SUMMARY
Drosophila Dicer-2 generates small interfering RNAs (siRNAs) from long double-stranded RNA (dsRNA), whereas Dicer-1 produces microRNAs from pre-microRNA. What makes the two Dicers specific for their biological substrates? We find that purified Dicer-2 can efficiently cleave pre-miRNA, but that inorganic phosphate and the Dicer-2 partner protein R2D2 inhibit pre-miRNA cleavage. Dicer-2 contains C-terminal RNase III domains that mediate RNA cleavage, and an N-terminal helicase motif whose function is unclear. We show that Dicer-2 is a dsRNA-stimulated ATPase that hydrolyzes ATP to ADP; ATP hydrolysis is required for Dicer-2 to process long dsRNA, but not pre-miRNA. Wild-type Dicer-2, but not a mutant defective in ATP hydrolysis, can generate siRNAs faster than it can dissociate from a long dsRNA substrate. We propose that the Dicer-2 helicase domain uses ATP to generate many siRNAs from a single molecule of dsRNA before dissociating from its substrate.
INTRODUCTION
In Drosophila melanogaster, distinct pathways produce 21 nt small interfering RNAs (siRNAs) and ~22 nt microRNAs (miRNAs). The RNase III enzyme Drosha, aided by its partner protein, Pasha, cleaves primary miRNAs to release pre-miRNAs, ~70 nt long stem-loop structures that contain a mature miRNA within their stems (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Han et al., 2006). The pre-miRNA is then cleaved by Dicer-1, acting with its dsRNA-binding domain (dsRBD) protein partner, Loquacious-PB (Loqs-PB), to liberate a duplex comprising the mature miRNA bound to its miRNA*, a partially complementary small RNA derived from the opposite arm of the pre-miRNA stem (Förstemann et al., 2005; Jiang et al., 2005; Saito et al., 2005; Ye et al., 2007). Mature miRNAs can derive from either the 5′ or 32 arm of the pre-miRNA stem.
In contrast to miRNAs, Drosophila siRNAs are generated by Dicer-2 (Lee et al., 2004), which forms a stable complex with the dsRNA-binding protein R2D2 (Liu et al., 2003). In vitro, Dicer-2 can produce siRNAs in the absence of R2D2, but both Dicer-2 and R2D2 are required to load siRNAs into Ago2 (Liu et al., 2003; Tomari et al., 2004; Pham and Sontheimer, 2005; Liu et al., 2006; Tomari et al., 2007). Exogenous siRNAs derive from long dsRNA molecules that are generated experimentally, from viral RNA genomes or intermediates of replication, whereas endo-siRNAs derive from convergent transcription of mRNAs or from RNA from mobile genetic elements (Yang and Kazazian, 2006; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a; Okamura et al., 2008b; Tam et al., 2008; Watanabe et al., 2008). A special class of endogenous siRNAs, hp-esiRNAs derive from partially self-complementary hairpin transcripts. Production of esiRNAs by Dicer-2 requires an alternative partner protein, Loqs-PD. This Loqs isoform contains only two of the three dsRBDs found in the Dicer-1 partner protein, Loqs-PB (Okamura et al., 2008b; Hartig et al., 2009; Zhou et al., 2009; Miyoshi et al., 2010; Hartig and Forstemann, 2011).
Dicer-1 and Dicer-2 each contain two RNase III domains, which form an intramolecular heterodimer whose dimer interface creates two active sites (Zhang et al., 2004; Macrae et al., 2006; Ye et al., 2007). Like other members of the Dicer family, Dicer-1 and Dicer-2 each contain a C-terminal dsRBD and a central PAZ domain, an RNA-binding motif specialized to recognize the two-nucleotide, 3′ single-stranded tails of Drosha and Dicer products (Bass, 2000; Cerutti et al., 2000; Lingel et al., 2003; Song et al., 2003; Yan et al., 2003; Lingel et al., 2004; Ma et al., 2004; Zhang et al., 2004; Gan et al., 2006; Macrae et al., 2006; MacRae et al., 2007). The structure of Giardia intestinalis Dicer and functional studies using human Dicer suggest that the distance between the PAZ domain and the active sites of the RNase III domains establishes the length of the small RNA product (Zhang et al., 2004; Gan et al., 2006; Macrae et al., 2006; MacRae et al., 2007; Takeshita et al., 2007).
Differences in the domain architecture of Dicer-1 and Dicer-2 (Figure S1) are unlikely to explain their distinct substrate specificities. Drosophila Dicer-2 shares its domain architecture—including N-terminal DExDc, Helicase C, PAZ, RNase IIIa, RNase IIIb, dsRBD domains—with human Dicer, which produces both siRNAs and miRNAs. In contrast, Dicer-1 lacks a DExDc domain, yet has a Helicase C domain. DExDc/H and DEAD box domains are found in a wide range of RNA “helicases,” proteins that couple ATP hydrolysis to RNA binding or unwinding (Pyle, 2008). In addition to unwinding nucleic acids, helicase domains can couple ATP hydrolysis to translocation along nucleic acid molecules, rearrange RNA:protein or protein: protein interactions, or act as RNA chaperones (Bianco and Kowalczykowski, 2000; Beran et al., 2006; Bowers et al., 2006; Dumont et al., 2006; Halls et al., 2007; Pyle, 2008; Franks et al., 2010).
In Drosophila and Caenorhabditis elegans, siRNA production from long dsRNAs requires ATP (Zamore et al., 2000; Bernstein et al., 2001; Ketting et al., 2001; Nykanen et al., 2001). Moreover, a point mutation (G31R) in the Drosophila Dicer-2 helicase domain blocks siRNA production in vivo, although the protein retains the ability to collaborate with R2D2 to load synthetic siRNAs (Lee et al., 2004; Pham et al., 2004) and highly paired miRNA/miRNA* duplexes (Forstemann et al., 2007) into Ago2. In contrast, a mutation predicted to inhibit nucleotide binding by the human Dicer helicase domain does not affect dicing (Zhang et al., 2002).
What restricts a given Dicer to a specific dsRNA substrate? We find that purified, recombinant Dicer-2 can cleave pre-miRNA, but that R2D2 inhibits processing of pre-miRNA by Dicer-2, while promoting use of its biologically relevant substrate by reducing the KM of Dicer-2 for long dsRNA. Moreover, physiological concentrations of inorganic phosphate block pre-miRNA processing by Dicer-2, but do not inhibit processing of long dsRNA by Dicer-2 or pre-miRNA processing by Dicer-1. Thus, the characteristic specificity of Dicer-2 for long dsRNA is not intrinsic to the enzyme, but rather emerges in the presence of inorganic phosphate and R2D2. We also find that Dicer-2 is a dsRNA-stimulated ATPase that hydrolyzes ATP to ADP; ATP hydrolysis is required for Dicer-2 to process long dsRNA but not pre-miRNA. Wild-type Dicer-2, but not a mutant defective in ATP hydrolysis, can generate siRNAs faster than it dissociates from its long dsRNA substrate. We envision that the Dicer-2 helicase domain uses ATP to drive the movement of Dicer-2 along dsRNA, enabling it to generate many siRNAs from a single molecule of substrate before dissociating from the dsRNA.
RESULTS
Dicer-2 Processes Pre-miRNA Inaccurately
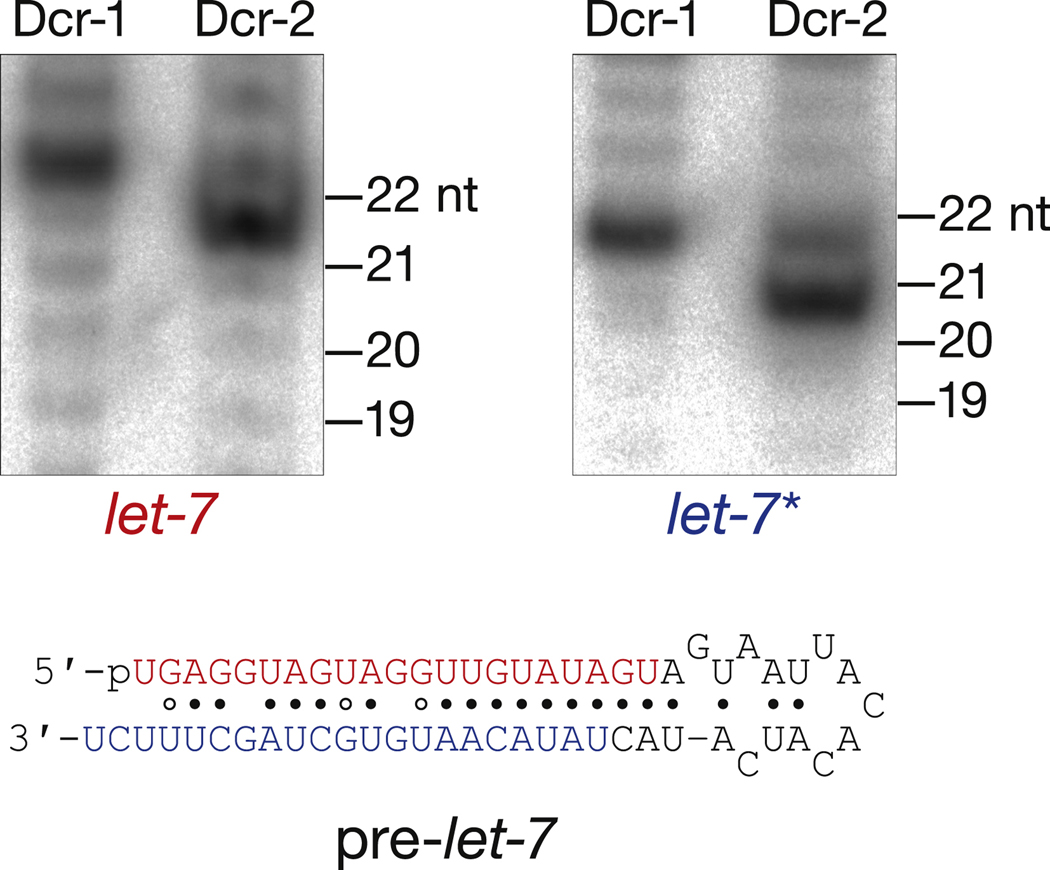
In vivo, Dicer-1 but not Dicer-2 is required to produce miRNAs from the stems of pre-miRNA. Surprisingly, purified, recombinant Dicer-2 cleaved pre-miRNA (Figure 1). However, the size of the miRNA and miRNA* products generated by Dicer-2 differed from those produced by Dicer-1: the predominant Dicer-2 product was one nucleotide shorter than that produced by Dicer-1. For miRNAs residing on the 5′ arm of their pre-miRNA, such a difference in size would not alter the miRNA seed sequence, but could promote their inappropriate loading into Argonaute2, which favors 21 nt RNAs, rather than Argonaute1, which prefers 22mers (Ameres et al., 2011). For the ~60% of D. melanogaster miRNAs derived from the 32 arm of their pre-miRNA, the seed sequence of the Dicer-2 product would differ from the authentic miRNA and would therefore regulate a repertoire of mRNAs different from that controlled by the authentic miRNA. The biological consequences of such misregulation are predicted to be dramatic, suggesting that processing of pre-miRNA by Dicer-2 is suppressed in vivo.
Figure 1. Dicer-1 and Dicer-2 Produce Different Products from Pre let-7.
Synthetic, 5′ monophosphorylated pre-let-7 (1.0 µM) was incubated with Dicer-1 (6.0 nM) or Dicer-2 (16.2 nM) for 1 h. Products were resolved by electrophoresis. let-7 and let-7* were detected by Northern hybridization.
Dicer-1 Does Not Efficiently Process Long dsRNA
In the absence of Dicer-2, flies do not accumulate siRNAs (Lee et al. 2004; Ghildiyal et al. 2008; Okamura et al. 2008; Czech et al. 2008). Why does Dicer-1 not make siRNAs in the absence of Dicer-2, especially since immunopurified Dicer-1 has been reported to dice long dsRNA (Saito et al., 2005)? Loqs-PB has been proposed to prevent Dicer-1 processing long dsRNA, restricting it to the miRNA pathway (Saito et al., 2005). However, we find that Dicer-1 is unable to catalyze multiple-turnover cleavage of long dsRNA (data not shown). In fact, 225 nM Dicer-1 was approximately as active at siRNA production from 25 nM long dsRNA as 5.4 nM Dicer-2. Unlike Dicer-2, Dicer-1 generated intermediates when processing long dsRNA (Figure S1B). Our data argue against Loqs-PB restricting Dicer-1 to the miRNA pathway. Instead, they suggest that the fundamental block to Dicer-1 processing long dsRNA is its inherent inefficiency in using this substrate.
R2D2 Inhibits Dicing of Pre-miRNA by Dicer-2
We compared the rate of pre-let-7 processing by Dicer-2 alone to the rate of purified Dicer-2/R2D2 heterodimer, Dicer-2 supplemented with equimolar Loqs-PD, and Dicer-2/R2D2 heterodimer supplemented with Loqs-PD. R2D2 significantly inhibited pre-let-7 processing by Dicer-2 when either enzyme (data not shown) or substrate was in excess (Figure 2A; p-value for excess substrate= 0.0009). Similar inhibition of Dcr-2 processing by R2D2 was observed for a 25 bp RNA duplex (Figure S2A), suggesting that R2D2 suppresses processing of short, double-stranded substrates irrespective of the extent of complementarity or the presence of a loop. In contrast, we did not detect any inhibition of pre-let-7 processing when Loqs-PD was added to Dicer-2, even though the same preparation of Loqs-PD lowered the KM of Dicer-2 for long dsRNA 10-fold (Table 1). These data suggest that, R2D2, but not Loqs-PD, helps suppress pre-miRNA processing by Dicer-2 in vivo.
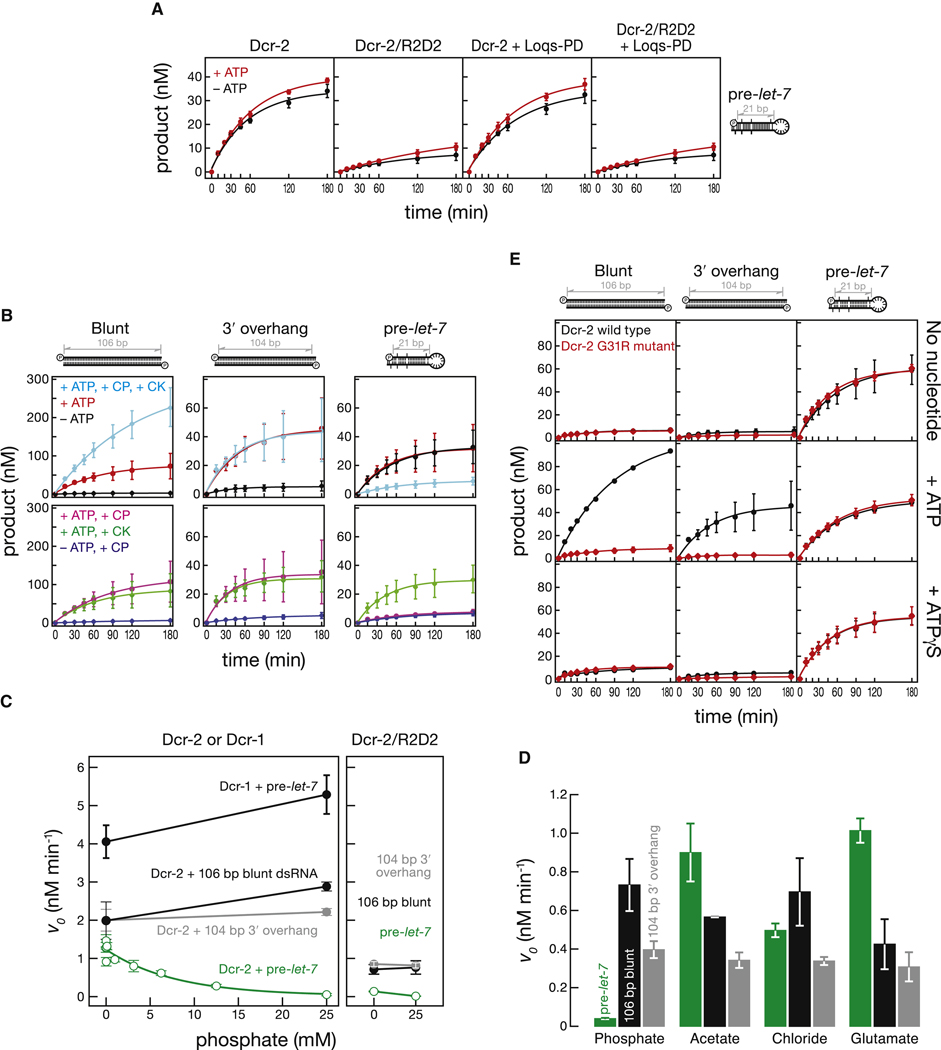
Figure 2. R2D2 and Phosphate Inhibit Dicer-2 Processing of Short Substrates.
(A) let-7 production was monitored using 5′ 32P-radiolabeled pre-let-7 (100 nM) with or without ATP for Dicer-2 alone (8 nM), Dicer-2/R2D2 (8 nM), Dicer-2 + Loqs-PD (8 nM + 8 nM) or Dicer-2/R2D2 + Loqs-PD (8 nM + 8 nM). (B) Processing of internally 32P-radiolabeled long dsRNA or 5′ 32P-radiolabeled pre-let-7 (100 nM) by Dicer-2 (8 nM). CK, creatine kinase; CP, creatine phosphate. (C) Initial velocities for the processing of 100 nM pre-let-7 or long dsRNA (106 bp blunt ended or 104 bp with 2 nt, 3′ overhanging ends) by 8 nM Dicer-2 or Dicer-2/R2D2 in the presence of increasing concentrations of potassium phosphate. Total potassium in the reaction was kept constant. (D) Initial velocities for Dicer-2 processing pre-let-7, 106 bp blunt ended dsRNA or 104 bp dsRNA with 2 nt, 3′ overhanging ends in the presence of 25 mM potassium phosphate, acetate, chloride, or glutamate. (E) Processing of internally 32P-radiolabeled long dsRNA or 5′ 32P-radiolabeled pre-let-7 substrate (100 nM) by wild-type or G31R mutant Dicer-2 (8 nM) with or without ATP or with ATPγS (1 mM). Values are mean ± standard deviation for three independent experiments.
Table 1. Michaelis-Menten Analysis of Dicer-2.
Dicer-2, Dicer-2/R2D2, or Dicer-2 supplemented with equimolar Loqs-PD was incubated with a 515 bp dsRNA and saturating ATP (1 mM) ATP. The initial rates of converting dsRNA into siRNA for increasing amounts of substrate were measured and fit to the Michaelis-Menten equation (Figure S5). The table reports mean ± standard deviation for three trials.
|
KM (nM) |
Change in KM |
Vmax (nM min−1) |
[ET] (nM) |
kcat (min−1) |
Change in kcat |
kcat/KM (nM−1 min−1) |
Change in kcat/KM |
|
|---|---|---|---|---|---|---|---|---|
| Dicer-2 | 6 ± 2.0 | 1 | 0.07 ± 0.01 | 2 | 0.03 ± 0.01 | 1 | 0.005 ± 0.003 | 1 |
| Dicer-2/R2D2 | 2 ± 1 | 0.3 | 0.09 ± 0.06 | 3 | 0.03 ± 0.02 | 1 | 0.02 ± 0.02 | 4 |
| Dicer-2 + Loqs-PD | 0.4 ± 0.1 | 0.07 | 0.06 ± 0.02 | 2 | 0.03 ± 0.01 | 1 | 0.08 ± 0.03 | 10 |
R2D2 and Loqs-PD Decrease the KM of Dicer-2 for Long dsRNA
Dicer-2 forms a stable heterodimer with R2D2 (Liu et al., 2003; Tomari et al., 2004; Liu et al., 2006), but it is unknown whether R2D2 modulates dicing rate. We measured the initial rate of dicing by Dicer-2 alone or by the Dicer-2/R2D2 heterodimer using increasing concentrations of long dsRNA substrate and saturating ATP (1 mM). For both Dicer-2 and Dicer-2/R2D2 the data fit well to the Michaelis-Menten kinetic scheme (Figure S5)
where kcat is the rate of complete conversion of substrate into siRNAs at saturating dsRNA concentration. The kcat of Dicer-2/R2D2 processing a 515 bp dsRNA (0.03 ± 0.02 min−1) was indistinguishable from that of Dicer-2 alone (0.03 ± 0.01 min−1; Table 1). In contrast, the KM for Dicer-2/R2D2 (2 ± 1 nM) was less than that of Dicer-2 alone (6 ± 2 nM, p-value = 0.04), suggesting that R2D2 increases the affinity of Dicer-2 for long dsRNA (Table 1). Similarly, supplementing Dicer-2 (2 nM) with purified recombinant Loqs-PD (2 nM) did not alter the kcat, but did decrease KM (0.4 ± 0.1 nM, p-value = 0.02), suggesting that Loqs-PD also increases the affinity of Dicer-2 for long dsRNA (Table 1).
Phosphate Inhibits Dicing of Pre-miRNA by Dicer-2
In flies, dicing of long dsRNA requires ATP (Nykanen et al., 2001). Typically, creatine kinase (CK) and creatine phosphate (CP) are included in dicing reactions to maintain high levels of ATP and constant levels of free Mg2+. Relative to ATP alone, the inclusion of CK and CP modestly enhanced Dicer-2 processing of both a 106 bp dsRNA bearing a 5′ monophosphorylated blunt end (Figure 2B) and a 316 bp dsRNA bearing a 5′ triphosphorylated, 2 nt, 5′ overhang (Figure S2B), but did not enhance processing of a 104 bp dsRNA with a 5′ monophosphorylated, 2 nt 3′ overhang (Figure 2B). In contrast, standard “ATP” conditions (+ATP, +CP, +CK) inhibited pre-let-7 processing by Dicer-2. More detailed analyses revealed that CP sufficed to inhibit pre-let-7 dicing.
CP can be hydrolyzed in water to creatine and phosphate. We therefore tested whether inorganic phosphate inhibited pre-let-7 processing by Dicer-2. We measured the initial rate of processing (v0) with increasing concentrations of potassium phosphate (KH2PO4/K2H PO4, pH 7.4) for pre-let-7, a 106 bp blunt end dsRNA and a 104 bp dsRNA with 2 nt 3′ overhanging ends. Physiological concentrations of phosphate (Burt et al., 1976; Erecinska et al., 1977; Auesukaree et al., 2004) inhibited processing of pre-let-7, but neither of the two dsRNAs (Figure 2C). We observed little or no inhibition of pre-miRNA processing by Dicer-2 with 25 mM acetate, chloride, or glutamate (Figure 2D). Moreover, none of the anions—including phosphate—had a significant effect on the dicing of long dsRNA. Phosphate further inhibited the low level of pre-let-7 processing of Dicer-2/R2D2 (Figure 2C) and Dicer-2/R2D2 + Loqs-PD, and suppressed pre-let-7 processing by Dicer-2 + Loqs-PD (data not shown). In contrast, processing of pre-let-7 by Dicer-1 was unaffected by phosphate (Figure 2C). We conclude that under physiological conditions—2–25 mM phosphate and a majority of Dicer-2 complexed with R2D2—Dicer-2 is unlikely to use pre-miRNA as a substrate.
ATP Hydrolysis and siRNA Production by Dicer-2
The Dicer-2 G31R point mutation, which lies in the protein’s DExDc motif, uncouples Argonaute2 loading from dsRNA dicing (Lee et al., 2004). The mutation is predicted to disrupt ATP binding. Consistent with the requirement for ATP in siRNA production by Dicer-2, dsRNA processing by purified recombinant Dicer-2G31R was significantly less than wild-type Dicer-2 for both substrates with blunt and 3′ overhanging ends (Figure 2D, p-value = 2.2 × 10−6 for blunt end substrate). While dsRNA processing by wild-type Dicer-2 was strongly stimulated by ATP, mutant Dicer-2G31R was not (Figure 2D). Nonetheless, Dicer-2G31R cleaved pre-let-7 as efficiently as the wild-type enzyme, consistent with the finding that ATP was not required for wild-type Dicer-2 to cleave pre-let-7 (Figure 2A). Moreover, processing of long dsRNA by wild-type Dicer-2 was inhibited by adenosine 5′-O-(3-thio)triphosphate (ATPγS), but processing of pre-let-7 was not (Figure 2D).
In agreement with ATPγS inhibiting dsRNA processing, Dicer-2 hydrolyzed ATP to ADP (Figure 3A). Both ATP and ATPαS supported the production of siRNA from long dsRNA, but ATPγS did not (Figure 3B). In the presence of 1 mM ATP, the rate of dicing long dsRNA declined exponentially with increasing ATPγS, suggesting that ATPγS competes with ATP for binding to Dicer-2 and that once bound, ATPγS is not efficiently hydrolyzed, preventing the production of siRNAs from long dsRNA.
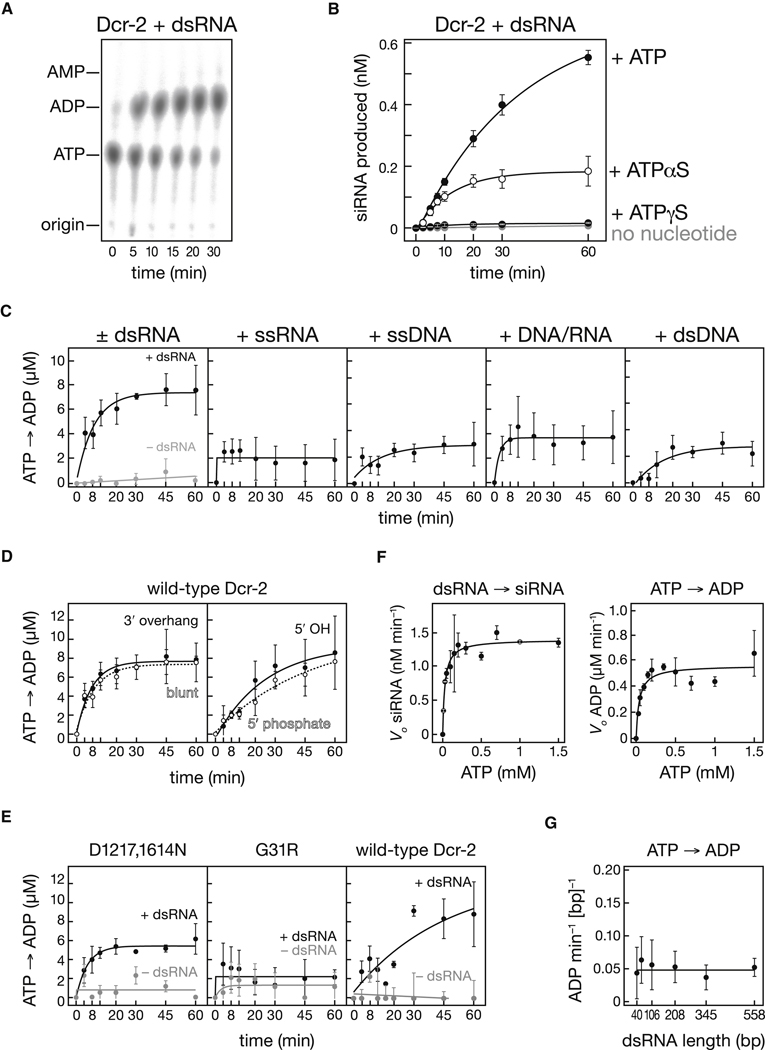
Figure 3. Dicer-2 Requires ATP Hydrolysis to Process Long dsRNA.
(A) Dicer-2 (10.8 nM) was incubated with 2.5 µM α-32P ATP and 150 nM 515 bp dsRNA, and ATP hydrolysis monitored by thin-layer chromatography. (B) siRNA production by Dicer-2 (5.4 nM) from a 515 bp dsRNA (25 nM) was monitored with or without ATP or ATPγS (1 mM). (C) ATP hydrolysis by Dicer-2 (5.4 nM) was monitored for 120 nt long nucleic acid substrates (20 nM): dsRNA, single-stranded RNA, single-stranded DNA, RNA/DNA heteroduplex or double-stranded DNA in the presence of ATP (1 mM). (D) ATP hydrolysis by Dicer-2 (5.4 nM) in the presence of ATP (1 mM) was measured for 20 nM dsRNA bearing a 5′ monophosphate and a 2 nt 3′ overhang or blunt ends; the blunt ended dsRNA bearing either 5′ monophosphate or 5′ hydroxy termini. (E) ATP hydrolysis by mutant Dicer-2D1217,1614N (3.3 nM) , Dicer-2G31R (2 nM) and wild-type Dicer-2 (2 nM) was measured in the absence or presence of 20 nM dsRNA bearing 5′ monophosphate, blunt ends. (F) Left panel, initial rates for the conversion of substrate into siRNA by Dicer-2 (2.7 nM) for an internally 32P-radiolabeled 515 bp dsRNA (150 nM) were measured at increasing concentrations of ATP. Right panel, initial rates for the hydrolysis of ATP to ADP by Dicer-2 in the presence of 150 nM, 515 bp dsRNA were measured for increasing ATP concentrations. The data were fit to the Michaelis-Menten equation. Table 2 reports the Michaelis-Menten parameters. (G) ATP consumption by Dicer-2 was measured in the presence of ATP (350 µM) and dsRNA substrates with blunt, 5′ triphosphorylated termini for six different lengths: 40 bp (420 nM), 60 bp (280 nM), 106 bp (158 nM), 208 bp (80.5 nM), 345 bp (49 nM), 558 bp (30 nM). Substrate concentrations were selected to ensure an equal number of base pairs in each reaction. Values are mean ± standard deviation for three independent experiments.
Dicer-2 is a dsRNA-Stimulated ATPase
ATP hydrolysis by Dicer-2 increased when dsRNA was added (Figure 3C, p-value= 0.024, Wilcoxon rank sum test). The inclusion of 120 nt single-stranded RNA or DNA or 120 bp DNA/RNA heteroduplex or double-stranded DNA stimulated ATP hydrolysis less than the 120 bp dsRNA (all 20 nM; Figure 3C). A 25 nt single-stranded RNA or DNA, a 21 bp dsRNA or a 21 bp DNA/RNA heteroduplex, all failed to stimulate the Dicer-2 ATPase activity above the rate observed when no substrate was present (Figure S3B). Neither the end structure of the dsRNA—blunt versus 3′ overhang—nor the presence of a 5′ monophosphate (v05′ PO4 = 0.4 ± 0.1 µM min−1 versus v05′ OH 0.3 ± 0.1 µM min1) significantly changed the ATP hydrolysis rate (Figure 3D).
ATP hydrolysis by Dicer-2 does not require dsRNA cleavage. A Dicer-2 mutant, D1217,1615N, in which a key aspartate in each of the two RNase III domain active sites was changed to asparagine, retained significant dsRNA-stimulated ATPase activity (p-value = 0.02; Figure 3E). Under multiple-turnover conditions, Dicer-2D1217,1615N was essentially inactive for dicing, and siRNA production was detected only when [Dicer-2D1217,1615N] >> [substrate] (data not shown). In contrast, Dicer-2G31R, which also does not support multiple-turnover dicing of long dsRNA, did not hydrolyze ATP in the presence of dsRNA (Figure 3E). We conclude that the Dicer-2 helicase domain is responsible for the enzyme’s dsRNA-stimulated ATPase activity.
ATP Consumption and Dicing are Coupled
We measured the initial rates of siRNA production and ATP hydrolysis using a 515 bp dsRNA in the presence of increasing concentrations of ATP. The dependence on ATP concentration of both siRNA production and ATP hydrolysis fit well to the Michaelis-Menten kinetic scheme (Figure 3F). When both substrate and ATP were saturating (i.e., ≥ 10 × KM), the kcat for ATP hydrolysis was 93 ± 14 min−1, whereas the kcat for siRNA production was 4 ± 1 min−1, as inferred from the kcat for the complete conversion of a molecule of substrate into siRNA. These values predict that 23 ± 8 molecules of ATP are hydrolyzed for each 21 nt siRNA formed (Figure 3F, Table 2). Such a high rate of ATP hydrolysis for each siRNA produced might suggest that ATP energy and siRNA production are poorly coupled. Alternatively, ATP might be hydrolyzed to power translocation of Dicer-2 along the dsRNA, with approximately one ATP molecule consumed for each base pair traversed, a rate similar to DNA and RNA translocases containing ATPase/helicase domains (Bianco and Kowalczykowski, 2000; Patel and Donmez, 2006; Seidel et al., 2008). Consistent with this idea, the rate of ATP consumption was essentially unchanged for different substrate lengths when the molar concentration of base pairs was kept constant (Pearson correlation, r = 0.98; Figure 3G, S3B).
Table 2. Michaelis-Menten Analysis of Dicer-2 incubated with a 515 bp dsRNA and ATP.
The initial rates of converting dsRNA into siRNA for increasing amounts of substrate were measured at saturating ATP (1 mM), and the initial rates of hydrolysis of ATP to ADP were measured at saturating dsRNA (150 nM) for increasing amounts of ATP. The table reports mean ± standard deviation for four trials. Different preparations of Dicer-2 were used here and in Table 1.
| KM(nM) | Vmax(nM min−1) | [ET] (nM) | kcat(min−1) | kcat/KM(nM−1 min−1) | |
|---|---|---|---|---|---|
| Substrate consumed | 6 ± 2 | 0.2 ± 0.1 | 1.2 | 0.2 ± 0.1 | 0.03 ± 0.02 |
| siRNA produced | 6 ± 2 | 5 ± 2 | 1.2 | 4 ± 1 | 0.7 ± 0.4 |
| ATP hydrolyzed | 14,000 ± 4,000 | 460 ± 70 | 5 | 93 ± 14 | 0.007 ± 0.002 |
Evidence that Dicer-2 is Processive
If ATP fuels the translocation of Dicer-2 along dsRNA, dicing should produce successive siRNAs along the substrate. In the presence of ATP, Dicer-2 would be predicted to produce the first siRNA—i.e., the terminal siRNA—at roughly the same rate as subsequent, internal siRNAs. In contrast, in the absence of ATP, the rate of production of siRNA should decline as its distance from 5′ end increases. To test these predictions, we synthesized three, identical 120 nt dsRNA substrates, each bearing a single 32P radiolabel at position 16, 35, or 104 from the 5′ end of one strand (Figure 4A). All substrates contained two deoxynucleotides at one end, forcing Dicer-2 to initiate processing from the opposite end (Figure S4A) (Rose et al., 2005). We used this “one-ended” substrate to examine the production of the first, second, or fourth siRNA (see Experimental Procedures) in the presence or absence of ATP (Figure 4A). With 1 mM ATP, the initial rates for the production of the first, second, and fourth siRNAs were essentially indistinguishable (~28 nM min−1); with no ATP, the rate for the first siRNA (1.4 ± 0.3 nM min−1) was greater than that of the second (0.9 ± 0.1 nM min−1), which was greater than the rate for the fourth siRNA (0.2 ± 0.1 nM min−1; Figure 4A).
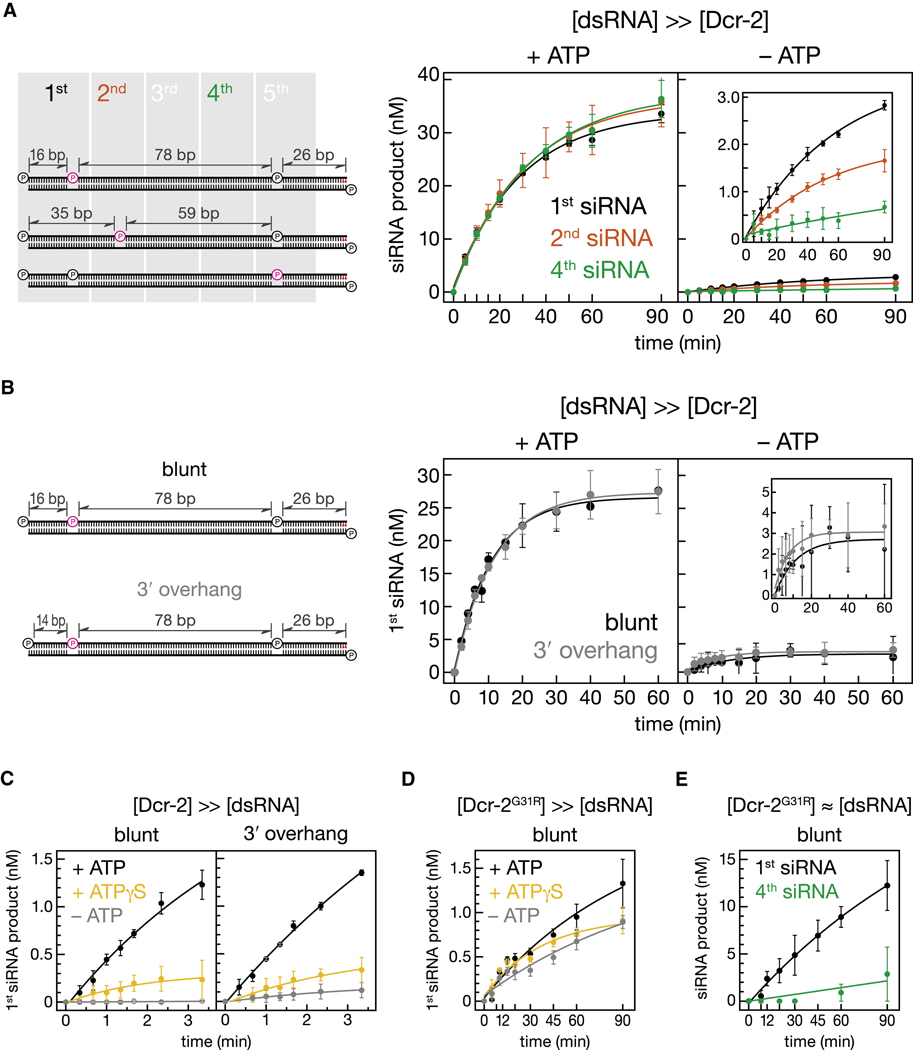
Figure 4. Evidence that Dicer-2 is Processive.
(A) Site-specifically 32P-radiolabeled 120 bp dsRNAs were used to measure the rate of production of the first, second and fourth siRNAs from the all-RNA end of the substrate; the 32 end of the sense strand contained two deoxynucleotides (red) to block entry of Dicer-2 from the other end (Figure S4A). Production of siRNA was monitored in the presence or absence of ATP using 100 nM dsRNA and 5.4 nM Dicer-2. Values correspond to the mean ± standard deviation from three independent experiments. The data were fit to a single exponential function. In (A) and (B), pink denotes the position of the 32P-radiolabel. (B) The rate of production of the initial siRNA was measured as in (A), but using a site-specifically 32P-radiolabeled dsRNA with either a blunt (black) or a 2 nt 3′ overhanging (red) end in the presence or absence of ATP. (C) The rate of production of the initial siRNA was monitored using 100 nM Dicer-2 and 10 nM dsRNA bearing either a blunt (left) or a 2 nt 3′ overhanging (right) end in the presence or absence of ATP or ATPγS. (D) The rate of production of the initial siRNA from a 120 bp blunt ended dsRNA (10 nM) was measured using 100 nM mutant Dicer-2G31R. (E) The rate of production of the first and fourth siRNAs from a 120 bp blunt ended dsRNA (100 nM) was measured using 100 nM Dicer-2G31R. Values are mean ± standard deviation for three independent experiments.
We can envision two explanations consistent with these results and previous studies on Dicer enzymes: either ATP fuels processive dicing of long dsRNA or dicing of long dsRNA in the presence of ATP comprises a slow initial binding step to the end of the substrate, followed by rapid but ATP-dependent production of subsequent siRNAs. Such a two-step process might occur if the rate of production of the first siRNA was slowed by the blunt structure of the substrate; the first dicing event would convert the substrate end to a 2 nt, 3′ overhang bearing a 5′ monophosphate, with all subsequent siRNAs produced rapidly. To test this idea, we prepared a substrate bearing one blocked end and a 2 nt, 3′ overhang bearing a 5′ monophosphate at the other end (Figure S4B). The rates of first siRNA production from both blunt and 3′ overhanging end substrates were indistinguishable when the reactions contained ATP—conditions where dicing was efficient. They were also similar when ATP was omitted—when dicing was slow (Figure 4B). The subsequent siRNAs were also produced at similar rates from the two different substrates (Figure S4B). Thus, we favor the hypothesis that ATP converts Dicer-2 from an inefficient, distributive enzyme into a processive enzyme.
Surprisingly, ATP also enhanced the production of the terminal siRNA from long dsRNA under single-turnover conditions ([Dicer-2] >> [dsRNA]); the enhancement by ATPγS was considerably weaker (Figure 4C). This suggests that ATP hydrolysis is required for the production of even the first siRNA. ATP was required irrespective of the terminal structure of the dsRNA (blunt versus 3′ overhang), excluding a role for ATP in the binding of Dicer-2 to a particular type of dsRNA end.
Moreover, when the [enzyme] > [dsRNA], helicase mutant Dicer-2G31R produced the first siRNA at similar rates in the presence or absence of either ATP or ATPγS (Figure 4D and S4C). For Dicer-2G31R, the rate of production of the terminal siRNA was faster than that of the fourth siRNA, consistent with the idea that ATP hydrolysis converts Dicer-2 from a distributive to a processive enzyme (Figure 4E).
Dicer-2 produces siRNAs without dissociating from the dsRNA
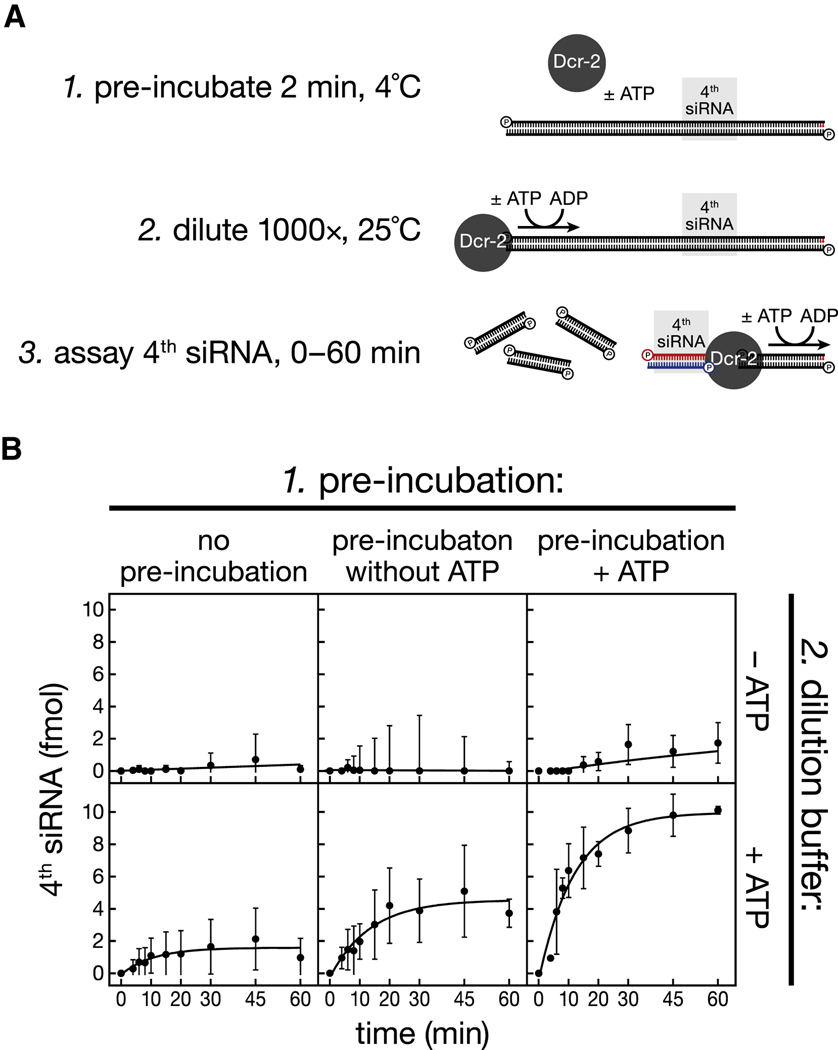
A processive enzyme can act multiple times on its substrate before dissociating. Thus, catalysis by processive enzymes resists dilution (Rivera and Blackburn, 2004). To test whether Dicer-2 remains physically associated with the long dsRNA after three subsequent dicing events, we used the 120 nt, site-specifically 32P-radiolabeled dsRNA to monitor the rate of production of the fourth siRNA following dilution (Figure 4A). The dsRNA substrate (200 fmol) was first pre-incubated with Dicer-2 (54 fmol) for 2 min at 4˚C to allow the enzyme to bind substrate. During this pre-incubation step, no siRNA was detected (data not shown). Next, the reaction was diluted 1,000-fold into buffer pre-warmed to 25˚C. The diluted reaction was incubated at 25°C, and production of the fourth siRNA measured over time (Figure 5A). During the pre-incubation, the substrate concentration (20 nM) was > 3-fold greater than the KM of Dicer-2 for long dsRNA; after dilution, the substrate concentration was ~300 times less than the KM. For both the pre-incubation and the dilution steps, the dsRNA substrate was present at ~4-fold higher concentration than Dicer-2. When the pre-incubation was omitted, little fourth siRNA was produced (Figure 5B), demonstrating that the conditions largely prevented reassociation of Dicer-2 with dsRNA once it dissociated from the substrate.
Figure 5. Dicer-2 remains associated with its substrate in the presence of ATP.
(A) Design of the experiment. The reaction contained 54 fmol Dicer-2 and 200 fmol dsRNA, corresponding to 5.4 nM enzyme and 20 nM dsRNA before dilution. (B) The rate of production of the fourth siRNA was measured for six different combinations of pre-incubation (no pre-incubation, without ATP, and with ATP) and dilution (without and with ATP). Values are mean ± standard deviation for three independent experiments.
When ATP was included in both the pre-incubation and the dilution buffer, 54 fmol of Dicer-2 produced 10 fmol of fourth siRNA in 1 h. About half as much fourth siRNA was produced when ATP was present in the dilution buffer but omitted from the pre-incubation. When ATP was omitted from the dilution buffer, essentially no fourth siRNA was produced, regardless of whether ATP was present during the pre-incubation, suggesting that ATP dissociates rapidly from Dicer-2. More fourth siRNA was made when ATP was present in both the pre-incubation and dilution buffers than when pre-incubation was carried out in the absence of ATP (p-value = 0.015). We conclude that the initial binding of Dicer-2 to the end of long dsRNA is enhanced by ATP and that in the presence of ATP Dicer-2 remains associated with the dsRNA. We propose that the stably bound Dicer-2 then cleaves successive siRNAs along the dsRNA.
DISCUSSION
Purified Dicer-1 and Dicer-2 both process pre-miRNAs, but generate different length products (22 versus 21 nt). Genetic analyses suggest that Dicer-1 and Dicer-2 are restricted to specific substrate classes in vivo (Lee et al. 2004). For example, Dicer-2 cannot replace Dicer-1 in the miRNA pathway. Similarly, dicer-2 mutants are defective for RNAi, even though they express normal levels of Dicer-1 (Lee et al., 2004). Despite structural similarities, Dicer-2 specifically processes esiRNA hairpins, while Dicer-1 cleaves pre-miRNAs (Lee et al., 2004; Förstemann et al., 2005; Jiang et al., 2005; Saito et al., 2005; Miyoshi et al., 2010). This observation suggests that the length of a dsRNA is the primary determinant of substrate choice.
Our data argue that the combination of R2D2 and cellular phosphate restricts Dicer-2 to its biologically relevant substrates by inhibiting the processing of short substrates such as pre-miRNA. Thus, a protein, R2D2, and a small molecule, phosphate, convert a promiscuous dsRNA endonuclease into one specific for the long dsRNA substrates that trigger RNAi. It is tempting to speculate that inorganic phosphate interferes with recognition of the 5′ monophosphate present on all pre-miRNAs, and that 5′ phosphate recognition is unnecessary for longer substrates, because their greater length allows additional protein-RNA contacts—perhaps by the dsRNA-binding and the helicase domains—between Dicer-2 and long dsRNA. We note that human Dicer has been reported to recognize a 5′ monophosphate on single-stranded RNA (Kini and Walton, 2007).
In flies, the Dicer-2 partner proteins Loqs-PD and R2D2 likely enhance substrate specificity by increasing the affinity of the enzyme for long dsRNA. The kcat for and Dicer-2 and Dicer-2/R2D2 were similar, but R2D2 decreased the KM of Dicer-2 for long dsRNA (Table 1 and Figure S5). We note that the specificity constant, kcat/KM, was ~4 fold higher for the Dicer-2/R2D2 heterodimer than for Dicer-2 alone. Similarly, Loqs-PD lowered the KM of Dicer-2 for long dsRNA without reducing the catalytic rate, resulting in a ~10-fold higher kcat/KM.
Processing of pre-miRNA by Dicer-1 was unaffected by phosphate. We find that the intrinsic properties of Dicer-1, which cannot efficiently catalyze multiple-turnover processing of long dsRNA, restrict that enzyme to process pre-miRNA. We do not yet know whether the transition of substrates from Dicer-1 to Dicer-2 is gradual, such that some substrates are processed equally well by both enzymes. In theory, such intermediate substrates might be selected against in evolution, enforcing the distinction between Dicer-1 and Dicer-2 substrates.
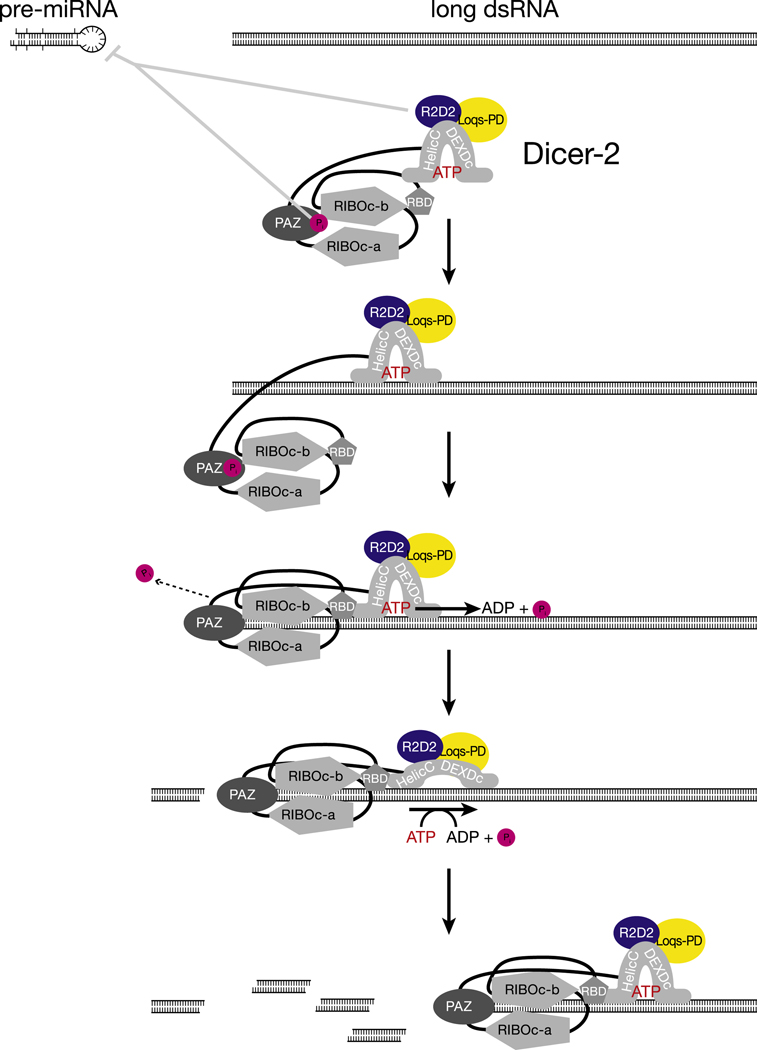
The Dicer-2 helicase domain is similar to that of RIG-I, a sensor in the mammalian innate immune system. The RIG-like ATPase/helicase domain is conserved among plant and animal Dicers. Yet, its function has remained unknown. Our data suggest that this domain of Dicer-2 is involved in ATP-dependent production of successive siRNAs from long dsRNA. Notably, two other members of this helicase family, DRH-3 and RIG-I are also bona fide ATPases: DRH-3, a C. elegans protein required for RNA silencing and germ-line development (Nakamura et al., 2007), is a dsRNA-stimulated ATPase (Matranga and Pyle, 2010); and the mammalian protein RIG-I, which recognizes viral 5′ triphosphorylated dsRNA and initiates an innate immune response, uses ATP to translocate along dsRNA (Myong et al., 2009). Our data are consistent with the idea that ATP hydrolysis fuels translocation of Dicer-2 along long dsRNA substrates. An alternative view—that the ATP-dependent binding of a molecule of Dicer at the end of the substrate promotes the complete and rapid oligomerization of Dicer-2 along the entire extent of the dsRNA—would require that the Dicer and RIG-I helicase domains share a conserved sequence but have highly divergent functions.
ATP was not required for Dicer-2 to process pre-miRNA, and a mutant Dicer-2 unable to hydrolyze ATP remained able to process pre-miRNA but not long dsRNA. These results help explain why in C. elegans, in which a single Dicer processes both long dsRNA and pre-miRNA, a mutation in the DCR-1 helicase domain disrupted endo-siRNA, but not miRNA, accumulation (Welker et al., 2010).
Four lines of evidence support a role for ATP hydrolysis in the production of successive siRNAs along the dsRNA by Dicer-2. First, Dicer-2 consumes a constant amount of ATP per base-pair. Second, ~23 molecules of ATP were consumed for each 21 nt siRNA produced. Third, the rate of production of the first, second, and fourth siRNAs from a long dsRNA substrate were indistinguishable in the presence of ATP, but in the absence of ATP, the rate of siRNA production declined with increasing distance from the end of the dsRNA. Finally, the association of Dicer-2 with a long dsRNA was resistant to dilution provided ATP was present, suggesting that after binding the end of its substrate, Dicer-2 remains bound to the dsRNA and uses ATP energy to reposition itself to produce the next 21 bp siRNA. Translocation along the dsRNA seems a likely mechanism.
Although helicase mutant Dicer-2G31R processed pre-miRNAs as efficiently as wild-type Dicer-2, the mutant was unable to produce even the terminal siRNA from a long RNA duplex under multiple turnover conditions. This suggests an inherent ability of the helicase domain of Dicer-2 to distinguish between long and short substrates. We note that the helicase domain of human Dicer auto-inhibits processing of an RNA duplex, and its dsRNA-binding protein partner TRBP, a homolog of R2D2 and Loqs, relieves this inhibition (Ma et al., 2008; Chakravarthy et al., 2010). We hypothesize that Drosophila Dicer-2 can occupy two distinct conformations. When inorganic phosphate is low, Dicer-2 assumes a conformation—perhaps similar to the auto-inhibited conformation of human Dicer—that can bind and load siRNA. This conformation is unaffected by ATP and, we presume, is involved in promiscuously processing pre-miRNA in vitro. When inorganic phosphate is higher and the enzyme’s helicase and dsRNA-binding domains engage its substrate, Dicer-2 assumes a conformation that requires ATP for binding and hydrolysis to process dsRNA.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Expression and purification of His6-Dicer-2 or His6-Dicer-2 and His6-R2D2 in Sf21 cells was as described (Liu et al., 2003; Tomari et al., 2004). His6-Dicer-2G31R, His6-Dicer-2D1217,1614N and His6-Dicer-1 were expressed in Sf9 insect cells using the BAC-to-BAC Baculovirus Expression System (Invitrogen, Carlsbad, CA) and purified from cell lysates by using Ni-NTA agarose (QIAGEN, Valencia, CA), HiTrap Q, HiTrap Heparin (GE Healthcare, Pittsburgh, PA), and Superdex 200 gel filtration. Loqs-PD was expressed in Escherichia coli Rosetta2(DE3), isolated using Ni-Sepharose (GE Healthcare), treated with HRV3C protease cleavage to remove the His-tag, and purified using HiTrap SP and HiTrap Heparin. Proteins were exchanged into 20 mM HEPES-KOH (pH 8.0), 100 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine hydrochloride. Protein concentrations were determined by quantitative amino acid analysis (Keck Biotechnology Resource Laboratory, New Haven, CT). Two different preparations of recombinant Dicer-2 were used in this study. Preparation 1 was used for Table 1 and Figures 2, 3E, 4C, S2B, S3A, S3D, and S5. Preparation 2 was used for Table 2 and Figures 1, 3A–D, 3F, 3G, 4A, 4B, 5, S1, S2A, S3B, S3C, and S4.
RNA Substrates
DsRNAs were prepared as described (Haley et al., 2003). PCR templates for transcription of sense and antisense RNAs were generated from the EGFP sequence of pN3-eGFP using primers listed in Table S1. Twenty-five and 29 bp dsRNAs (Figures S2A, S3B, S4A, Table S2) were as previously described (Rose et al., 2005). Synthetic RNAs and synthetic Drosophila pre-let-7 (Dharmacon, Lafayette, CO) were 5′ 32P-radiolabeled using γ-32P ATP (6000 Ci/mmol; PerkinElmer, Waltham, MA) and T4 polynucleotide kinase (NEB, Ipswich, MA). After gel purification, RNA strands or pre-let-7 were incubated at 65°C for 5 min and then at 25°C for 30 min. Site-specifically radiolabeled 120 nt dsRNAs were prepared by DNA-splinted ligation (Table S3 and S4) (Moore and Sharp, 1993; Moore and Query, 2000). To monitor formation of the fourth siRNA in Figure 4B, we used a 120 nt substrate in which the 5th siRNA (the last siRNA generated by Dicer-2 from this substrate) was site-specifically 32P-radiolabeled. The fourth siRNA produced corresponds to the sum of the two 32P-radiolabeled cleavage products produced when the dsRNA was cleaved to generate the fourth and fifth siRNAs.
In Vitro RNA Processing
For Dcr-2 +Loqs-PD, Loqs-PD was first mixed with Dicer-2 or Dicer-2/R2D2 and incubated 10 min on ice followed by 5 min at room temperature. Dicing reactions contained 7.5 mM DTT, 3.3 mM magnesium acetate, 0.25% v/v glycerol, 100 mM potassium acetate, 18 mM HEPES-KOH (pH 7.4), 15 mM CP, 2.25 µg CK, and 1 mM ATP; −ATP reactions contained 1 mM EDTA but no CK or CP; ATPγS reactions contained 1 mM ATPγS only. In Figure 2A–D, 3, 4B–E, 5 , S3, and S4, +ATP reactions contained no CP or CK; +CP and +CK reactions in Figure 2B and S2B contained 20 mM CP or 2.25 µg CK. Reactions were assembled on ice and pre-incubated at 25°C for 5 min before adding RNA. In Figure S4, dilution buffer contained 0.1% NP-40.
Aliquots (1 µl) of reactions with radiolabeled RNA substrate were quenched by the addition of 25 volumes of formamide loading buffer (98% v/v formamide, 0.1% w/v bromophenol blue and xylene cyanol, 10 mM EDTA), incubated 5 min at 95°C, and analyzed by electrophoresis through a denaturing polyacrylamide 7 M urea gel using 0.5× Tris-borate-EDTA buffer (National Diagnostics, Atlanta, GA). In Figure 5, 200 µL aliquots from dilution reactions were stopped with 300 mM sodium acetate and 25 mM EDTA, isopropanol precipitated, and dissolved in formamide loading buffer before gel analysis. Gels were exposed to image plates and analyzed with an FLA-5000 and ImageGauge 3.0 software (Fujifilm, Tokyo, Japan). In Figure 1, let-7 and let-7* strands were detected by Northern hybridization with 5′ 32P-radiolabeled DNA probes (Table S1).
ATP Hydrolysis
α-32P-ATP (250 nM, 3000 mmol/Ci; PerkinElmer) was used to monitor hydrolysis. Reactions were stopped with a 25 vol formamide loading dye and spotted onto 20 cm × 20 cm cellulose plates (EMD, Darmstadt, Germany) and chromatographed in 0.75 M KH2PO4 (adjusted to pH 3.3 with H3PO4) until the solvent reached the top of the plate. The plate was dried and analyzed by phosphorimagery. For Figure S3B, ATP hydrolysis was monitored using the ATP Bioluminescent Assay Kit (Sigma, St. Louis, MO). The reaction was stopped by diluting the sample 10 times in H2O and immediately flash-freezing in liquid nitrogen. Samples were stored at −80°C until they were measured. A standard curve spanning at least 100-fold less than and greater than the experimental values was used to determine ATP concentrations.
Rate Analyses
Substrate converted to siRNA versus time was fit to y = y0 + A(1 – e−kt), where dy/dt = Ake−kt. When t = 0, dy/dt = Ak (Lu and Fei, 2003). Data were fit to the Michaelis-Menten scheme using Visual Enzymics 2008 (Softzymics, Princeton, NJ) for Igor Pro 6.11 (WaveMetrics, Lake Oswego, OR). The rate of substrate consumption was fit for each replicate separately, and two-tailed, two sample equal variance t-test used to compare rates (Excel, Microsoft, Seattle, WA). R 2.6.0 software was used for other statistical analyses.
HIGHLIGHTS.
Dicer-2 cleaves pre-miRNA, but its product is shorter than the authentic miRNA
The protein R2D2 and inorganic phosphate block pre-miRNA processing by Dicer-2
Dicer-2 is a double-stranded RNA-stimulated ATPase that hydrolyzes ATP to ADP
Wild-type, but not mutant, Dicer-2 makes siRNAs faster than it dissociates from dsRNA
Supplementary Material
Figure 6.
A model for Drosophila Dicer-2
ACKNOWLEDGMENTS
We thank Yukihide Tomari for help purifying recombinant Dicer-2, members of the Zamore and Hall laboratories and Can Cenik for advice and comments on the manuscript, and Sean Ryder for suggesting we test the effect of phosphate on dicing of pre-miRNA. This work was supported in part by grants from the National Institutes of Health to PDZ (GM62862 and GM65236), the Intramural Research Program of the National Institute of Environmental Health Sciences to TMTH, and a JSPS Research Fellowship for Research Abroad and a Charles A. King Trust Postdoctoral Fellowship to RF.
REFERENCES
- Ameres SL, Hung JH, Xu J, Weng Z, Zamore PD. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA. 2011;17:54–63. doi: 10.1261/rna.2498411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auesukaree C, Homma T, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J Biol Chem. 2004;279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- Bass BL. Double-Stranded RNA as a Template for Gene Silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- Beran RK, Bruno MM, Bowers HA, Jankowsky E, Pyle AM. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J Mol Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Kowalczykowski SC. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- Bowers HA, Maroney PA, Fairman ME, Kastner B, Luhrmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt CT, Glonek T, Barany M. Analysis of phosphate metabolites, the intracellular pH, and the state of adenosine triphosphate in intact muscle by phosphorus nuclear magnetic resonance. J Biol Chem. 1976;251:2584–2591. [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends in Biochemical Sciences. 2000:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco IJ, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erecinska M, Stubbs M, Miyata Y, Ditre CM. Regulation of cellular metabolism by intracellular phosphate. Biochim Biophys Acta. 1977;462:20–35. doi: 10.1016/0005-2728(77)90186-4. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. doi: 10.1016/j.cell.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Haley B, Tang G, Zamore PD. In vitro analysis of RNA interference in Drosophila melanogaster. Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Bottcher R, Saito K, Forstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Forstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini HK, Walton SP. In vitro binding of single-stranded RNA by human Dicer. FEBS Lett. 2007;581:5611–5616. doi: 10.1016/j.febslet.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a Bridge Between the Initiation and Effector Steps of the Drosophila RNAi Pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WP, Fei L. A logarithmic approximation to initial rates of enzyme reactions. Anal Biochem. 2003;316:58–65. doi: 10.1016/s0003-2697(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Matranga C, Pyle AM. The double stranded RNA-dependent ATPase DRH-3: insight into its role in RNA silencing in Caenorhabditis elegans. J Biol Chem. 2010 doi: 10.1074/jbc.M110.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010 doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ando R, Nakazawa T, Yudazono T, Tsutsumi N, Hatanaka N, Ohgake T, Hanaoka F, Eki T. Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells. 2007;12:997–1010. doi: 10.1111/j.1365-2443.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Rivera MA, Blackburn EH. Processive utilization of the human telomerase template: lack of a requirement for template switching. J Biol Chem. 2004;279:53770–53781. doi: 10.1074/jbc.M407768200. [DOI] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106–120. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HHJ. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007 doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.