Abstract
Exome sequencing is a powerful tool for discovery of the Mendelian disease genes. Previously, we reported a novel locus for autosomal recessive non-syndromic mental retardation (NSMR) in a consanguineous family [Nolan, D.K., Chen, P., Das, S., Ober, C. and Waggoner, D. (2008) Fine mapping of a locus for nonsyndromic mental retardation on chromosome 19p13. Am. J. Med. Genet. A, 146A, 1414–1422]. Using linkage and homozygosity mapping, we previously localized the gene to chromosome 19p13. The parents of this sibship were recently included in an exome sequencing project. Using a series of filters, we narrowed the putative causal mutation to a single variant site that segregated with NSMR: the mutation was homozygous in five affected siblings but in none of eight unaffected siblings. This mutation causes a substitution of a leucine for a highly conserved proline at amino acid 182 in TECR (trans-2,3-enoyl-CoA reductase), a synaptic glycoprotein. Our results reveal the value of massively parallel sequencing for identification of novel disease genes that could not be found using traditional approaches and identifies only the seventh causal mutation for autosomal recessive NSMR.
INTRODUCTION
Mental retardation (MR) is among the most common disabilities among children, affecting 1–3% of the population (1,2). The majority of MR cases have isolated MR without other associated abnormalities, a condition that is referred to as non-syndromic mental retardation (NSMR) (3). The genetic etiology of NSMR has been characterized in only ∼10% of reported cases, with just 19 X-linked and six autosomal genes reported to date (4–10). It has been suggested that genes implicated in NSMR may also harbor mutations that are associated with other developmental abnormalities, such as autism (11,12) and schizophrenia (12,13), so identifying mutations for NSMR could identify candidate genes for these common conditions.
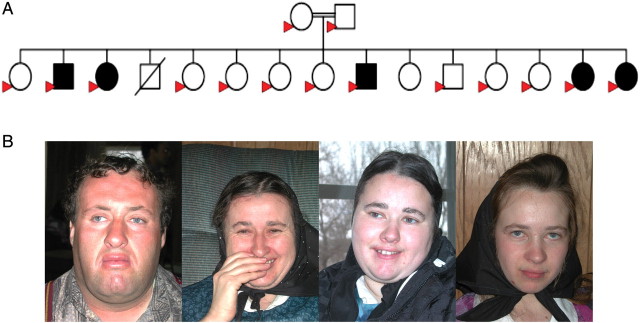
We previously described a consanguineous family (referred to as Family G) with five adult children with NSMR (14) (Fig. 1A). This family belongs to a religious community that traces its ancestry to Europe. The known pedigree that includes all members of this community is 15 generations deep. Four of the affected siblings (a brother and three sisters) were examined by D.W. (Fig. 1B). Growth parameters, including head circumference, were normal and there were no dysmorphic features other than a narrow palate in all four siblings. The neurological exam showed normal muscle bulk and tone, and normal cranial nerves and peripheral reflexes. Three of the affected individuals had an intention tremor when asked to perform finger–nose–finger exam or any other task requiring fine motor coordination and had slow rapid finger movements. There were no resting tremors, signs of ataxia or other abnormal movements. One affected brother, who was not available for examination, was reported to also have an intention tremor. One of the sisters was not considered as affected in our earlier study (14) because her complications were attributed by the parents to a difficult delivery and fevers in the newborn period. Her developmental delay was milder than the other three affected individuals who were examined, but she had similar speech, language and fine motor skill abnormalities. However, because she was homozygous for the same shared haplotype as her affected siblings, we thought it likely that she had the same autosomal recessive condition. Because this condition was segregating in only a single sibship and the locus mapped to one of the most gene rich regions of the human genome, we were previously unsuccessful in identifying the specific gene or mutation in this family (14).
Figure 1.
(A) Pedigree of the Family G. DNA was available for the individuals marked with a filled red arrow. The kinship coefficient, θ, for the parents is 0.059. The 545C → T mutation in the TECR gene (Pro182Leu) was homozygous in the five affected siblings and was heterozygous in the parents and seven of eight unaffected siblings. One deceased son (affection status unknown) and one unaffected daughter were not included in this study. (B) Four of the siblings with NSMR are shown. Photographs reprinted with permission.
RESULTS
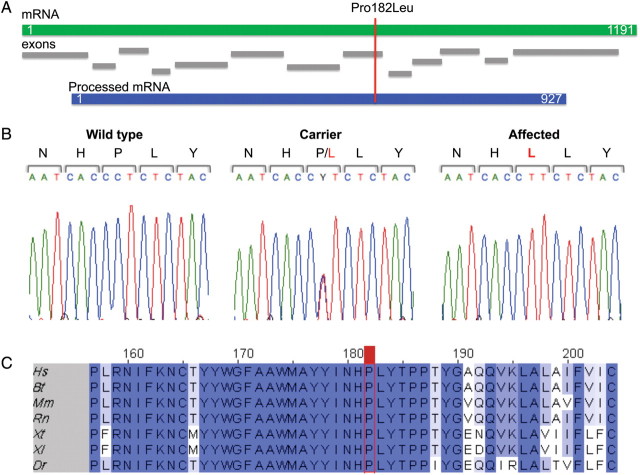
Following our initial linkage study (14), we performed whole-genome genotyping of the affected individuals and identified a shared homozygous segment that spans >2 Mb on chromosome 19p13 (Chr 19: 13 610 401–15 645 116 bp in hg Build 36.3). To facilitate the discovery of the causal mutation and gene, we included both obligate carrier parents in an exome sequencing study. We identified 71 variants (33 missense, 37 synonymous, 1 non-protein coding) in the critical region in the parents. Given the pattern of homozygosity in the critical region in Family G, we assumed that the NSMR in this family was due to a fully penetrant autosomal recessive mutation (14). Therefore, we considered 18 of the 71 variants that were heterozygous in both parents as candidates for the NSMR mutation (Supplementary Material, Table S1). Seventeen of these 18 variants were present in the dbSNP (v131) and were, therefore, further excluded as NSMR mutations (Supplementary Material, Table S2). The remaining novel mutation occurred in exon 8 of the TECR (trans-2,3-enoyl-CoA reductase) gene (Chr 19: 14 536 653 in hg Build 36.6), causing a Pro to Leu substitution at amino acid 182 in the TECR protein (Fig. 2A). Using Sanger sequencing, we confirmed that the mutation in the TECR gene segregated with NSMR in Family G: the non-reference allele was homozygous in the five affected siblings, heterozygous in the parents and seven unaffected siblings and absent in one unaffected sibling (Fig. 2B).
Figure 2.
(A) Schematic representation of the TECR primary transcript, exons and processed transcript. The location of the mutation affecting Pro182 in exon 8 of the TECR gene is indicated with a red line. (B) Results of sequencing studies. The nucleotide sequences corresponding to amino acids 180–184 in an affected, a carrier and a non-carrier sibling. (C) Cross-species sequence alignment of this segment of the TECR protein. The red line indicates the location of the highly conserved proline at amino acid 182 that is mutated to leucine in NSMR. Sequences were obtained from NCBI. Hs, Homo sapiens; Bt, Bos taurus; Mm, Mus musculus; Rn, Rattus norvegicus; Xt, Xenopus tropicalis; Xl, X. laevis; Dr, Danio rerio.
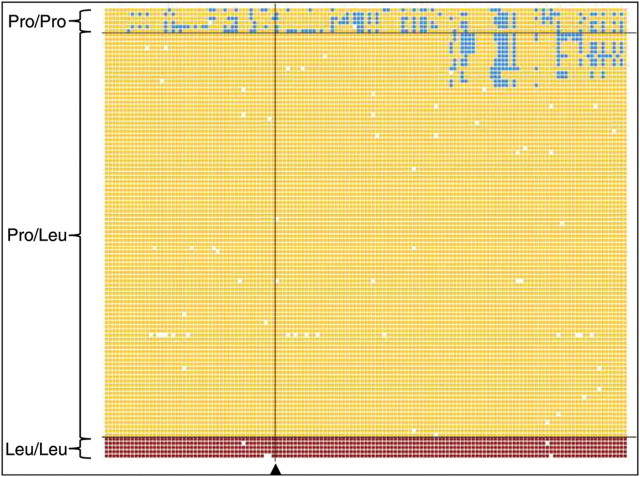
Additional genotyping of this mutation in 1523 individuals that belong to the same religious community as Family G revealed no homozygotes other than those in the affected sibship. Overall, there were 109 carriers of the TECR Leu182 mutation (including those in Family G), yielding a carrier frequency of 7.1% and mutation frequency of 3.9% in this population. The observed genotype distributions fit with the Hardy–Weinberg expectations (P > 0.10). Despite the high carrier frequency of the TECR mutation (1 in 14), none of the other carriers other than the parents of Family G were married to each other. Ninety-seven of 109 carriers were also genotyped with the Affymetrix 500k or 6.0 SNP arrays. In those individuals, the mutation was present on the same haplotype that was homozygous in the affected individuals in Family G, indicating a single ancestral founder haplotype carrying the mutation in this population (Fig. 3). The combined segregation and population data suggest that the Pro182Leu mutation in the TECR gene is the NSMR causal mutation in Family G.
Figure 3.
Haplotype analysis of the Pro182Leu mutation; the arrowhead shows the position of the mutation. The x-axis shows the 141 SNPs that define the 2.2 Mb homozygous segment in the affected siblings; individuals representing each genotype are shown in each row, grouped by the three Pro182Leu genotypes. All Pro/Leu carriers shared at least one allele (orange) with the affected individuals at every SNP within a 1.6 Mb interval surrounding the TECR mutation; blue shows SNP genotypes that do not share either allele with the homozygous segment in the affected siblings.
We next applied bioinformatic approaches to predict the potential effect of the Pro182Leu mutation. First, we used ClustalW algorithm (15) to align the human TECR protein sequence with the amino acid sequences of the orthologous proteins in six other species. The proline at amino acid 182 was highly conserved, being present in all species as distantly related to humans as zebrafish and Xenopus laevis (Fig. 2C). We then used two in silico methods: Polymorphism Phenotyping (PolyPhen) (16) and Sorting Intolerant from Tolerant (SIFT) (17). The effect of Pro182Leu mutation was predicted to be ‘probably damaging’ by PolyPhen (PSIC score difference 2.97) and ‘not tolerated’ by SIFT with a substitution score of 0.01. Thus, two in silico approaches predict that the substitution of a leucine for a highly conserved proline at amino acid 182 in the TECR protein is a deleterious/damaging mutation that very likely alters protein function and, therefore, downstream phenotypes.
DISCUSSION
TECR, also referred to as GPSN2 (synaptic glycoprotein 2), is a synaptic glycoprotein that is involved in the synthesis of very long-chain fatty acids (VLCFA) in a reduction step of the microsomal fatty acyl-elongation process (18). The mouse ortholog of TECR is highly expressed in the nervous system (19). Diseases involving perturbations to normal synthesis and degradation of VLCFA (e.g. adrenoleukodystrophy and Zellweger syndrome) have significant neurological consequences. For example, mutations in FACL4, a long-chain acyl-CoA synthetase gene that is important in the degradation of VLCFA and the production of key intermediates in the synthesis of complex lipids, cause X-linked NSMR (20–22). The role of TECR in fatty acyl elongation and its high expression in nervous system suggest that the Pro182Leu mutation might perturb similar pathways and result in NSMR. On the other hand, it is also possible that TECR, as a synaptic glycoprotein, may have a specialized, as yet unknown, function in the nervous system that affects communication between neurons (23) or synaptic plasticity (24,25).
We describe here for the first time the discovery of a mutation underlying an autosomal recessive condition by performing exome sequencing in obligate carrier parents of an affected sibship. The success of this study was further facilitated by having previously localized the disease gene to an ∼2 Mb region on chromosome 19 and by conducting these studies in a founder population in which most recessive conditions will be due to mutations that are homozygous by descent in affected individuals. However, this approach should also be applicable to mapping disease genes in the absence of prior localization, although there will likely be more candidates to screen after the initial filtering, and in non-founder populations in which affected individuals could be compound heterozygotes for different mutations in the same gene. Lastly, the Pro182Leu mutation in Family G might be private to this population. However, it is possible that other mutations in the TECR gene underlie NSMR, or even related phenotypes, such as schizophrenia and autism, in other populations. In fact, TECR was previously identified as a candidate schizophrenia gene with high probability (P < 0.001) by modeling genome-wide linkage and molecular interaction data (26) and was among the top-ranked nervous system genes identified as a candidate for neurological and psychiatric diseases in a study of tissue-specific gene expression patterns (19). Thus, further studies of the TECR gene in patients with neurological and psychiatric diseases, as well as with NSMR, may reveal additional mutations in this gene and a common biochemical pathway for these common conditions.
We report here the discovery of only the seventh autosomal gene harboring a putatively pathogenic mutation for NSMR and only the second involving fatty acid metabolism. Although the precise mechanism through which the Pro182Leu mutation in TECR leads to MR is not known, the role of this gene in fatty acid synthesis and the phenotypes of the affected individuals suggest that NSMR observed in Family G is an inborn error of metabolism in which fatty acids are not properly synthesized, highlighting the importance of normal lipid homeostasis for proper nervous system development.
MATERIALS AND METHODS
Phenotype studies
Family G was identified during the studies of common diseases in this population (27). The affected siblings were examined by D.W. as described previously (14). These studies were approved by the University of Chicago Institutional Review Board. Informed consents were obtained from the parents of Family G.
Exome sequencing
Genomic DNA was extracted from peripheral blood using the standard procedures. Five micrograms of DNA from each of the parents of Family G was used to construct a shotgun sequencing library. DNA oligonucleotides, corresponding to 170 bp of target sequence flanked by 15 bp of universal primer sequence, were synthesized in parallel on an Agilent 244K microarray, and then cleaved from the array. The oligonucleotides were PCR amplified, then transcribed in vitro in the presence of biotinylated UTP to generate single-stranded RNA ‘bait’. Genomic DNA was sheared, ligated to Illumina sequencing adapters and selected for lengths between 200 and 350 bp. This ‘pond’ of DNA was hybridized with an excess of bait in solution. The ‘catch’ was pulled down by magnetic beads coated with streptavidin, then eluted. Each sample was sequenced on three lanes of an Illumina Genome Analyzer II (GAII) sequencer using 76 bp end reads.
The exome target array covered 32.8 Mb of human genome, which includes 79.2 kb of coding sequence in our ∼2 Mb critical region. The latter corresponds to 100% of the NCBI Consensus Coding Sequence (CCDS) in the chromosome 19p13 critical region in Family G. We obtained an average of 103× coverage in the target region and achieved coverage of ≥4× for 91% of the CCDS bases and ≥8× for 85.6% of the CCDS bases in the critical region in at least one parent (Supplementary Material, Fig. S1 and Table S3).
Base quality scores were recalibrated and reads were locally realigned with the Genome Analysis Toolkit (GATK; M. DePristo et al., unpublished data). SNPs were called with the GATK with a minimum quality score of 50 and then filtered to remove those with SNP quality score over sequencing depth <5 (low-quality SNP sites), allelic balance among heterozygotes >75% (indicative of low-frequency errors), and neighboring repeated homopolymers of length >3 (often problematic in exome sequencing). Annotations of variants were performed based on NCBI and UCSC databases.
Sanger sequencing of the candidate variant in Family G
A 506 bp fragment harboring the candidate mutation on Chr 19: 14 536 653 (hg Build 36.6) was amplified using 5′-GCCTGCCTATCCACCCTAGT-3′ and 5′-CCAGCCTCCTTACCACAAAG-3′ primers. Sequencing was performed using ABI PRISM 3.0 BigDye terminator chemistry. Sequences were analyzed using FinchTV 1.4 (Geospiza Inc., Seattle, WA, USA).
Genotyping of the candidate variant in a population sample
The candidate mutation in the TECR gene was genotyped using TaqMan assay-by-design (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's protocols. Primer and probe sequences were designed using Primer Express 3.0 software (Applied Biosystems), and genotypes were analyzed using the allelic discrimination software of the 7900 HT Fast Real-Time PCR system (Applied Biosystems). There were no Mendelian errors in the sample, and the genotypes of 20 duplicated samples were 100% concordant. Observed genotype frequencies were compared with expected frequencies using a Hardy–Weinberg test that corrects for the relatedness between individuals in the population (28).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by R01 HD21244, R01 HL085197 and the NHLBI-funded sequencing center at the Broad Institute.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the members of Family G for their participation in this study; Dr Daniel Nolan for previous mapping studies in Family G.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Curry C.J., Stevenson R.E., Aughton D., Byrne J., Carey J.C., Cassidy S., Cunniff C., Graham J.M., Jr, Jones M.C., Kaback M.M., et al. Evaluation of mental retardation: recommendations of a consensus conference: American College of Medical Genetics. Am. J. Med. Genet. 1997;72:468–477. doi: 10.1002/(sici)1096-8628(19971112)72:4<468::aid-ajmg18>3.0.co;2-p. doi:10.1002/(SICI)1096-8628(19971112)72:4<468::AID-AJMG18>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 2.Chelly J., Khelfaoui M., Francis F., Cherif B., Bienvenu T. Genetics and pathophysiology of mental retardation. Eur. J. Hum. Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. doi:10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- 3.Ropers H.H., Hamel B.C. X-linked mental retardation. Nat. Rev. Genet. 2005;6:46–57. doi: 10.1038/nrg1501. doi:10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 4.Inlow J.K., Restifo L.L. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1534/genetics.166.2.835. doi:10.1534/genetics.166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisik M.Z., Sieron A.L. X-linked mental retardation. Med. Sci. Monit. 2008;14:RA221–RA229. [PubMed] [Google Scholar]
- 6.Basel-Vanagaite L. Genetics of autosomal recessive non-syndromic mental retardation: recent advances. Clin. Genet. 2007;72:167–174. doi: 10.1111/j.1399-0004.2007.00881.x. doi:10.1111/j.1399-0004.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 7.Garshasbi M., Hadavi V., Habibi H., Kahrizi K., Kariminejad R., Behjati F., Tzschach A., Najmabadi H., Ropers H.H., Kuss A.W. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. doi:10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molinari F., Foulquier F., Tarpey P.S., Morelle W., Boissel S., Teague J., Edkins S., Futreal P.A., Stratton M.R., Turner G., et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am. J. Hum. Genet. 2008;82:1150–1157. doi: 10.1016/j.ajhg.2008.03.021. doi:10.1016/j.ajhg.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motazacker M.M., Rost B.R., Hucho T., Garshasbi M., Kahrizi K., Ullmann R., Abedini S.S., Nieh S.E., Amini S.H., Goswami C., et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2007;81:792–798. doi: 10.1086/521275. doi:10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamdan F.F., Gauthier J., Spiegelman D., Noreau A., Yang Y., Pellerin S., Dobrzeniecka S., Cote M., Perreau-Linck E., Carmant L., et al. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N. Engl. J. Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. doi:10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tidmarsh L., Volkmar F.R. Diagnosis and epidemiology of autism spectrum disorders. Can. J. Psychiatry. 2003;48:517–525. doi: 10.1177/070674370304800803. [DOI] [PubMed] [Google Scholar]
- 12.Kirov G., Gumus D., Chen W., Norton N., Georgieva L., Sari M., O'Donovan M.C., Erdogan F., Owen M.J., Ropers H.H., et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. doi:10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 13.Friedman J.I., Vrijenhoek T., Markx S., Janssen I.M., van der Vliet W.A., Faas B.H., Knoers N.V., Cahn W., Kahn R.S., Edelmann L., et al. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol. Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. doi:10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 14.Nolan D.K., Chen P., Das S., Ober C., Waggoner D. Fine mapping of a locus for nonsyndromic mental retardation on chromosome 19p13. Am. J. Med. Genet. A. 2008;146A:1414–1422. doi: 10.1002/ajmg.a.32307. doi:10.1002/ajmg.a.32307. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunyaev S., Ramensky V., Koch I., Lathe W., 3rd, Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. doi:10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 17.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. doi:10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon Y.A., Horton J.D. Identification of two mammalian reductases involved in the two-carbon fatty acyl elongation cascade. J. Biol. Chem. 2003;278:7335–7343. doi: 10.1074/jbc.M211684200. doi:10.1074/jbc.M211684200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J., Moseley A., Jegga A.G., Gupta A., Witte D.P., Sartor M., Medvedovic M., Williams S.S., Ley-Ebert C., Coolen L.M., et al. Neural system-enriched gene expression: relationship to biological pathways and neurological diseases. Physiol. Genomics. 2004;18:167–183. doi: 10.1152/physiolgenomics.00220.2003. doi:10.1152/physiolgenomics.00220.2003. [DOI] [PubMed] [Google Scholar]
- 20.Meloni I., Muscettola M., Raynaud M., Longo I., Bruttini M., Moizard M.P., Gomot M., Chelly J., des Portes V., Fryns J.P., et al. FACL4, encoding fatty acid-CoA ligase 4, is mutated in nonspecific X-linked mental retardation. Nat. Genet. 2002;30:436–440. doi: 10.1038/ng857. doi:10.1038/ng857. [DOI] [PubMed] [Google Scholar]
- 21.Piccini M., Vitelli F., Bruttini M., Pober B.R., Jonsson J.J., Villanova M., Zollo M., Borsani G., Ballabio A., Renieri A. FACL4, a new gene encoding long-chain acyl-CoA synthetase 4, is deleted in a family with alport syndrome, elliptocytosis, and mental retardation. Genomics. 1998;47:350–358. doi: 10.1006/geno.1997.5104. doi:10.1006/geno.1997.5104. [DOI] [PubMed] [Google Scholar]
- 22.Longo I., Frints S.G., Fryns J.P., Meloni I., Pescucci C., Ariani F., Borghgraef M., Raynaud M., Marynen P., Schwartz C., et al. A third MRX family (MRX68) is the result of mutation in the long chain fatty acid-CoA ligase 4 (FACL4) gene: proposal of a rapid enzymatic assay for screening mentally retarded patients. J. Med. Genet. 2003;40:11–17. doi: 10.1136/jmg.40.1.11. doi:10.1136/jmg.40.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin P.T. Glycobiology of the synapse. Glycobiology. 2002;12:1R–7R. doi: 10.1093/glycob/12.1.1r. doi:10.1093/glycob/12.1.1R. [DOI] [PubMed] [Google Scholar]
- 24.Smalla K.H., Matthies H., Langnase K., Shabir S., Bockers T.M., Wyneken U., Staak S., Krug M., Beesley P.W., Gundelfinger E.D. The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. Proc. Natl Acad. Sci. USA. 2000;97:4327–4332. doi: 10.1073/pnas.080389297. doi:10.1073/pnas.080389297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleene R., Schachner M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004;5:195–208. doi: 10.1038/nrn1349. doi:10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- 26.Iossifov I., Zheng T., Baron M., Gilliam T.C., Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–1162. doi: 10.1101/gr.075622.107. doi:10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ober C., Abney M., McPeek M.S. The genetic dissection of complex traits in a founder population. Am. J. Hum. Genet. 2001;69:1068–1079. doi: 10.1086/324025. doi:10.1086/324025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourgain C., Abney M., Schneider D., Ober C., McPeek M.S. Testing for hardy-weinberg equilibrium in samples with related individuals. Genetics. 2004;168:2349–2361. doi: 10.1534/genetics.104.031617. doi:10.1534/genetics.104.031617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.