Abstract
This report outlines and applies a high performance sequencing technology to evaluate the glycome of a common model glycoprotein, ovalbumin. The targets were the N-linked glycans enzymatically released from the protein, the N-glycoproteome. These product glycans were reduced, methylated and directly infused into the MS using a chip-based nanoelectrospray with the ions structurally characterized by sequential disassembly. Ten major ions were selected for detailed analysis. Isomer topologies (glycan connectivity) were determined from ion pathways of disassembly. Linkage information was revealed by specific cross-ring cleavage fragments within smaller oligomers. Both connectivity and linkage features were assisted with described bioinformatic tools and details confirmed with a standards library of fragments. The number of isomeric structures found within these 10 parent ions were 37, more than double earlier reports, and setting a new goal for developing technology. In this non-chromatographic, high performance spatial approach, the focus has been patterned to be comprehensive, and stay within the bounds of a plausible high throughput strategy consistent with automation. Selective structures are described in the text to appraise readers of the general approach; a more comprehensive coverage has been included in supplemental material.
Keywords: isomers, linkage, ovalbumin, ion trap MS, high performance glycan, sequencing
1. Introduction
Structural glycobiology has an endemic sequencing problem with a great need to focus and contrast analytical protocols. Compounding this is a huge diversity of structures, so the hope for finding analytical continuity for all glycoconjugates (GAGs, LPS, etc.) would appear unlikely. In that regard however, a more defined set of structures might be considered feasible, for instance, the N- and O-linked glycans of a protein, the glycoproteome. Readily available glycoprotein standards would provide opportunities for in-house validation and possible benchmarks for future instrumental development, both fundamental cornerstones for progress. Such a list would be a remarkable achievement for this important and evolving field. With that consideration, ovalbumin has served in that role for a number of years. Although commercial ovalbumin lots lack protein purity, N-glycan characterization does provide an operational opportunity to evaluate interresidue linkage, monomer connectivity, branching and, in this case, a preponderance of structural isomers. Establishing strategies to address these components of structure would advance this discipline immeasurably and give credence to the meaning of comprehensive.
Stereo and structural isomers are largely absent in glycoproteome reports, as they are hidden to many analytical approaches and transparent to mass measurement alone. This inability to detect or observe such structures often projects to false conclusions that an analysis is complete. This absence of evidence is not evidence of absence, and structures envisioned from biological inference[1], higher mass resolution[2, 3], or methylation linkage analysis[4], include measures of self interpretation[5], not data to define an exacting structure. In this study we omit the challenge of anomeric structure, not because this detail is not available, but we have no precise standards to contrast and define spectral data. The need for a detailed characterization that includes linkage, branching, stereo and structural isomers, for all monomers brings enormous analytical challenge, but omitting such detail exposes a missed opportunity to define functional relationships. From a functional standpoint, R. D. Cummings (Emory University) has reported the glycome may contain some 3000 determinants[6]. This was considered to be an achievable target for new chemical and/or enzymatic syntheses, but validation brings an enormous analytical challenge, especially when structural protocols for characterization are incomplete. Many reported determinants are grouped behind brackets and assumed on the basis of a composition, biological insight, or a bioinformatic projection. The default strategy for resolving structural complexity is chromatography, with separations led by nanoLC and miniaturization on chips. Some schemes have shown exceptional resolution under exacting parameters[7], but the resolving power required to identify all components in a tissue glycome would appear unlikely. With the complication of both resolving and identifying structures, we have considered attacking the problems collectively utilizing the elegant features of the ion trap to mass select and disassemble. With characteristic signature ions, scars of former linkage, and a searchable library of fragments, patterns of monomer connectivity become accessible, [8–11], and with adequate ion current, most structural features can be assigned. Fragment ions inconsistent with a single precursor (isomer) can be detected, isolated and resolved somewhere along a disassembly pathway. Spatial resolution has been shown to be largely a result of two features, stereospecific metal ion binding (which yields unique collision products), and methylation which places components of structure in a glycan array. These features are assisted with a searchable library of small oligomers. In applying this technology, we introduce the term high-performance glycan sequencing to signify an approach germane to most structural features, somewhat analogous to high-performance protein sequencing recently introduced[12].
Spatial resolution and direct infusion gives concern for ion suppression in the competitive electrospray plume, but sample workup (enzymatic release, SPE, derivatization, and lipophilic extraction) contributes to the critical need for enrichment. It remains clear however, that charge competition sets the limitations to structural interrogation by direct infusion, and further improvements in this area would be most helpful[13]. In practice, this sequencing technology has resolved over 80 isomers within the N-linked core of C. elegans [8], reported multiple new isomers in the common standard glycans of ribonuclease B[14], and doubled the number of N-glycans previously reported in human plasma[10]. In this account we extend the application to ovalbumin with comparable results. Our motives remain the same, to bring attention to the limitations of current analytical MS/MS efforts and to raise the bar of analysis for others to challenge and improve.
2. Experimental
2.1 Materials
Chicken egg ovalbumin (grade V), sodium hydroxide (pellets 99.998%), Dowex 50wx8-400 ion-exchange resin, dimethylsulfoxide, anhydrous (99%), sodium borohydride(99%) and iodomethane (99.5%) were obtained from Sigma-Aldrich(St. Louis, MO). Peptide-N-glycosidase F (PNGase F) deglycosylation kits were purchased from New England Biolabs Inc. (Ipswich, MA). All solvents (acetonitrile, acetate, methanol, water, toluene, methylene chloride and ethanol) were HPLC grade and purchased from Fisher Scientific (Fair Lawn, NJ). SPE C18, reservoir and Carbogragh SPE (120/400 mesh) were purchased from Alltech Associates Inc. (Deerfield, IL).
2.2 Sample Preparation
N-linked glycans from 2 mg of chicken ovalbumin were released by PNGase overnight, following the manufacturer’s instruction. The isolated glycans were reduced with sodium borohydride[9] and methylated[15]. The sample was re-suspended in 75% (v/v) methanol aqueous solution for MS analysis. For differential lithium adduct studies, a portion of the sample was dissolved in 10 mM lithium acetate previous to MS analysis.
2.3 Mass Spectrometry
Mass spectra were obtained with an LTQ (Thermo Fisher Scientific, Waltham, MA) equipped with a nanospray ion source. Samples were infused at flow rates ranging between 0.30 and 0.60 µL/min, and spectra were collected using Xcalibur 1.4 and 2.0 software (Thermo Fisher Scientific). Direct infusion ESI was carried out with a Triversa Nanomate (Advion, Ithaca, NY). Signal averaging was accomplished by 5 micro-scans within each scan and adjusting between 50 – 300 scans for each spectrum depending on signal intensity. Collision parameters were left at default values 35% or to a value leaving a minimal precursor ion peak. Activation Q was set at 0.25 and activation time at 30 ms. Ions were generated by adduction of indigenous levels of sodium. Two software tools have been written to process spectral data from this instrument. They have been identified as Glycoscreen and Composition Finder (Cfinder)[11]. GlycoScreen has been designed to filter out background contaminants and consider only those ions possessing carbohydrate compositions. Common carbohydrates, N-, O-linked glycans and their modifications are considered as input data. The output defines specific isotopic distributions to rebuild MS profiles, thereby facilitating data analysis. The general features include fitting profile data to glycan and carbohydrate-specific ions and assigning compositions to each ion. The fitting score is generated to quantify how well an isotopic distribution fits the theoretical one. The following formula was used, where n is the number of peaks within the isotopic envelop considered; Oi and Ei are the signal intensities of the i th isotopic observed and expected (theoretical) peaks respectively.
Cfinder generates compositions of each N-linked or O-linked precursor ion and is able to derive parent ion topology using logic reasoning. For MSn data, the software considers the constraints of the precursor ion and returns only substructure products. For determining topology, both the observed ion and neutral loss compositions are reported.
Product ion compositions from disassembly of the precursor, m/z 1799.7 (Fig. 1) are presented in linear notation (Table 2). The syntax established for coding Cfinder is presented in the format, [Type of monomer] [Number of monomer]–[Type of scar] [Number of scar(s)].
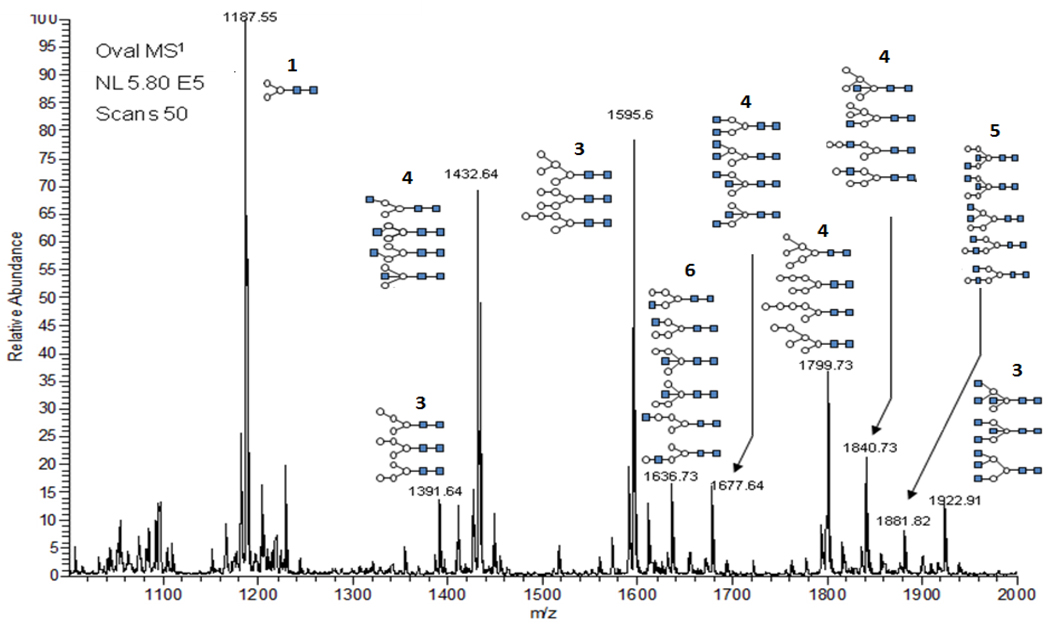
Figure 1.
Mass ion profile (IT-MS1) of N-glycans released from commercial grade hen ovalbumin following SPE purification, reduction and methylation. The non-chromatographic, direct nanoESI-MS1 analysis identified twenty-five ion compositions that were confirmed to be N-glycans by MS2 analysis. Ten ions were selected for 'in space' IT-MSn characterization which identified and additional twenty-six isomers for a total N-glycome count of thirty-six independent glycan structures.
Table 2.
Screenshot provided by software tool (Cfinder) for calculating fragment ion comp ositions, and by difference, the neutral loss fragments. Compositions are abbreviated in linear notation with asterisks (scars) identifying points of former linkage. Depth of MSn inquire (left column) indicating disassembly sequence showing the pathway for the ion m/z 1799.6, H6N1NR1 from ovalbumin m/z 1800.0→1506.7→1084.5→839.4→621.3→431.2→259.1.
| Observed Peak (Na+) | Neutral Loss | |||
|---|---|---|---|---|
| Depth | Mass | Composition | Mass | Composition |
| MS1 | 1800.00 | H6N1NR1 | n/a | n/a |
| MS2 | 1506.73 | *H6N1 -(Pos1DB)1 | 293.27 | *NR1 -(Pos2/3/4/6OH)1 |
| MS3 | 1084.55 | **H4N1 -(Pos2/3/4/6OH)1 -(Pos1DB)1 | 422.18 | *H2 -(Pos1DB)1 |
| MS4 | 839.45 | **H4 -(Pos2/3/4/6OH)1 -(Pos1DB)1 | 245.1 | **N1 -(Pos2/3/4/6OH)1 -(Pos1DB)1 |
| MS5 | 621.27 | ***H3 -(Pos2/3/4/6OH)2 -(Pos1DB)1 | 218.18 | *H1 -(Pos1DB)1 |
| MS6 | 431.18 | **H2 -(Pos2/3/4/6OH)1 -(Pos1DB)1 | 190.09 | ***H1 -(Pos2/3/4/6OH)2 -(Pos1DB)1 |
| MS7 | 259.09 | *H1 -(Pos1OH)1 | 172.09 | no match |
3. Results and discussion
Reduced, methylated N-glycans released from ovalbumin were mass profiled (800–2000 Da) (Fig. 1) providing twenty-five prominent ions, more than half of which were doubly charged (Table 1). Ten of the most abundant ions (asterisk, Table 1) were selected for disassembly to ascertain isomer, linkage and branching details as described. The selected ions had the following compositions; H3N2, H4N2, H3N3, H5N2, H4N3, H3N4, H6N2, H5N3, H4N4 and H3N5; (H, hexose and N, HexNAc). Compositions were also determined using the ExPASy GlycoMod tool (http://ca.expasy.org/tools/glycomod/) with the following parameter limits: (i) monosaccharide residues: Hex ≤ 9, HexNAc ≥ 8; (ii) monoisotopic mass tolerance ± 0.5 Da, (iii) ions existed as [M + Na]+ ; and (iv) on the basis of enzymatic release all returned compositions were assumed to have a core structure of Manα1-6(Manα1-3)Manβ-4GlcNAcβ1-4GlcNAc. Using these delimiters compositions were established that satisfied all profiled ions (Table 1). Commercial samples of ovalbumin may vary between lots, with mixtures of other proteins, [16, 17]. Thus, an exacting comparison of structures could not be considered; however, the resolution of isomers within each glycan represents a more precise measure of glycosylation detail, a prime consideration of this study.
Table 1.
Listing of 25 N-glycan ion compositions released from ovalbumin (Commercial Grade) processed by GlycoScreen[Zhang, 2005 #38]. Abbreviations, H = Hexose, N = HexNAc, NR = HexNAcol. Ion derivative, PM-R = methylated and reduced. Asterisks identifies ions selected for study by sequential disassembly following direct injection by nanoESI-MSn. The following parameters were used for processing; relative intensity threshold, 0.025; max charge state, 3; minimum number of isotopic peaks, 3; m/z range, 500–2000; glycan type, N-Linked; positive ions by Na+ adduction; error tolerance, 200 ppm; derivative, methylated and reduced. A fitting score represents a numeric measurement reported by GlycoScreen to quantify how well an experimental isotopic distribution fits the theoretical value. This score ranges from 0.00 to 1.00. A fitting score of 1.00 means that the experimental and theoretical distributions are identical.
| Composition | Ion Type | Observed m/z | Fitting Score | Relataive Intensity | Theoretical m/z | Absolute Error (mu) | Relative Error (ppm) |
|---|---|---|---|---|---|---|---|
| H5N2 | PM-R [M+2Na]2+ | 809.4546 | 0.91 | 0.04 | 809.5070 | 0.0524 | 65 |
| H4N3 | PM-R [M+2Na]2+ | 829.9091 | 0.97 | 0.03 | 829.9028 | −0.0063 | −8 |
| H3N4 | PM-R [M+2Na]2+ | 850.4546 | 0.94 | 0.03 | 850.4805 | 0.0259 | 30 |
| H6N2 | PM-R [M+2Na]2+ | 911.4546 | 0.97 | 0.07 | 911.4571 | 0.0025 | 3 |
| H5N3 | PM-R [M+2Na]2+ | 931.9091 | 0.98 | 0.11 | 931.8529 | −0.0562 | −60 |
| H4N4 | PM-R [M+2Na]2+ | 952.4546 | 0.94 | 0.04 | 952.4306 | −0.0240 | −25 |
| H3N5 | PM-R [M+2Na]2+ | 973.0000 | 0.97 | 0.09 | 973.0082 | 0.0082 | 8 |
| H5N4 | PM-R [M+2Na]2+ | 1054.4546 | 0.96 | 0.19 | 1054.3807 | −0.0739 | −70 |
| H4N5 | PM-R [M+2Na]2+ | 1075.0000 | 1.00 | 0.09 | 1074.9583 | −0.0417 | −39 |
| H3N6 | PM-R [M+2Na]2+ | 1095.5455 | 0.97 | 0.26 | 1095.5360 | −0.0095 | −9 |
| H5H5 | PM-R [M+2Na]2+ | 1177.0000 | 0.97 | 0.08 | 1176.9084 | −0.0916 | −78 |
| H3N2 * | PM-R [M+Na]+ | 1187.4546 | 0.97 | 0.82 | 1187.2942 | −0.1604 | −135 |
| H4N6 | PM-R [M+2Na]2+ | 1197.5455 | 1.00 | 0.05 | 1197.4861 | −0.0594 | −50 |
| H3N7 | PM-R [M+2Na]2+ | 1218.0000 | 0.97 | 0.11 | 1217.8819 | −0.1181 | −97 |
| H4N7 | PM-R [M+2Na]2+ | 1320.0000 | 0.96 | 0.03 | 1319.8320 | −0.1680 | −127 |
| H3N8 | PM-R [M+2Na]2+ | 1340.5455 | 0.98 | 0.03 | 1340.4097 | −0.1358 | −101 |
| H4N2 * | PM-R [M+Na]+ | 1391.5455 | 0.97 | 0.20 | 1391.3762 | −0.1693 | −122 |
| H3N3 * | PM-R [M+Na]+ | 1432.5455 | 0.96 | 0.76 | 1432.3497 | −0.1958 | −137 |
| H5N2 * | PM-R [M+Na]+ | 1595.5455 | 0.97 | 1.00 | 1595.2764 | −0.2691 | −169 |
| H4N3 * | PM-R [M+Na]+ | 1636.5455 | 0.97 | 0.26 | 1636.2499 | −0.2956 | −181 |
| H3N4 * | PM-R [M+Na]+ | 1677.6364 | 0.98 | 0.23 | 1677.4052 | −0.2312 | −138 |
| H6N2 * | PM-R [M+Na]+ | 1799.6364 | 0.99 | 0.48 | 1799.3584 | −0.2780 | −154 |
| H5N3 * | PM-R [M+Na]+ | 1840.6364 | 0.98 | 0.35 | 1840.3319 | −0.3045 | −165 |
| H4N4 * | PM-R [M+Na]+ | 1881.6364 | 0.98 | 0.15 | 1881.3054 | −0.3310 | −176 |
| H3N5 * | PM-R [M+Na]+ | 1922.7273 | 0.97 | 0.24 | 1922.4608 | −0.2666 | −139 |
3.1 Topology and linkage determination of m/z 1677.6 (Fig. 1, #4, H3N4)
Structural details that resolved both topology and linkage isomers have been summarized for the ion, m/z 1677.7, (H3N4), (Fig. 2). When studied by IT-MSn this glycan was found to exist in three topologies (Fig. 2D) that included four different linkage isomers (Fig. 2E). Previous reports had indicated one structure for this composition using ion mobility-MS [18] and two reports indicated two isomers using MALDI-MS[16] and MALDI-PSD and CID [19]. Oddly, in a recent study using nanoLC/ESI/MS/MS all compositions listed (Table 1) were detected, but no isomers were reported [20]. These disparities may reflect some lot-to-lot variations in commercial ovalbumin preparations (as mentioned above), but demonstrating isomer presence could reflect a more perceptive analytical approach much needed for glycosylation surveillance.
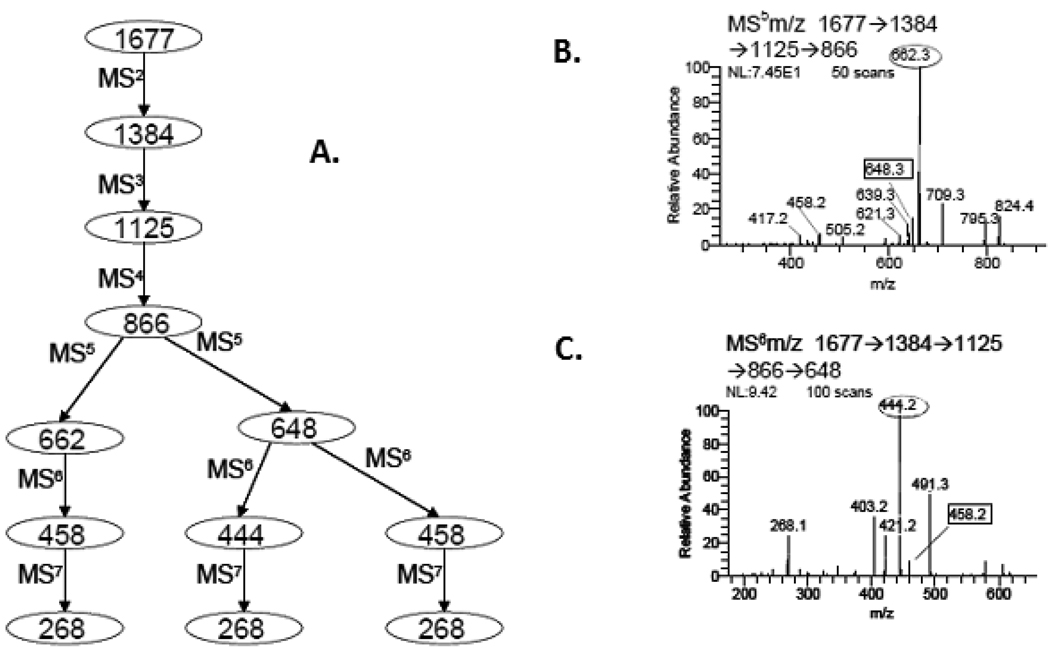
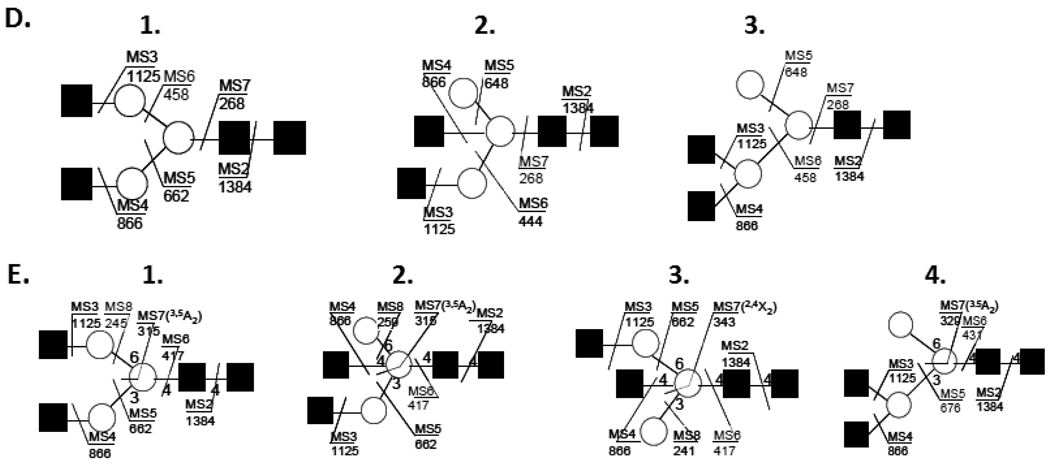
Figure 2.
(A.) Pathway of sequential disassembly (MSn) for the ion m/z 1677.64, (Fig. 1, #4) that resolved three isomeric topologies (D.) and four linkage isomers (E.) determined within the ion composition Hex3HexNAc4. Spectra of m/z 866, MS5 (B.) and m/z 648, MS6 (C.) exposed ion fragments inconsistent with a single structure, identified with boxed annotation, m/z 648.3 and m/z 458.2.
This detailed characterization started with isolation and collision (MS2) of the profiled ion m/z 1677.6, (Fig. 1 #4) which provided a prominent and specific B-ion, (m/z 1384.5, Fig. 2A), the result of a facile neutral mass loss equal to the reducing-end terminus, GlcNAcol; fixing its position in the structure. The composition written in linear notation would be, H3N3-(Pos1DB)1; three hexoses, three HexNAc, with one single scar at position 1. Selecting this product, m/z 1384.5, for further disassembly, MS3, provided a fragment ion, m/z 1125.4, *H3N2-(Pos1DB) with two scars indicating the loss of a fully methylated terminal HexNAc, (Fig. 2A). Without further inquiry one could conclude a linear oligomer with loss of its non-reducing termini. However, further analysis, MS4, (Fig. 2B) showed an additional loss of another fully methylated (terminal) HexNAc which can only mean a branched structure, supporting the topology, (Fig. 2D1). The spectrum of this product, MS5, (Fig. 2B) provided an expected loss of an extending hexose, (204Da, m/z 662.3), but a satellite fragment at m/z 648, indicated the loss of an additional terminal residue, (218Da). This could only arise if a core mannose was also a termination residue. To confirm such a product, the ion m/z 648 was isolated for MS6 analysis which provided a unique signature fragment as the base ion, m/z 444.2, (Fig. 2C) indicating a bisected GlcNAc, ***Hex1N1-(Pos1DB1) with four scars, (Fig. 2D2). This spectrum also exhibited another signature ion, m/z 458 ion, Man-GlcNAc with three scars **(Hex1N1-(Pos1DB1) and the penultimate GlcNAc monomer with two scars, *(N1-(Pos1DB1), (m/z 268.2, Fig. 3D3). By tracking the changes in ion composition and using logical reasoning, the putative glycan topology can be derived from these pathways. Branches in the disassembly scheme, m/z 866, 648 (Fig. 2A) indicate the presence of aberrant ions in the spectrum that were subsequently isolated and further disassembled for structural understanding. Such isomers are frequently suggested when satellite fragment ions are detected 14Da above or below a major fragment. This was observed in the MS5 and MS6 product spectrum of m/z 866 (Fig. 2B) and 648 (Fig. 2C). The paired 14Da satellite ions, m/z 662.3/648.3, 444.2/458.2, indicated the loss of a terminal core H and a bi-sected marker fragment ion which were keys to defining their topology. Importantly, these proposed isomers can be confirmed by isolating them, (m/z 648.3, 458.2, Fig. 2B and 2C) for further study by MSn. Thus, isolating m/z 648 via the pathway (m/z 1677→1384→1125→866→648→444→268) defined the topology presented (Fig. 2D2), and this pathway exposed an additional isomer in the spectrum (Fig. 2C). Selecting the 14Da satellite ion m/z 458 (signature ion) within the last step (Fig. 2C) (m/z 1677→1384→1125→866→648→458→268) indicated its structure. These signature ions are often signals for pathway selection, notably; such ions may not arise until the preceding spectrum, and hence are data dependent. However, it is highly probably that with data accumulation, including libraries of fragments and pathways will be resolved by automation.
Figure 3.
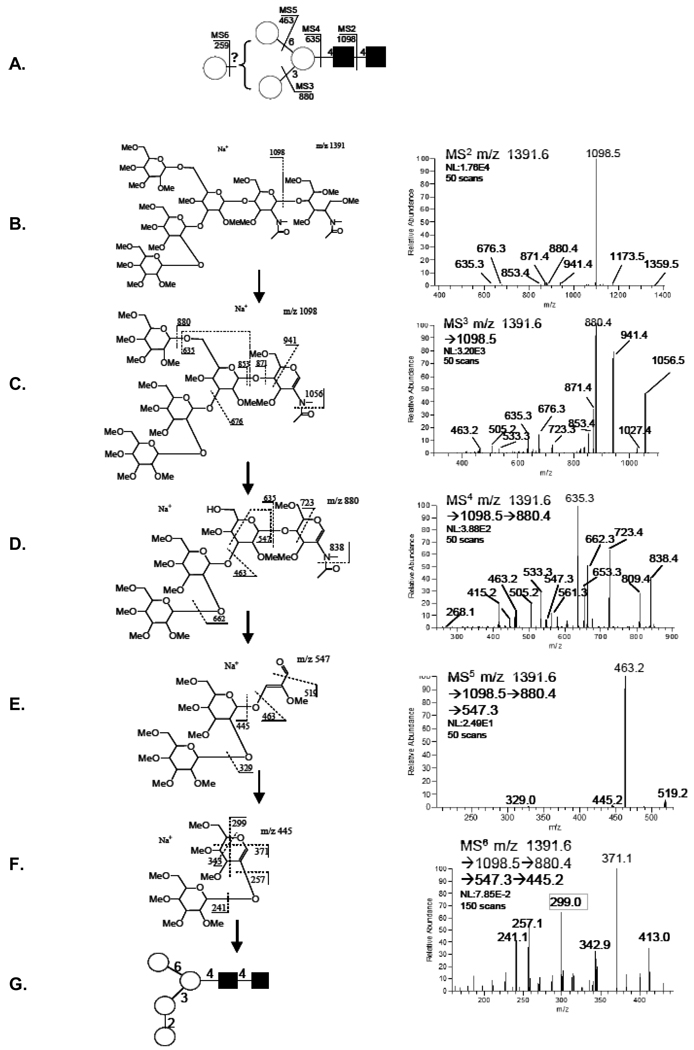
(A). Summary structure representing the single topology that included three linkage isomers detected for the composition H4N2 (Fig. 1, #3), annotated with fragments and pathways pursued. Proposed structure from resulting spectral data (B.–F.) for the assignment of linkage isomer (G.).
A linkage determination for all intervening monomers requires adequate sample signal, and in a complex glycome the ion current may dissipate before all linkages are assigned. Under such conditions, depletion of extraneous materials before analysis remains fundamental. In these studies sample workup using SFE, methylation, and lipophilic extraction provides significant clean-up so a direct infusion approach can be considered, steps not readily available for peptides. Thus, for the glycans of proteins, ion profiles (MS1) provide adequate signal for disassembly to the MS5–7 level for spectral evaluation and library searching, (see Fig. 2, B and C). Repetitive steps of isolation eliminate much biological noise allowing considerable sample signal amplification, but quantification is not possible in the absence of known ionization efficiencies.
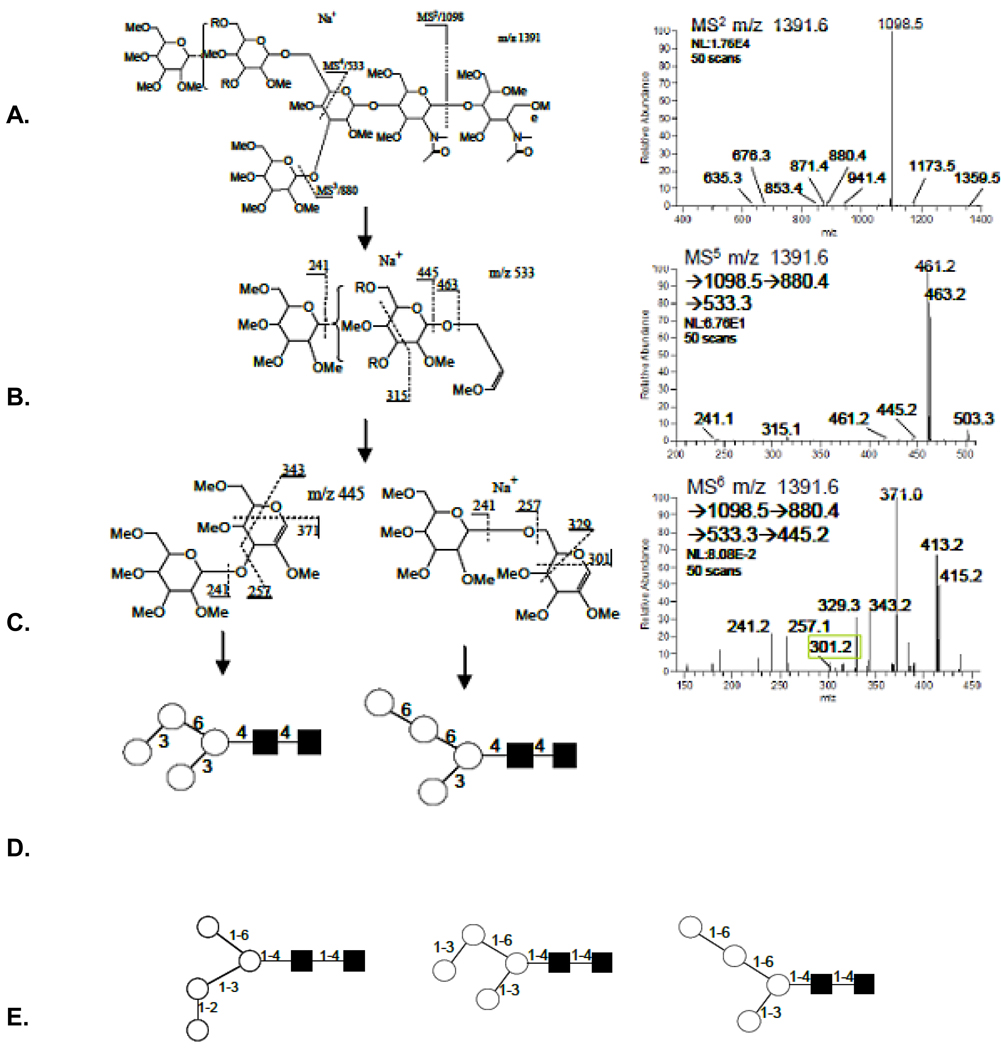
3.2 Topology and linkage determination, m/z 1391.6, (H4N2)
This simple ion composition H4N2 (Fig. 1, Table 1) was shown to be of a single topology in earlier reports (Fig. 3A) with linkage of the fourth hexose assigned enzymatically. Sequential IT-MSn analysis supported the single topology but resolved three linkage isomers. These details were exposed using known marker fragments that specified antennal details to the respective 6-, and 3-linked central core mannose through cross-ring cleavage ions detected upon MS3 analysis where comparable bond lability renormalizes spectra for improved detection. Tracking fragments of the 3-linked antenna illustrates these features; m/z 1391→1098→880→635→547→445 (Fig. 3, B–F). Neutral loss of the reduced GlcNAcol, provided the expected major fragment, m/z 1098 (Fig. 3B), which upon isolation and activation left the remaining linkages more energetically equal (Fig. 3C). Although dwarfed by the ions representing a non-terminal loss of H (m/z 880.4, 218Da, which could arise from either antennae), the spectra exhibited ions indicative of a disaccharide linked to both the 6-, and 3-positions of the central N-linked core mannose, (m/z 533.3, and 547.3). These marker fragments were specific for each with the 6-linked antennal exhibiting a 0,4A-type fragment and 3-linked antennal a sequential B- and 3,5A-type ion. This was amplified further upon isolation and collision of the m/z 880.4 fragment (MS4, Fig. 3D). It should be pointed out again that this target ion, m/z 880.4, was likely to consist of two isomeric fragments, one exhibiting a 3-, and the other a 6-linked H loss (Fig. 3C), and both linkage isomers were detected. The details of hexose linkage to the 3-linked mannose were available through the m/z 547.3 marker ion which upon isolation and fragmentation (Fig. 2E) provided a B-type ion (Fig. 2F) with all the cross-ring fragments expected from a 1–2 linked disaccharide. But no standard was available to confirm this expectation. A branched step in the pathway through the m/z 533.3 ion, a 3,5A-cross-ring cleavage fragment (Fig. 4b), (m/z 1391→1098→880→533→445) provides fragments linked from the 6-linked mannose core and the linking disaccharide (Fig. 4A). Disassembly of the m/z 533.3 fragment, MS5, provided disaccharide B-type ions (Fig. 4C), strongly suggesting a mixture of two linkage types, a 1–6 linkage characterized by the 3,5A- and 4,0A-type fragments, (m/z 329.3/301.2) and a prominent 1–3 linkage characterized by the 4,0A-type ion, (m/z 371.0) coupled with the absence of a 1–2 linkage fragments, 2,5A-type ions (m/z 299.0). These data support both linkage isomers one from the 6-antennae and one from the 3-antennae for the total of three (Fig. 4B).
Figure 4.
Spectra and proposed structures (A–C.) that resolved the two linkage isomers of the 6-linked central core mannose (D.). Absence of chemical standards prevented anomeric assignment from the spectra. Summed linkage isomers resolved from the single topology with composition H4N2 (E.).
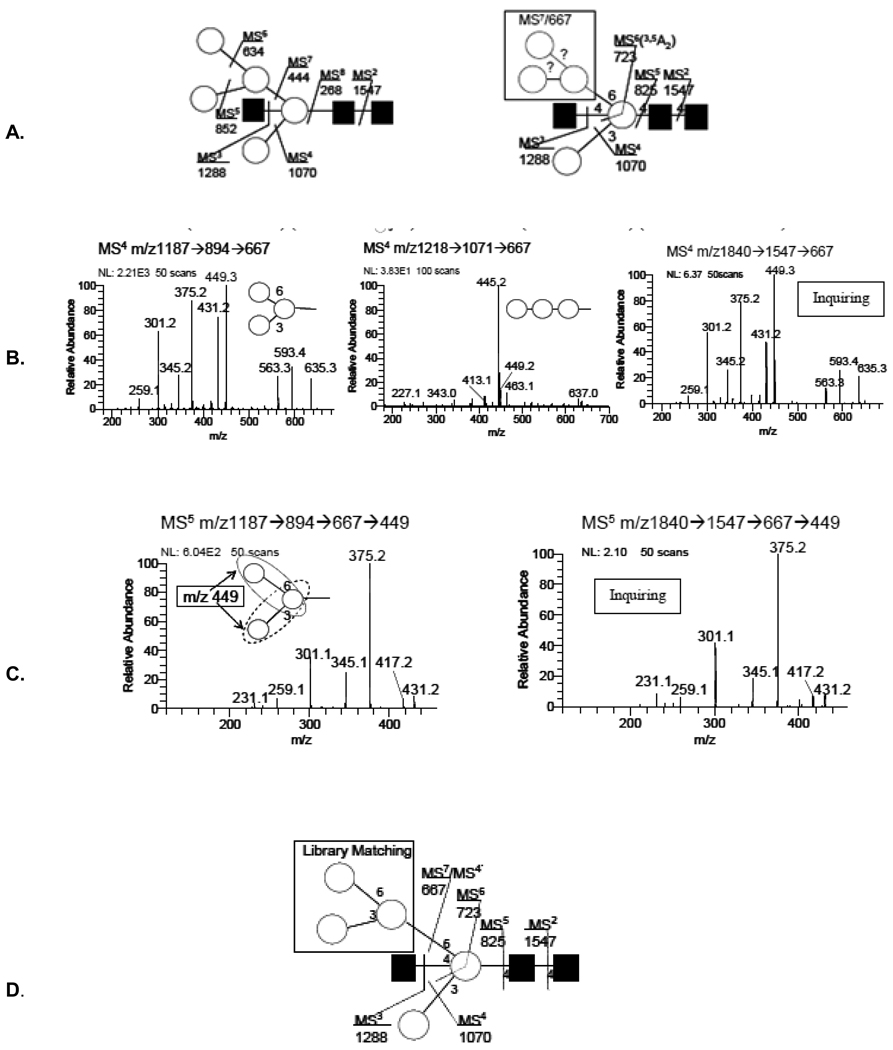
3.3 Topology and linkage determination, m/z 1840.8 (H5N3)
Within this profiled ion, m/z 1840.8, (Fig. 1, #4) four isomeric topologies were identified by IT-MSn with no linkage isomers apparent. Two isomers have been previously reported. MALDI-MS and enzymology [16] were used to define one topology and more recently an additional topology was reported using MALDI-MS post source decay and CID-MS/MS[19]. One of those structures has been detailed in this report, (Fig. 5). The glycosidic fragments and sequential disassembly pathways are annotated on one structure (a.) with substructure ions selected for detecting possible linkage isomers within the 6-linked antennae (b.)
Figure 5.
(A.) Pathways and fragments considered for the determination of structure for the composition (H5N3) (m/z 1840.7) (Fig. 1, #4). A total of 4 topologies were determined, two reported previously. [16, 19]
(B.) Trisaccharide substructure determination using library spectra. Comparison of the branched, and linear trimer with that fragment obtained in this study (inquiring) following the pathway, m/z 1840→1547→667 of H5N3. Results indicate the 6-linked antennae to be a branched structure.
(C.) Further disassembly of the C-type ion, m/z 667, from the library spectrum with that obtained from the ovalbumin, m/z 1840, following the pathway m/z 1840→1547→667→449. This latter fragment again shows spectral identity even though XXXXXXcould arise from two different parts of the precursor ion, m/z 667, (Fig. 5C), and the spectrum is a composite, (see circled disaccharide substructure) but their spectral identity strongly suggests a structural identity.
(D.) Final structure derived from a combination of IT-MSn disassembly and substructure library matching.
Different pathways can be utilized to isolate component substructures and their spectral identity provides additional confirming information. The core fragment, m/z 444, Man-GlcNAc with four scars, (3 on the Man) was a product of a number of pathways identifying a bi-sected substructure, m/z 1840→1547→1288→1070→852→634→444. An additional substructure of interest was available following the pathway, m/z 1288 →1070→825→723→667. This provided a C-ion trisaccharide connected to the core, through a 3,5A3-type cleavage ion (m/z 723), (b). Identical ions may appear in different pathways and the more direct (shorter) provides an enhanced spectrum for a library match, m/z 1840→1547→667. An inquiry of this ovalbumin trisaccharide C-type ion with the library revealed a branched (left spectrum) and linear (middle spectrum) structures confirmed the ovalbumin m/z 1840.73 ion (inquiring) to be a branched trisaccharide, (Fig. 5B). The source of the precursor ion (pathway) has no influence on the product ion spectrum. Further disassembly of this m/z 667 ion and isolating the product m/z 449.3 in MS4 showed them to provide an identical spectrum, (Fig. 5C). Contrasting and confirming substructures provides an opportunity to approach larger oligomers, as with this simple example, (Fig. 5D).
4. Conclusions
It is reasonable to expect that a glycome analysis of any discrete tissue (serum, saliva, organelle), would be a distribution of isomeric structures that would extend beyond the limits of detection. But, if biological function lies within such structures, a comprehensive analysis remains fundamental. To understand these products by MS analysis, it may be simplest to consider glycans as components of two structural features; monomer connectivity, (resolved by B/Y-, and C/Z-type fragment ions), and their interresidue linkages, (resolved by A-, and X-type fragment ions, Fig. S1). Without both ion types, a comprehensive sequence cannot be established. Structural isomers showing a different connectivity pattern (topology) can be differentiated, as in the case of trifucosyllacto-N-hexaose (TFLNH) and trifucosyl-paralacto-N-hexaose (TFpLNH),[21] but, in the absence of linkage information this evaluation remains incomplete. A biological response dependent on single structural feature needs little documentation. A review of blood typing epitopes demonstrates the remarkable specificity cast in minor structural details.
Products of IT-MSn disassembly have compositions, methylation patterns, hydroxyl groups and/or points of unsaturation that are important sequence identifiers. Fully methylated, unsaturated B-ions are exemplars of non-reducing termini, common in glycans with multi-antennae, and a fragment 14 Da lower in mass (open hydroxyl) indicating a former branch. Scar location on a monomer positions the fragment in its precursor structure, as exemplified by identifying a 6-hydroxyl on a galactosyl residue that specifically defined biological function in a 6-linked neuraminyl residue.[23] A fragment ion suggesting B-/Y-type glycosidic rupture should retain a composition that accounts for a pyranosyl-1-ene and/or a respective hydroxyl scar. Compositions possessing single scars represent chain termini. Multiple scars, induced by higher (effective) energies, arise near the end of a disassembly pathway, especially within fragments of tri-, di-, and monosaccharides. Such simple reasoning, the placement of termini, branching and extending fragments provides topology insight. Product ion confirmation can be supported by selecting different pathways that lead to identical spectral products. Ions smaller in mass (fewer oscillators) must dissipate the same collisional energy, and in so doing induce rupture of more stable and/or multiple bonds. These fragments are the A-/X-type cross-ring fragments utilized for linkage assignment (Fig. S1). The only limitation to a comprehensive structural understanding is insufficient ion current. Anomericity remains an open question, and although spectral differences have been observed when appropriate standards were available, (known disaccharides), such details were not considered in this study.
Rarely do glycans of proteins exist as single structures, most frequently they occur as a distribution of topologies and linkage isomers. Thus, MS/MS analysis alone would be unsuitable for a comprehensive analysis, nor does higher resolution provide any significant advantage. As outlined in this application and earlier studies, the coupling of ion isolation and spatial resolution exposes features of topology and linkage without the need for chromatography. Tissue glycomes may seem insurmountable, but the technology discussed here has been focused with those thoughts in mind, (direct infusion, spatial resolution, and library confirmation), and such considerations may offer some reprieve from this two century old problem of carbohydrate sequencing.
Supplementary Material
Acknowledgements
The authors wish to acknowledge the excellent technical support provided by David Ashline, the supporting role of the Chemistry Department while located in their environs and support provided for student scholars. Financial support was provided by NIH, NIGMS, GM-054045; NCRR, RP016459, and Glycan Connections, Lee, NH.
Abbreviations
- SPE
solid phase extraction
- IT-MSn
Ion trap mass spectrometry
- IM-MS
ion mobility mass spectrometry
- LC-MS
liquid chromatography mass spectrometry
- LPS
lipopolysaccharides
- GAG
glycosaminoglycans
- GlcNAcol
reduced terminal residue
- H
hexose
- N
HexNAc
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clark GF, Dell A. Molecular models for murine sperm-egg binding. J Biol Chem. 2006;281(20):13853–13856. doi: 10.1074/jbc.R600001200. [DOI] [PubMed] [Google Scholar]
- 2.Desaire H. When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? International J. of Mass Spectrometry. 2009;287:21–26. [Google Scholar]
- 3.Wang X, Emmett MR, Marshall AG. Liquid chromatography electrospray ionization Fourier transform ion cyclotron resonance mass spectrometric characterization of N-linked glycans and glycopeptides. Anal Chem. 2010;82(15):6542–6548. doi: 10.1021/ac1008833. [DOI] [PubMed] [Google Scholar]
- 4.Canis K, et al. The plasma von Willebrand factor O-glycome comprises a surprising variety of structures including ABH antigens and disialosyl motifs. J Thromb Haemost. 2010;8(1):137–145. doi: 10.1111/j.1538-7836.2009.03665.x. [DOI] [PubMed] [Google Scholar]
- 5.Packer NH. Introducing Glycoproteomics, a new section of PROTEOMICS. Proteomics. 2006;6:6121–6123. [Google Scholar]
- 6.Cummings RD. The repertoire of glycan determinants in the human glycome. Molecular BioSystems. 2009;5(10):1077–1248. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 7.Prien JM, Prater BD, Cockrill SL. A multi-method approach toward de novo glycan characterization: A Man-5 case study. Glycobiology. 2010;20:629–647. doi: 10.1093/glycob/cwq012. [DOI] [PubMed] [Google Scholar]
- 8.Hanneman AJ, et al. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 2006;16(9):874–890. doi: 10.1093/glycob/cwl011. [DOI] [PubMed] [Google Scholar]
- 9.Ashline D, et al. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal Chem. 2005;77(19):6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpo KAR, Vernon N. The N-Glycome of Human Plasma. Journal of Proteome Research. 2010;9(8):4823–4830. doi: 10.1021/pr100528k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhold Vernon N, Ashline David J, Zhang Hailong. Unraveling the Structural Details of the Glycoproteome by Ion Trap Mass Spectrometry. In: March RE, Todd JFJ, editors. Practical Aspects of Trapped Ion Mass Spectrometry. Boca Raton, Florida: CRC Press, Taylor & Francis Group; 2010. pp. 707–738. [Google Scholar]
- 12.Meyer HE, Stuhler K. High-performance proteomics as a tool in biomarker discovery. Proteomics. 2007;7 Suppl 1:18–26. doi: 10.1002/pmic.200700183. [DOI] [PubMed] [Google Scholar]
- 13.Tu C, et al. Depletion of Abundant Plasma Proteins and Limitations of Plasma Proteomics. Journal of Proteome Research. 2010 doi: 10.1021/pr100646w. e-file Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prien JM, et al. The high mannose glycans from bovine ribonuclease B isomer characterization by ion trap MS. J Am Soc Mass Spectrom. 2009;20(4):539–556. doi: 10.1016/j.jasms.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciucanu I, Kerek F. A Simple and Rapid Method for the Permethylation of Carbohydrates. Carbohydrate. Res. 1984;131(2):209–217. [Google Scholar]
- 16.Harvey DJ, et al. Composition of N-linked carbohydrates from ovalbumin and co-purified glycoproteins. J Am Soc Mass Spectrom. 2000;11(6):564–571. doi: 10.1016/S1044-0305(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 17.Larsen MR, Hojrup P, Roepstorff P. Characterization of gel-separated glycoproteins using two-step proteolytic digestion combined with sequential microcolumns and mass spectrometry. Mol Cell Proteomics. 2005;4(2):107–119. doi: 10.1074/mcp.M400068-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Isailovic D, et al. Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. J Proteome Res. 2008;7(3):1109–1117. doi: 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lattova E, Perreault H, Krokhin O. Matrix-assisted laser desorption/ionization tandem mass spectrometry and post-source decay fragmentation study of phenylhydrazones of N-linked oligosaccharides from ovalbumin. J Am Soc Mass Spectrom. 2004;15(5):725–735. doi: 10.1016/j.jasms.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 20.Luo Q, et al. Hydrophilic interaction 10 microm I.D. porous layer open tubular columns for ultratrace glycan analysis by liquid chromatography-mass spectrometry. J Chromatogr A. 2009;1216(8):1223–1231. doi: 10.1016/j.chroma.2008.09.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohmer M, et al. 3-Aminoquinoline acting as matrix and derivatizing agent for MALDI MS analysis of oligosaccharides. Anal Chem. 2010;82(9):3719–3726. doi: 10.1021/ac1001096. [DOI] [PubMed] [Google Scholar]
- 22.Ashline DJ, et al. Carbohydrate structural isomers analyzed by sequential mass spectrometry. Anal Chem. 2007;79(10):3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anthony RM, et al. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320(5874):373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domon B, Costello CE. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconjugate Journal. 1988;5:397–409. 1988. 5: p. 397–409. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.