Abstract
The facial photographs of 81 individuals with Noonan syndrome, from infancy to adulthood, have been evaluated by two dysmorphologists (JA and MZ), each of whom has considerable experience with disorders of the Ras/MAPK pathway. Thirty-two of this cohort have PTPN11 mutations, 21 SOS1 mutations, 11 RAF1 mutations, and 17 KRAS mutations. The facial appearance of each person was judged to be typical of Noonan syndrome or atypical. In each gene category both typical and unusual faces were found. We determined that some individuals with mutations in the most commonly affected gene, PTPN11, which is correlated with the cardinal physical features, may have a quite atypical face. Conversely, some individuals with KRAS mutations, which may be associated with a less characteristic intellectual phenotype and a resemblance to Costello and cardio-facio-cutaneous syndromes, can have a very typical face. Thus, the facial phenotype, alone, is insufficient to predict the genotype, but certain facial features may facilitate an educated guess in some cases.
Keywords: Noonan syndrome, PTPN11, SOS1, RAF1, KRAS, facial phenotype, genotype–phenotype correlation
INTRODUCTION
One of the most intriguing stories in developmental biology today is that of the Ras/MAPK pathway, a pathway critical for cell growth, differentiation, senescence, and death. Genes in this pathway are frequently mutated in cancer and germline mutations have been linked to Noonan syndrome (NS), cardio-facio-cutaneous syndrome (CFC), Costello syndrome (CS) and Legius syndromes, multiple lentigines or LEOPARD syndrome, and neurofibromatosis type 1. Mutations in PTPN11 were the first to be described in NS [Tartaglia et al., 2001]. Almost half of all individuals with NS will have a PTPN11 mutation, and hotspots are reported. In 2007, mutations in SOS1 were shown to cause NS in about 20% of affected persons without a PTPN11 mutation [Roberts et al., 2007; Tartaglia et al., 2007]. Mutations in KRAS only occasionally cause NS (1–3%) [Carta et al., 2006; Schubbert et al., 2006]. In 2007, a fourth gene responsible for causing NS was reported [Pandit et al., 2007; Razzaque et al., 2007]. Mutations in RAF1 are found in just 3–10% of affected individuals. The mutations causing NS are supposed to have gain-of-function effects on Ras/MAPK signaling. Two individuals have been reported with NS and mutations in MEK2; however, this genotype–phenotype correlation has yet to be reproduced [Nava et al., 2007]. Recently, a recurrent mutation in SHOC2 was demonstrated in 25 individuals with NS, all occurring de novo where parental samples were available for testing [Cordeddu et al., 2009]. Subsequently, five individuals with NS, three sporadic and a mother–son dyad, were found to have mutations in NRAS [Cirstea et al., 2010]. Nyström et al. [2008] reported an individual with NS and a BRAF mutation, however, the clinical diagnosis may be more in keeping with CFC syndrome [Neri et al., 2008]. Another individual with NS and a BRAF mutation is known to the authors. PTPN11 and RAF1 mutations also cause multiple lentigines syndrome [Legius et al., 2002]. Over 95% of individuals with CS have a mutation in HRAS, with the majority in codon 12 [Aoki et al., 2005]. CFC syndrome is associated with mutations in BRAF, MEK1, MEK2, and KRAS [Niihori et al., 2006; Rodriguez-Viciana et al., 2006; Narumi et al., 2007; Zenker et al., 2007a].
Knowledge of the phenotype can help to predict the likely causative gene. For example, PTPN11 mutations are more likely to be found in persons with pulmonary stenosis than those with hypertrophic cardiomyopathy [Tartaglia et al., 2002; Zenker et al., 2004]. They are positively correlated with short stature, pectus deformity, and factor VIII deficiency, and negatively correlated with hypertrophic cardiomyopathy and factor XI deficiency [Sarkozy et al., 2003; Yoshida et al., 2004; Limal et al., 2006]. The presence of florid ectodermal features is suggestive of SOS1 mutations [Roberts et al., 2007; Tartaglia et al., 2007]. These mutations also are associated with normal stature and intellectual functioning. Mutations in RAF1 show a very strong correlation with hypertrophic cardiomyopathy, present in 95% [Pandit et al., 2007; Razzaque et al., 2007]. KRAS mutations seem to cause a more severe intellectual handicap and may predispose to a phenotype similar to CFC syndrome or CS [Zenker et al., 2004]. The majority has short stature, webbed neck, pectus deformity, and few skin problems. Individuals with SHOC2 mutations have a distinctive hair phenotype described as loose anagen hair [Cordeddu et al., 2009]. We have carried out this study to determine if there are clues to mutation status in the face.
METHODS AND RESULTS
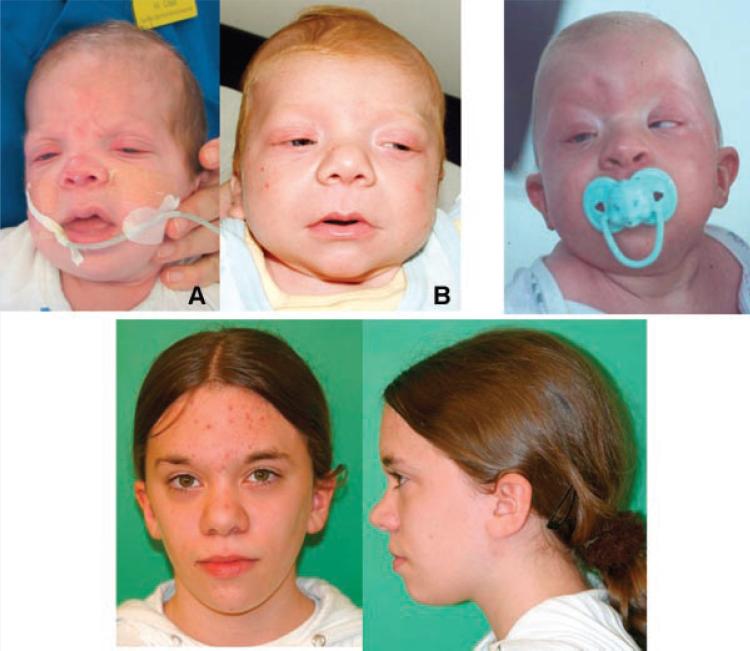
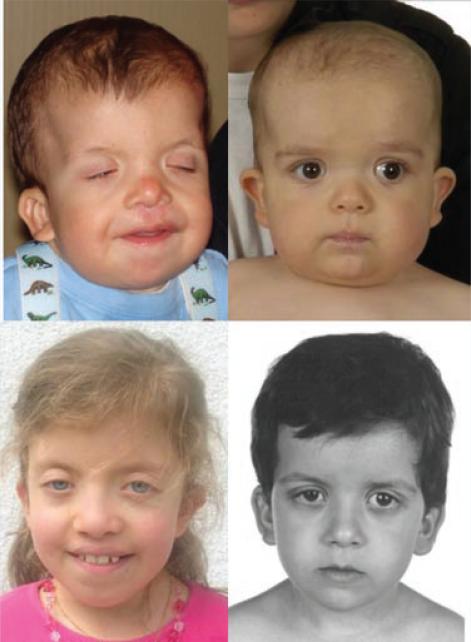
We have evaluated the facial photographs of 81 individuals with NS, from infancy to adulthood. Thirty-two have PTPN11 mutations, 21 SOS1 mutations, 11 RAF1 mutations, and 17 KRAS mutations. In each mutation category more than half of the individuals have classical facial features of NS [Allanson et al., 1985; Allanson, 1987]. The faces of children and adults with PTPN11 mutations and a typical gestalt are illustrated in Figure 1. However, many faces of individuals with PTPN11 mutations are unusual and diagnosis would be challenging (Fig. 2). Some faces are broader and coarser, reminiscent of CFC syndrome. Others have a long nose with low-hanging columella. Occasional individuals lack wide-spaced eyes and show close-spaced features and a narrow face. Coarseness of facial features is also seen in infants with SOS1 mutations, and ptosis is very common. In some infants, the face is similar to the face of CS or CFC syndrome (Fig. 3A,B). Later in life, the lips are often full and the nose fleshy. Curly or sparse hair may be present. In other individuals, facial features are quite characteristic of NS (Fig. 4). Two brothers with RAF1 mutations have unremarkable features (Fig. 5), while other children have quite a characteristic appearance of NS (Fig. 6). A few individuals have facial or cranial asymmetry that would be unusual for NS (Fig. 7). Lastly, those with mutations in KRAS can have a very typical NS face (Fig. 8). However, there are some individuals with KRAS mutations and coarser features, or a markedly prominent and wide nasal root and base, in whom diagnosis would be challenging (Fig. 9).
FIG. 1.
Individuals with PTPN11 mutations, from infancy to adolescence, and a typical face of Noonan syndrome.
FIG. 2.
PTPN11 mutations and a facial gestalt uncharacteristic of Noonan syndrome.
FIG. 3.
Individuals with SOS1 mutations and a coarse appearance, which, in infancy, can resemble the face of Costello syndrome (A,B). Reproduced from Zenker et al. [2007b] with permission from BMJ Publishing Group Ltd.
FIG. 4.
Very typical features of Noonan syndrome, seen from early childhood to adulthood, in individuals with SOS1 mutations.
FIG. 5.
Brothers with a RAF1 mutation and essentially unremarkable facial features.
FIG. 6.
Typical facial features of Noonan syndrome in four children of varying ages, each with a RAF1 mutation.
FIG. 7.
RAF1 mutations and cranial asymmetry, a long narrow nose, and markedly downslanting palpebral fissures.
FIG. 8.
KRAS mutations associated with typical facies of Noonan syndrome. Reproduced from Zenker et al. [2007a] with permission from BMJ Publishing Group Ltd.
FIG. 9.
KRAS mutations and unusual facial features: in childhood a coarseness suggestive of Costello syndrome; in adulthood a broadening of features with marked widespacing of eyes and broad nasal root and tip somewhat reminiscent of G/BBB and Teebi hypertelorism syndromes. Reproduced from Zenker et al. [2007a] with permission from BMJ Publishing Group Ltd.
DISCUSSION
The classical facial features of NS change with age [Allanson et al., 1985]. In the newborn, typical features include tall forehead, hypertelorism, downslanting palpebral fissures, epicanthal folds, a short and broad nose with a depressed root with upturned tip, deeply grooved philtrum with high, wide peaks of the vermilion, high palate, micrognathia, low-set and posteriorly angulated ears with thick helices, and excessive nuchal skin with low posterior hairline. During infancy, the head is relatively large with a tall and prominent forehead. Hypertelorism, ptosis, or thick hooded eyelids are characteristics. The nose is short and wide with a depressed root. During childhood, the face may appear coarse or myopathic. Facial contour becomes more triangular with age, as the face lengthens. The upper face is broad while the chin is narrow and pointed. During adolescence and young adulthood, the nose has a thin, high bridge and a wide base. The neck is longer with accentuated webbing (pterygium colli) or prominent trapezius. In older adults, the nasolabial folds are prominent and the skin appears thin and transparent [Allanson et al., 1985; Allanson, 1987]. Features present regardless of age include blue-green irides, arched and diamond-shaped eyebrows, and low-set posteriorly angulated ears with thickened helices [Allanson, 1987; Sharland et al., 1992]. The hair may be wispy during infancy and curly or woolly in later childhood and adolescence.
In this study, we demonstrate that some individuals with mutations in the most commonly affected gene, PTPN11, which is correlated with the cardinal physical features, including the characteristic facial phenotype, can have a quite atypical face. There is no one alternate gestalt, and several atypical features may be found, including a broader face and coarser appearance, reminiscent of the face in CFC syndrome, which is characterized by rounder and more bulbous nasal tip with wider nasal base, and fuller lips. Other atypical faces demonstrate a long nose with low-hanging columella. Occasional individuals lack the typical wide-spaced eyes and show close-spaced features and a narrow face. At the other end of the spectrum, some individuals with KRAS mutations, which may be associated with a more severe intellectual disability with severe and longstanding feeding problems and failure to thrive, resembling Costello and CFC syndromes, can have a very typical NS facial appearance. In each gene category, as reviewed in the Methods and Results Section, both typical and unusual faces may be found.
This study was carried out before knowledge of causative mutations in SHOC2 and NRAS was available. However, review of facial appearance in illustrations that are part of the supplemental information accompanying the publication of NRAS mutations suggests that both typical and atypical facial appearance may be found in this cohort as well [Cirstea et al., 2010]. The single recurrent SHOC2 mutation was found to be associated with a characteristic hair phenotype [Cordeddu et al., 2009].
If one considers the question posed by the title: “Does phenotype predict genotype?” the answer is a clear “No.” Characteristic NS facial appearance does not point to a specific genotype. It would be most useful to know how often a particular genotype is found with an atypical face; however, it is only reasonable to expect that level of detail by studying a cohort with PTPN11 mutations as each of the other causative genes is responsible for such a small proportion of affected individuals that any one study is likely to be biased by small numbers. Such a study of PTPN11-mutation-positive individuals has not been carried out to date.
However, it may be possible to “make an educated guess” about genotype based on clues present in the face. The child with rounded features, sparse eyebrows and eyelashes, curly or sparse hair, and skin erythema may have a higher likelihood of a SOS1 mutation. Loose anagen hair makes a SHOC2 mutation more likely. Cranial or facial asymmetry may point to higher odds of a RAF1 mutation, while coarse features or a markedly prominent and wide nasal root and base may suggest a KRAS mutation. In reality, one is more likely to make a correct prediction of genotype if other factors, such as height, IQ, and type of cardiac defect are added to impressions of facial gestalt. Fortunately, with the advent of newer molecular approaches using a combination of DNA sequencing techniques to evaluate the coding regions and splice sites of all known genes, a cost-efficient testing approach does not require the physician managing the patient to predict which gene to evaluate first.
How to Cite this Article.
Allanson JE, Bohring A, Dörr H-G, Dufke A, Gillessen-Kaesbach G, Horn D, König R, Kratz CP, Kutsche K, Pauli S, Raskin S, Rauch A, Turner A, Wieczorek D, Zenker M. 2010. The face of Noonan syndrome: Does phenotype predict genotype.
Am J Med Genet Part A 152A:1960–1966.
ACKNOWLEDGMENTS
C.P.K.'s work was supported by the Intramural Research Program of the US National Cancer Institute, National Institutes of Health. M.Z. was supported by the German Research Foundation (DFG; ZE 524/4-1).
Grant sponsor: US National Cancer Institute, National Institutes of Health
Grant sponsor: German Research Foundation (DFG); Grant number: ZE 524/4-1.
REFERENCES
- Allanson JE. Noonan syndrome. J Med Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, Hall JG, Hughes HE, Preus M, Witt RD. Noonan syndrome: The changing phenotype. Am J Med Genet. 1985;21:507–514. doi: 10.1002/ajmg.1320210313. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Carta C, Pantaleoni F, Bocchinfuso G, Stella L, Vasta I, Sarkozy A, Digilio C, Palleschi A, Pizzuti A, Grammatico P, Zampino G, Dallapiccola B, Gelb BD, Tartaglia M. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirstea IC, Kutsche K, Dvorsky R, Gremer L, Carta C, Horn D, Roberts AE, Lepri F, Merbitz-Zahradnik T, König R, Kratz CP, Pantaleoni F, Dentici ML, Joshi VA, Kucherlapati RS, Mazzanti L, Mundlos S, Patton MA, Silengo MC, Rossi C, Zampino G, Digilio C, Stuppia L, Seemanova E, Pennacchio LA, Gelb BD, Dallapiccola B, Wittinghofer A, Ahmadian MR, Tartaglia M, Zenker M. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42:27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeddu V, Di Schiavi E, Pennacchio LA, Ma'ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, Lipzen A, Zampino G, Mazzanti L, Digilio MC, Martinelli S, Flex E, Lepri F, Bartholdi D, Kutsche K, Ferrero GB, Anichini C, Selicorni A, Rossi C, Tenconi R, Zenker M, Merlo D, Dallapiccola B, Iyengar R, Bazzicalupo P, Gelb BD, Tartaglia M. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legius E, Schrander-Stumpel C, Schollen E, Pulles-Heintzberger C, Gewillig M, Fryns J-P. PTPN11 mutations in LEOPARD syndrome. J Med Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limal JM, Parfait B, Cabrol S, Bonnet D, Leheup B, Lyonnet S, Vidaud M, Le Bouc Y. Noonan syndrome: Relationships between genotype, growth, and growth factors. J Clin Endocrinol Metab. 2006;91:300–306. doi: 10.1210/jc.2005-0983. [DOI] [PubMed] [Google Scholar]
- Narumi Y, Aoki Y, Niihori T, Neri G, Cave H, Verloes A, Nava C, Kavamura MI, Okamoto N, Kurosawa K, Hennekam RC, Wilson LC, Gillessen-Kaesbach G, Wieczorek D, Lapunzina P, Ohashi H, Makita Y, Kondo I, Tsuchiya S, Ito E, Sameshima K, Kato K, Kure S, Matsubara Y. Molecular and clinical characterization of cardio-facio-cutaneous (CFC) syndrome: Overlapping clinical manifestations with Costello syndrome. Am J Med Genet Part A. 2007;143A:799–807. doi: 10.1002/ajmg.a.31658. [DOI] [PubMed] [Google Scholar]
- Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, Aoki Y, Matsubara Y, Arveiler B, Lacombe D, Pasmant E, Parfait B, Baumann C, Heron D, Sigaudy S, Toutain A, Rio M, Goldenberg A, Leheup B, Verloes A, Cave H. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signaling pathway: Genotype–phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri G, Allanson JE, Kavamura MI. No reason yet to change diagnostic criteria for the Noonan, CFC and Costello syndrome. J Med Genet. 2008;45:832. doi: 10.1136/jmg.2008.063263. [DOI] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cavé H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio -cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- Nyström A-M, Ekvall S, Berglund E, Björkvist M, Braathen G, Duchen K, Enell H, Holmberg E, Holmlund U, Olsson-Engman M, Annerén G, Bondeson M-L. Noonan and cardio-facio-cutaneous syndromes: Two clinically and genetically overlapping disorders. J Med Genet. 2008;45:500–506. doi: 10.1136/jmg.2008.057653. [DOI] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, López Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, Fujiwara Y, Matsushima M, Mizuno K, Tokuyama M, Hirota H, Muneuchi J, Higashinakagawa T, Matsuoka R. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- Sarkozy A, Conti E, Seripa D, Digilio MC, Grifone N, Tandoi C, Fazio VM, Di Ciommo V, Marino B, Pizzuti A, Dallapiccola B. Correlation between PTPN11 mutations and congenital heart defects in Noonan and LEOPARD syndromes. J Med Genet. 2003;40:704–708. doi: 10.1136/jmg.40.9.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KY, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Sharland M, Burch M, McKenna WM, Patton MA. A clinical study of Noonan syndrome. Arch Dis Child. 1992;67:178–183. doi: 10.1136/adc.67.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffrey S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffrey S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. PTPN11 mutations in Noonan syndrome: Molecular spectrum, genotype–phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70:1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, Ustaszewska A, Martin J, Bristow J, Carta C, Lepri F, Neri C, Vasta I, Gibson K, Curry CJ, Siguero JP, Digilio MC, Zampino G, Dallapiccola B, Bar-Sagi D, Gelb BD. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hasegawa T, Hasegawa Y, Nagai T, Kinoshita E, Tanaka Y, Kanegane H, Ohyama K, Onishi T, Hanew K, Okuyama T, Horikawa R, Tanaka T, Ogata T. Protein-tyrosine phosphatase, non-receptor type 11 mutation analysis and clinical assessment in 45 patients with Noonan syndrome. J Clin Endocrinol Metab. 2004;89:3359–3364. doi: 10.1210/jc.2003-032091. [DOI] [PubMed] [Google Scholar]
- Zenker M, Buheitel G, Rauch R, Koenig R, Bosse K, Kress W, Tietze HU, Doerr HG, Hofbeck M, Singer H, Reis A, Rauch A. Genotype-–phenotype correlations in Noonan syndrome. J Pediatr. 2004;144:368–374. doi: 10.1016/j.jpeds.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Zenker M, Lehmann K, Schulz AL, Barth H, Hansmann D, Koenig R, Korinthenberg R, Kreiss-Nachtsheim M, Meinecke P, Morlot S, Mundlos S, Quante AS, Raskin S, Schnabel D, Wehner LE, Kratz CP, Horn D, Kutsche K. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007a;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Horn D, Wieczorek D, Allanson J, Pauli S, van der Burgt I, Doerr H-G, Gaspar H, Hofbeck M, Gillessen-Kaesbach G, Koch A, Meinecke P, Mundlos S, Nowka A, Rauch A, Reif S, von Schnakenburg C, Seidel H, Wehner L-E, Zweier C, Bauhuber S, Matejas V, Kratz CP, Thomas C, Kutsche K. SOS1 is the second most common Noonan gene but plays no major role in cardio-facio-cutaneous syndrome. J Med Genet. 2007b;44:651–656. doi: 10.1136/jmg.2007.051276. [DOI] [PMC free article] [PubMed] [Google Scholar]