Abstract
Phenylalanine hydroxylase (PheH) is a iron(II)-dependent enzyme that catalyzes the hydroxylation of aromatic amino acid l-phenylalanine (l-Phe) to l-tyrosine (l-Tyr). The enzymatic modification has been demonstrated to be highly regiospecific, forming proteinogenic para-Tyr (p-Tyr) exclusively. Here we biochemically characterized the first example of a phenylalanine-3-hydroxylase (Phe3H) which catalyzes the synthesis of meta-Tyr (m-Tyr) from Phe. Subsequent mutagenesis studies revealed that two residues in the active site of Phe3H (Cys187 and Thr202) contribute to C-3 rather than C-4 hydroxylation of the phenyl ring. This work sets a stage for mechanistic and structural study of regiospecific control of the substrate hydroxylation by PheH.
Phenylalanine hydroxylase (PheH) is a member of the highly homologous family of aromatic amino acid hydroxylases which also include tyrosine hydroxylase (TyrH) and tryptophan hydroxylase (1, 2). They are iron(II)-dependent enzymes that typically utilize molecular oxygen and tetrahydrobiopterin (BH4) as cosubstrates. In accord with their essential functions in human aromatic amino acid metabolism, this family of enzymes has been studied extensively (3). All PheH enzymes characterized to date are phenylalanine-4-hydroxylases (Phe4Hs) yielding the 4-hydroxyphenylalanine regioisomer (p-Tyr) as the product. TyrH carries out a hydroxylation of p-Tyr to yield l-DOPA; it also slowly generates p-Tyr and m-Tyr (ratio of 25:1) when presented with the alternate substrate l-Phe (4). Despite several available crystal structures of PheH (5–9), the mechanism of regiospecific hydroxylation of Phe to form p-Tyr is poorly understood. No enzyme has been characterized to be dedicated in the catalysis of m-Tyr formation.
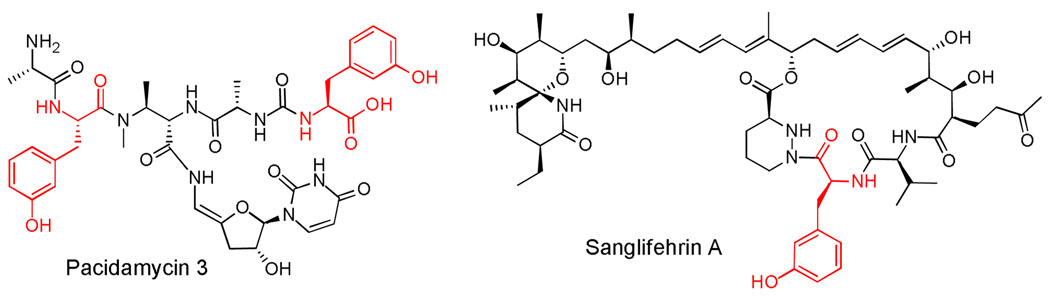
m-Tyr has been found in humans which could be mis-incorporated into proteins through phenylalanine-tRNA synthetase (10–12). It is presumably formed from phenylalanine by hydroxyl radicals. In addition, m-Tyr has been identified to be a natural herbicidal compound produced by fine fescue grasses to displace neighboring plants, thus it might be potentially useful in biorational weed control (13). Several bacterial secondary metabolites also contain m-Tyr as one of the building blocks, including uridyl peptide antibiotics such as pacidamycins and a potent cyclophilin inhibitor sanglifehrin A (SFA) (Figure 1) (14, 15). Our recent characterization of the biosynthetic gene cluster for pacidamycins indicated that m-Tyr could be formed from Phe, through the function of a phenylalanine hydroxylase homolog PacX (16, 17). A close protein homolog SfaA (49% identity to PacX) has recently been identified in the SFA biosynthetic gene cluster, and inactivation of SfaA abolished SFA production which could be restored by feeding of m-Tyr (15). This finding strongly indicated that these phenylalanine hydroxylases could act on Phe to introduce a hydroxyl moiety at the meta-position. In parallel with the sfaA knockout studies and to probe the regiospecific control mechanism of PheH, we have purified and characterized PacX from the pacidamycin producer Streptomyces coeruleoribudus.
Figure 1.
Examples of m-Tyr (highlighted in red) containing bacterial secondary metabolites.
The catalytic core of PacX more closely resembles that of the human Phe4H (31% identity) than that of the bacterial Phe4H from Chromobacterium violaceum (23% identity), yet PacX lacks the N-terminal regulatory domain and the C-terminal tetramerization domain found in human Phe4H (18, 19) (Figure S1). PacX was cloned as an N-terminal 6XHis-tagged protein, expressed and purified from E. coli with a yield of 6.7 mg/L (Figure S2). To directly visualize the formation of Tyr, an HPLC-based assay was performed using both p-Tyr and m-Tyr as standards. The purified enzyme was mixed with l-Phe and 6-methyltetrahydropterin (6-MePH4) in sodium phosphate buffer, and the reaction was incubated at room temperature, quenched by 10% trichloroacetic acid (TCA) and subjected to HPLC analysis (Figure 2). m-Tyr was detected as the product, confirmed by UV profile, HRMS and compared to the standard (Figure S3). Only trace amounts of p-Tyr were formed from the reaction mixture (<3% compared to m-Tyr). Control experiments demonstrated that both substrates l-Phe and 6-MePH4 were essential for m-Tyr formation. In addition to l-Phe, the substrate specificity of PacX towards d-Phe, l-Tyr and l-Trp was tested: PacX was not able to hydroxylate any of these substrates. The addition of ferrous ammonium sulfate to the reaction mixture increased the product yield more than three-fold, strongly indicating that PacX is a Fe-dependent phenylalanine hydroxylase. DTT was also found to increase the product yield more than 72%, consistent with a previous report (20). The pH effect on PacX was tested using phosphate buffer of pH values ranging from 5.8 to 7.8 (Figure S4). The activity of PacX reached maximum at pH 6.0, and the enzyme tended to precipitate gradually below pH 6.5. We thus used a pH value of 6.2 throughout the studies.
Figure 2.
Characterization of PacX as a phenylalanine-3-hydroxylase. a) Schematic of PheH catalyzed reactions. b) Fluorescence chromatograms (λex 270 nm and λem 310 nm) showing PacX catalyzed production of m-Tyr with various control reactions omitting one of the components indicated. c) Determination of PacX kinetic parameters by HPLC assays monitoring the formation of m-Tyr. Parameters were determined for l-Phe (top panel) by fixing the 6-MePH4 concentration at 0.5 mM, while parameters were determined for 6-MePH4 (bottom panel) by fixing the l-Phe concentration at 5 mM.
The kinetic parameters of PacX were determined using HPLC assays monitoring the formation of m-Tyr (Figure 2c). The Km values of the enzyme were determined to be 26 µM for 6-MePH4 and 1.1 mM for l-Phe, which were comparable to reported values for Phe4H originating from different organisms (9, 20–22). The turnover number was determined to be ~1.3 min−1, which was more than 100-fold less than those of typical Phe4H. This low kcat value could reflect a low fraction of properly folded enzyme from heterologous expression, or more likely, it could be representative of low kcat values seen with other enzymes in secondary metabolic pathways. In particular it may be one way of reducing the flux of the key metabolite l-Phe to nonproteinogenic m-Tyr rather than proteinogenic p-Tyr.
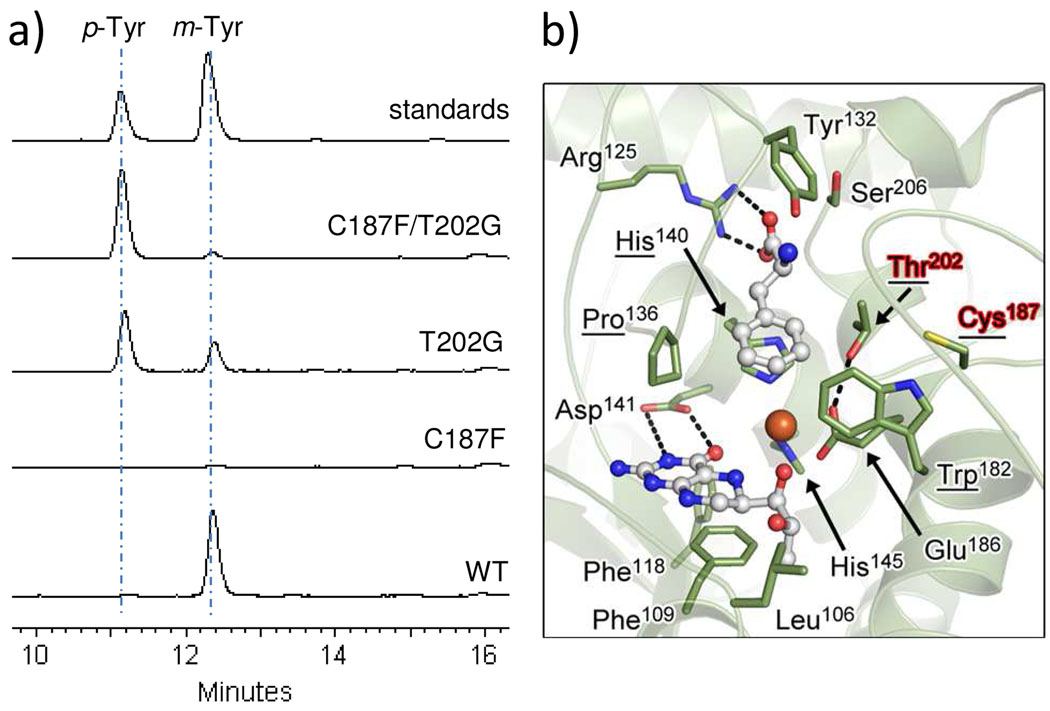
The enzymatic control of regiospecific hydroxylation of l-Phe was probed by site-directed mutagenesis. The residues predicted to make up the hydrophobic cage surrounding the aromatic side chain of l-Phe in PacX are Trp182, Cys187, Pro136, His140, and Thr202 (Figure 3), which are conserved among putative Phe3Hs with the minor exception that Thr202 is Ser in SfaA. Compared to Phe4H and TyrH, three out of these five residues are highly conserved, while Cys187 and Thr202 are replaced by Phe and Gly respectively (Figure S1 and S5). Thus we constructed the single C187F, T202G and the double C187F/T202G PacX mutants to examine their effects on the activity of PacX in forming m-Tyr versus p-Tyr. All three mutants were purified from E. coli with decreased yields of 0.4 mg/L, 0.5 mg/L and 3.9 mg/L respectively (Figure S2). The overall hydroxylation rate was comparable for the T202G single mutant, the C187F/T202G double mutant and the wild-type enzyme, while it was reduced to <4% of the rate of wild-type for the C187F single mutant (Figure 3a). We suspect that the inactivity is due to the steric effects of the bulky side chain of Phe187 in the presence of Thr202, leading to possible rearrangement of the active site architecture. The ratio of product p-Tyr to m-Tyr was changed to ~2:1 for the T202G mutant, which represents a 60-fold increase in the preference for p-Tyr over m-Tyr formation. A more dramatic change on the product profile was found in the C187F/T202G double mutant reaction, where only < 8% of m-Tyr compared to p-Tyr was formed (Figure 3a). These results demonstrated that both residues Cys187 and Thr202 are critical in controlling the regiospecific hydroxylation of the phenyl ring. It is likely that the two residues work in concert to clear possible hindrance at meta-position and to promote hydroxylation by hydrogen-bonding. Elucidation of the exact mechanism will require future structural studies of Phe/m-Tyr bound Phe3H.
Figure 3.
Mutagenesis studies on the regiospecific hydroxylation control of PacX. a) Fluorescence chromatograms showing production of m-Tyr and p-Tyr catalyzed by wild-type PacX and mutants. b) Three-dimensional structural model of PacX active site (Phe3H). The structure of PacX was modeled based on Phe4H (PDB 1KW0) (23) using the online program SWISS-MODEL. The five residues that form the hydrophobic cage around the aromatic side chain of l-Phe are underlined.
In summary, we have characterized in vitro for the first time a phenylalanine-3-hydroxylase which catalyzes the synthesis of m-Tyr from l-Phe. The reaction presumably proceeds through a consensus PheH mechanism in which a high-spin Fe(IV)=O species is the immediate donor of oxygen to the substrate phenylalanine bound in the active site. Subsequent electrophilic aromatic substitution generates a cationic diene species that undergoes a low energy hydride shift forming the cyclohexadienone, followed by aromatization to yield the product phenol (2, 24, 25) (Figure S6). We furthermore identified a role for residues Cys187 and Thr202 in determining the regiospecific hydroxylation of l-Phe substrate. Phe3H is presumably evolved from ancient Phe4H, with first mutation of Phe to Cys which would have been a one-base event. This is also indicated by the inactivity of the C187F mutant of PacX. The second mutation of Gly to Thr/Ser further increased the selectivity for the formation of m-Tyr over p-Tyr. It would be interesting to test if Phe4H could be converted to Phe3H by mutating the corresponding residues.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH Grant GM49338 (C.T.W.) and NIH Grant F32GM090475 (B.D.A).
Footnotes
Supporting Information Available: Experimental procedures, sequence alignment of PheHs and TyrHs, SDS-PAGE analysis of purified proteins, UV and HRMS characterization of m-Tyr, pH effect on PacX activity, modeled three-dimensional structure of PacX and the comparison of the active site of PacX to Phe4H, proposed reaction mechanism of PacX. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Hufton SE, Jennings IG, Cotton RG. Structure and function of the aromatic amino acid hydroxylases. Biochem. J. 1995;311(Pt 2):353–366. doi: 10.1042/bj3110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzpatrick PF. Mechanism of aromatic amino acid hydroxylation. Biochemistry. 2003;42:14083–14091. doi: 10.1021/bi035656u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flatmark T, Stevens RC. Structural insight into the aromatic amino acid hydroxylases and their disease-related mutant forms. Chem. Rev. 1999;99:2137–2160. doi: 10.1021/cr980450y. [DOI] [PubMed] [Google Scholar]
- 4.Fitzpatrick PF. Kinetic isotope effects on hydroxylation of ring-deuterated phenylalanines by tyrosine hydroxylase provide evidence against partitioning of an arene oxide intermediate. J. Am. Chem. Soc. 1994;116:1133–1134. [Google Scholar]
- 5.Erlandsen H, Bjorgo E, Flatmark T, Stevens RC. Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase. Biochemistry. 2000;39:2208–2217. doi: 10.1021/bi992531+. [DOI] [PubMed] [Google Scholar]
- 6.Andersen OA, Flatmark T, Hough E. High resolution crystal structures of the catalytic domain of human phenylalanine hydroxylase in its catalytically active Fe(II) form and binary complex with tetrahydrobiopterin. J. Mol. Biol. 2001;314:279–291. doi: 10.1006/jmbi.2001.5061. [DOI] [PubMed] [Google Scholar]
- 7.Erlandsen H, Kim JY, Patch MG, Han A, Volner A, Abu-Omar MM, Stevens RC. Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates. J. Mol. Biol. 2002;320:645–661. doi: 10.1016/s0022-2836(02)00496-5. [DOI] [PubMed] [Google Scholar]
- 8.Andersen OA, Stokka AJ, Flatmark T, Hough E. 2.0A resolution crystal structures of the ternary complexes of human phenylalanine hydroxylase catalytic domain with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine or L-norleucine: substrate specificity and molecular motions related to substrate binding. J. Mol. Biol. 2003;333:747–757. doi: 10.1016/j.jmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Leiros HK, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J. Biol. Chem. 2007;282:21973–21986. doi: 10.1074/jbc.M610174200. [DOI] [PubMed] [Google Scholar]
- 10.Nair UJ, Nair J, Friesen MD, Bartsch H, Ohshima H. Ortho- and meta-tyrosine formation from phenylalanine in human saliva as a marker of hydroxyl radical generation during betel quid chewing. Carcinogenesis. 1995;16:1195–1198. doi: 10.1093/carcin/16.5.1195. [DOI] [PubMed] [Google Scholar]
- 11.Gurer-Orhan H, Ercal N, Mare S, Pennathur S, Orhan H, Heinecke JW. Misincorporation of free m-tyrosine into cellular proteins: a potential cytotoxic mechanism for oxidized amino acids. Biochem. J. 2006;395:277–284. doi: 10.1042/BJ20051964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klipcan L, Moor N, Kessler N, Safro MG. Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc. Natl. Acad. Sci. U S A. 2009;106:11045–11048. doi: 10.1073/pnas.0905212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin C, Weston LA, Huang T, Jander G, Owens T, Meinwald J, Schroeder FC. Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. U S A. 2007;104:16964–16969. doi: 10.1073/pnas.0707198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winn M, Goss RJ, Kimura K, Bugg TD. Antimicrobial nucleoside antibiotics targeting cell wall assembly: recent advances in structure-function studies and nucleoside biosynthesis. Nat. Prod. Rep. 2010;27:279–304. doi: 10.1039/b816215h. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, Jiang N, Xu F, Shao L, Tang G, Wilkinson B, Liu W. Cloning, sequencing and characterization of the biosynthetic gene cluster of sanglifehrin A, a potent cyclophilin inhibitor. Mol. Biosyst. 2011;7:852–861. doi: 10.1039/c0mb00234h. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, Ostash B, Walsh CT. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc. Natl. Acad. Sci. U S A. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Ntai I, Bolla ML, Malcolmson SJ, Kahne D, Kelleher NL, Walsh CT. Nine enzymes are required for assembly of the pacidamycin group of peptidyl nucleoside antibiotics. J. Am. Chem. Soc. 2011;133:5240–5243. doi: 10.1021/ja2011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobe B, Jennings IG, House CM, Michell BJ, Goodwill KE, Santarsiero BD, Stevens RC, Cotton RG, Kemp BE. Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 19.Fusetti F, Erlandsen H, Flatmark T, Stevens RC. Structure of tetrameric human phenylalanine hydroxylase and its implications for phenylketonuria. J. Biol. Chem. 1998;273:16962–16967. doi: 10.1074/jbc.273.27.16962. [DOI] [PubMed] [Google Scholar]
- 20.Ledley FD, Grenett HE, Woo SL. Biochemical characterization of recombinant human phenylalanine hydroxylase produced in Escherichia coli. J. Biol. Chem. 1987;262:2228–2233. [PubMed] [Google Scholar]
- 21.Onishi A, Liotta LJ, Benkovic SJ. Cloning and expression of Chromobacterium violaceum phenylalanine hydroxylase in Escherichia coli and comparison of amino acid sequence with mammalian aromatic amino acid hydroxylases. J. Biol. Chem. 1991;266:18454–18459. [PubMed] [Google Scholar]
- 22.Daubner SC, Melendez J, Fitzpatrick PF. Reversing the substrate specificities of phenylalanine and tyrosine hydroxylase: aspartate 425 of tyrosine hydroxylase is essential for L-DOPA formation. Biochemistry. 2000;39:9652–9661. doi: 10.1021/bi000493k. [DOI] [PubMed] [Google Scholar]
- 23.Andersen OA, Flatmark T, Hough E. Crystal structure of the ternary complex of the catalytic domain of human phenylalanine hydroxylase with tetrahydrobiopterin and 3-(2-thienyl)-L-alanine, and its implications for the mechanism of catalysis and substrate activation. J. Mol. Biol. 2002;320:1095–1108. doi: 10.1016/s0022-2836(02)00560-0. [DOI] [PubMed] [Google Scholar]
- 24.Pavon JA, Fitzpatrick PF. Insights into the catalytic mechanisms of phenylalanine and tryptophan hydroxylase from kinetic isotope effects on aromatic hydroxylation. Biochemistry. 2006;45:11030–11037. doi: 10.1021/bi0607554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panay AJ, Lee M, Krebs C, Bollinger JM, Fitzpatrick PF. Evidence for a high-spin Fe(IV) species in the catalytic cycle of a bacterial phenylalanine hydroxylase. Biochemistry. 2011;50:1928–1933. doi: 10.1021/bi1019868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.