Zhou H, LoBrutto R, Reed GH, Liu H-w. Characterization of a unique coenzyme B6 radical in the ascarylose biosynthetic pathway. J Am Chem Soc. 2000;122:4239–4240. [Google Scholar]
- 85.Frey PA. Lysine 2,3-aminomutase: is adenosylmethionine a poor man’s adenosylcobalamine? FASEB J. 1993;7:662–670. doi: 10.1096/fasebj.7.8.8500691. [DOI] [PubMed] [Google Scholar]
- 86.Lieder KW, Booker S, Ruzicka FJ, Beinert H, Reed GH, Frey PA. S-adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance. Biochemistry. 1998;37:2578–2585. doi: 10.1021/bi972417w. [DOI] [PubMed] [Google Scholar]
- 87.Lepore BW, Ruzicka FJ, Frey PA, Ringe D. The x-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. Proc Natl Acad Sci USA. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang SC, Frey PA. Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme. Biochemistry. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith P, Szu P-h, Bui C, Liu H-w, Tsai S-C. 2.4 Å structure and mutagenic retroevolution of the E1 dehydrase: at the cross road of dehydration, aminotransferation and racemization. Biochemistry. 2008;47:6329–6341. doi: 10.1021/bi702449p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cook PD, Thoden JB, Holden HM. The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme. Protein Sci. 2006;15:2093–2106. doi: 10.1110/ps.062328306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cook PD, Holden HM. A structural study of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: caught in the act of geminal diamine formation. Biochemistry. 2007;46:14215–14224. doi: 10.1021/bi701686s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cook PD, Holden HM. GDP-4-keto-6-deoxy-D-mannose 3-dehydratase, accommodating a sugar substrate in the active site. J Biol Chem. 2008;283:4295–4303. doi: 10.1074/jbc.M708893200. [DOI] [PubMed] [Google Scholar]
- 93.Popovic B, Tang X, Chirgadze DY, Huang F, Blundell TL, Spencer JB. Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis. Proteins: Struc Func Bioinfo. 2006;65:220–230. doi: 10.1002/prot.21076. [DOI] [PubMed] [Google Scholar]
- 94.Wu Q, Liu Y-n, Chen H, Molitor EJ, Liu H-w. A retro-evolution study of CDP-6-deoxy-D-glycero-L-threo-4-hexulose-3-dehydrase (E1): implications for C-3 deoxygenation in the biosynthesis of 3,6-dideoxyhexoses. Biochemistry. 2007;46:3759–3767. doi: 10.1021/bi602352g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alam J, Beyer N, Liu H-w. Biosynthesis of colitose: expression, purification, and mechanistic characterization of GDP-4-keto-6-deoxy-D-mannose-3-dehydrase (ColD) and GDP-L-colitose synthase (ColC) Biochemistry. 2004;43:16450–16460. doi: 10.1021/bi0483763. [DOI] [PubMed] [Google Scholar]
- 96.Kim HJ, White-Phillip JA, Ogasawara Y, Shin N, Isiorho EA, Liu H-w. The biosynthesis of spinosyn in Saccharopolyspora spinosa: synthesis of the methylated rhamnose and characterization of the functions of SpnH, SpnI, and SpnK. J Am Chem Soc. 2010;132:2901–2903. doi: 10.1021/ja910223x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hong L, Zhao Z, Liu Hw. Characterization of SpnQ from the spinosyn biosynthetic pathway of Saccharopolyspora spinosa: mechanistic and evolutionary implications for C-3 deoxygenation in deoxysugar biosynthesis. J Am Chem Soc. 2006;128:14262–14263. doi: 10.1021/ja0649670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen H, Guo Z, Liu H-w. The enzyme catalyzing the branched-chain coupling in the biosynthesis of yersiniose is a thiamine 5′-phosphate dependent flavoprotein. J Am Chem Soc. 1998;120:11796–11797. [Google Scholar]
- 99.Chen H, Zhao Z, Hallis TM, Guo Z, Liu H-w. Insights into the branched-chain formation of mycarose: methylation catalyzed by an S-adenosylmethionine-dependent methyltransferase. Angew Chem Int Ed. 2001;40:607–610. doi: 10.1002/1521-3773(20010202)40:3<607::AID-ANIE607>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 100.Takahashi H, Liu Y-n, Chen H, Liu H-w. Biosynthesis of TDP-mycarose: the specificity of a single enzyme governs the outcome of the pathway. J Am Chem Soc. 2005;127:9340–9341. doi: 10.1021/ja051409x. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi H, Liu Y-n, Liu H-w. A two-stage one-pot enzymatic synthesis of TDP-L-mycarose from thymidine and glucose-1-phosphate. J Am Chem Soc. 2006;128:1432–1433. doi: 10.1021/ja0562144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams N, Wander J. Deoxy and branched-chain sugars. In: Pigman W, Horton D, editors. The Carbohydrates: Chemistry and Biochemistry. 1B. Academic Press; New York: 1980. pp. 761–798. [Google Scholar]
- 103.Lehwald P, Richter M, RÖhr C, Liu H-w, Müller M. Asymmetirc intermolecular crossed aldehyde-ketone condensation through enzymatic carboligation reaction. Angew Chem Int Ed. 2010;49:2389–2392. doi: 10.1002/anie.200906181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaysser L, Lutsch L, Siebenberg S, Wemakor E, Kammerer B, Gust B. Identification and manipulation of the caprazamycin gene cluster lead to new simplified liponucleoside antibiotics and give insights into the biosynthetic pathway. J Biol Chem. 2009;284:14987–14996. doi: 10.1074/jbc.M901258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaysser L, Siebenberg S, Kammerer B, Gust B. Analysis of the liposidomycin gene cluster leads to the identification of new caprazamycin derivatives. Chembiochem. 2010;11:191–196. doi: 10.1002/cbic.200900637. [DOI] [PubMed] [Google Scholar]
- 106.Schirch VDM, Szebenyi E. Serine hydroxymethyltransferase revisited. Curr Opin Chem Biol. 2005;9:482–487. doi: 10.1016/j.cbpa.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 107.Florio R, Luigi di Salvo M, Vivoli M, Contestabile R. Serine hydroxymethyltransferase: A model enzyme for mechanistic, structural and evolutionary studies. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbapap.2010.10.010. online. [DOI] [PubMed] [Google Scholar]
- 108.Makart S, Bechtold M, Panke S. Towards preparative asymmetric synthesis of β-hydroxy-α-amino acids:l-allo-threonine formation from glycine and acetaldehyde using recombinant GlyA. J Biotechnol. 2007;130:402–410. doi: 10.1016/j.jbiotec.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 109.Takayama S, McGarvey GJ, Wong CH. Microbial aldolases and transketolases: new biocatalytic approaches to simple and complex sugars. Annu Rev Microbiol. 1997;51:285–310. doi: 10.1146/annurev.micro.51.1.285. [DOI] [PubMed] [Google Scholar]