Abstract
Recent studies have shown UV vision and markings to be important in vertebrates, particularly birds, where behavioral experiments have demonstrated its potential importance in sexual selection. However, there has been no genetic evidence that UV markings determine patterns of evolution among natural populations. Here we report molecular evidence that UV markings are associated with the pattern of gene flow in the Tenerife lizard (Gallotia galloti). This species has vicariance-induced, approximate east–west lineages in Tenerife closely congruent with the primary lineages of the sympatric gecko species. Against expectations, these molecular phylogeographic lineages (representing geological history) and isolation-by-distance do not appear to influence gene flow. Sexually mature males from populations either side of a latitudinal ecotone have different UV markings and gene flow appears to be linked to this difference in UV markings. It may be that these groups with different UV sexual markings mate assortatively, restricting the gene flow between them. This has implications for debate on the relative importance of vicariance and biotopes in influencing biodiversity, with this evidence supporting the latter.
Keywords: microsatellite, speciation, assortative mating, Tenerife, Gallotia galloti
Visual perception in the UV range, (sometimes including tetrachromatic vision), has been found in fishes (1, 2), amphibians (3), both iguanian (4–6) and scleroglossan lizards (7, 8), some mammals (9), and many birds (10). Visual cues are important in sexual selection in some lizards and birds and behavioral experiments with birds suggest that these cues include UV markings (10–15). This suggests that UV markings may be important in determining patterns of population differentiation via assortative mating and may have a role in speciation and maintaining reproductive isolation in natural populations. However, there is no empirical genetic evidence of this from patterns of population differentiation in nature.
The western Canary Island lacertid, Gallotia galloti, is a scleroglossan lizard that uses visual cues in sexual selection (16, 17). Females and immature males are dull in color and relatively cryptic, whereas sexually mature males in breeding condition may display in prominent positions. On Tenerife the sexually mature males always have bright lateral markings that will tend to be visible to conspecifers rather than overhead avian (kestrel) predators (18). The dorsum, which will tend to be seen by avian predators, may have disruptive cross bars. Previous work has shown distinct geographic variation in color pattern linked to biotope (Fig. 1), rather than vicariance-induced mtDNA lineages (19). However, there has been no investigation of the nature of the bright lateral sexual markings and no investigation of the extent of gene flow between mtDNA lineages or color-pattern types to determine their relative importance in influencing genetic differentiation.
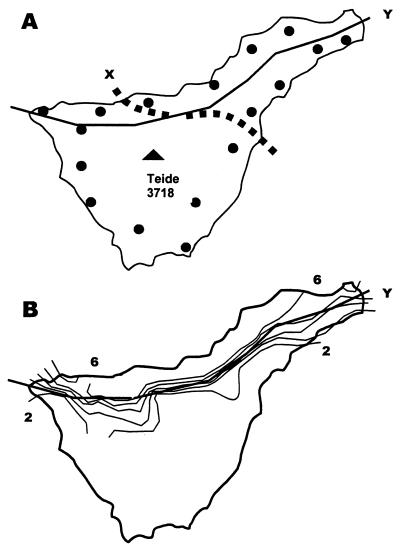
Figure 1.
(A) Map of Tenerife showing study localities and approximate distribution of eastern and western lineages (X; although opposing haplotypes interdigitate either side of this line), and northern and southern biotopes (Y) with populations that have different reflective lateral markings. (B) Geographic variation in the reflective shoulder markings is a stepped cline coinciding with the ecotone (Y) where the contours 2–6 (on a 0–10 range) represent an increase in size with a decrease in mean value (derived from 18).
The aim of this study is to investigate the nature of these sexual markings and to test whether they, or vicariance patterns, are associated with gene flow determined by tandem repeat nuclear markers (microsatellites) specifically designed for this study.
Methods
Measurement of UV Markings.
Reflectance of a marking, as a percentage of a Spectralon (Labsphere, Poynton, U.K.) white tile standard was measured by using an S2000 spectrometer (Ocean Optics Europe, Eerbeek, The Netherlands) with deuterium and halogen light sources and SPECTRAWIN 4.1 software (Top Sensor Systems, Eerbeek, The Netherlands). We recorded reflectance from three body regions that may have UV markings (cheek, first shoulder mark, and midtrunk) from both modal northern and modal southern specimens together with the associated three background areas adjacent to these marks. The spectra were averaged across ten independent recordings. Average percent reflectance was plotted against the 325–700-nm segment of the spectrum.
The size and distribution of UV markings were recorded via high-resolution macrophotography. A prefocused 100-mm Canon macro lens on an EOS1 body (mounted on a tripod) was fitted with a UV pass filter and the subject illuminated against a UV reflective background by a Metz flash modified for UV output, exposing 1600 ASA Fuji Neopan monochrome negative film. This gave sufficiently sharp resolution for individual scales to be clearly defined. The size (in number of scales covered) was recorded for the lateral cheek mark, the two lateral shoulder marks, and the total number of lateral trunk marks posterior to the shoulder marks. This was recorded for 105 sexually mature males from 17 localities across Tenerife [Fig. 1; localities 1, 6, 9, 10, 12, 17, 19, 20, 21, 24, 29, 39, 48, 51, 53, 61, and 63 in Thorpe and Brown (20)].
Molecular Gene Flow.
Five highly polymorphic dinucleotide microsatellite loci (AC19, CA10, TC19, AC12, and GT3(AT)GT10; GenBank accession nos. AF070978–AF070982), developed specifically for this study, were screened across 30 specimens from each of the 17 localities above. The laboratory methods for DNA extraction, PCR amplification, and screening, together with a description of the microsatellites, are in Richard and Thorpe (21). The five loci were tested for linkage equilibrium (22), departure from Hardy–Weinberg equilibrium, and null alleles (23, 24). Gene flow among sampled populations was estimated by FST(refs. 25 and 26; see Discussion).
Hypothesis Testing by Matrix Correspondence.
In geographic variation analysis, hypotheses may be tested by matrix correspondence (Mantel) tests where the association between actual or hypothesized patterns, represented by dissimilarity matrices among populations, is measured as a correlation, or absolute standardized regression, and the probability on the null hypothesis of no association is assessed by repeatedly randomizing (10,000 times) the dependant matrix to produce a distribution of association coefficients (19, 27). This allows multidimensional hypotheses, such as isolation-by-distance, to be considered. When several alternative hypotheses have nonindependent patterns these may be considered simultaneously as independent variables in a partial regression matrix correspondence test, with the observed pattern matrix (estimated gene flow in this case) as the dependent variable (19, 28–30).
The dependent variable, estimated gene flow among the 17 geographic localities, is represented by a matrix of linearized FSTvalues computed across five microsatellite loci. Mitochondrial DNA lineage (19) membership is represented by a 0,1 matrix, with the former representing membership of the same lineage and the latter membership of the different lineage. Isolation-by-distance is represented by a matrix of geographic distances among localities. The type of UV marking pattern was then generalized across the three characters (size of cheek, shoulder, and trunk marks) by using a principal coordinate analysis on locality means (characters standardized to zero wean, unit variance) to generate a Euclidean taxonomic distance matrix representing the degree of (dis)similarity in UV marks among localities. This was repeated by using a Mahalonobis distance derived from canonical analysis of the three UV characters that takes into account their within-locality covariance. First, pair-wise analyses were computed between estimated gene flow on the one hand and mtDNA lineage, isolation-by-distance, and UV marking type on the other. Then partial analyses were computed so that the association between gene flow and type of UV marking could be assessed while taking into account the effect of lineage and isolation-by-distance.
Results and Discussion
UV Patterns.
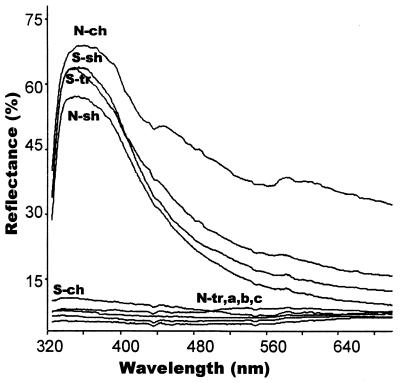
Spectrometer analysis shows that peak reflectance of the highly reflective lateral blotches of sexually mature males, whether from the north or south, are in the UVA segment of the electromagnetic spectrum (ca. 360 ± 20 nm) with substantially less in the human visible spectrum (Fig. 2). These UV markings are comparable with other “UV” marks with published spectrograms (4, 5, 11, 12, 13, 15, 31–34). In sharp contrast, the background of the lateral surface of the head and trunk has extremely low reflectance across all wavelengths as have the cheek and midtrunk areas when a UV mark is absent.
Figure 2.
Percentage reflectance of UV markings and associated background. A modal northern specimen has a large highly UV reflective cheek marking (N-ch) and a small UV reflective first shoulder marking (N-sh) but with a midtrunk marking too small to record such that the area that it would have occupied has very low reflectance (N-tr). A modal southern specimen has a large highly UV reflective first shoulder (S-sh) and midtrunk marking (S-tr), and a nonreflective cheek (S-ch). Areas adjacent to the cheek (a), shoulder (b), and trunk (c) are also nonreflective (and may appear black to human vision) to give a sharp contrast between the UV reflective marking and its background.
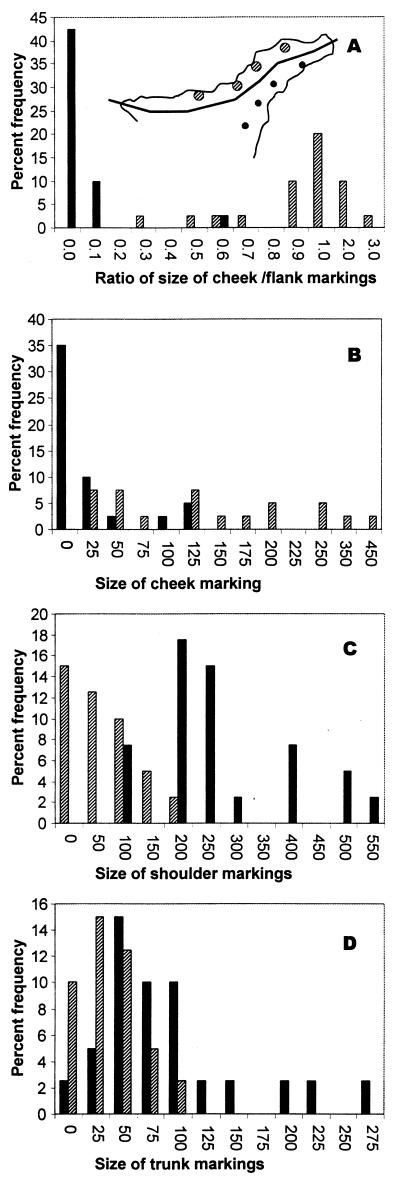
Previous work (Fig. 1) has shown that geographic variation in these lateral reflective markings (previously, incorrectly referred to as blue markings) is related to the north–south climatic/vegetation biotopes (18, 19, 35). The hot, arid, and barren southern biotope tends to have lizards with large reflective blotches on the lateral trunk and shoulder, whereas the cloudy, warm, well vegetated part of the island along the northern coastal strip below 1,500 m tends to have lizards with reflective markings on the ventro-lateral surface of the cheeks rather than on the shoulder and trunk (Fig. 3). The variation in reflective markings not only tends to be categorical across geographic space (Fig. 1), but also distinctly bimodal with a tendency to either look like the northern or southern type in Fig. 3. This is shown in Fig. 4, where a comparison of individuals from four pairs of eastern populations either side of the ecotone shows there is widely separated modes with almost no overlap in the ratio of the size of cheek to flank UV markings. Northern biotope forms modally have UV cheek markings up to twice the size of their total UV flank markings, whereas southern biotope forms modally have UV flank markings at least ten times larger than their UV cheek markings (which they may lack entirely). The basic situation is one of variation in size (or even presence or absence) of highly UV reflective markings on a given part of the lateral surface rather than of subtle variation in hue and reflectivity (Fig. 2).
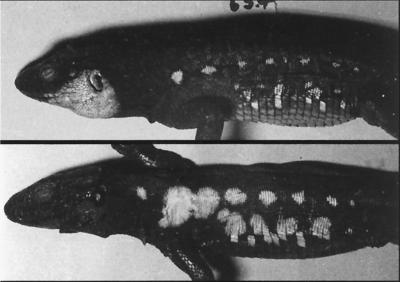
Figure 3.
UV photographs. The northern specimen (Upper) has a well developed cheek UV mark and the southern specimen (Lower) has well developed shoulder and trunk UV marks.
Figure 4.
Histograms showing the bimodality of UV reflective markings based on four pairs of populations either side of the ecotone (Inset) in largely the range of the eastern lineage (southern, solid bars; northern, hatched bars). (A) The ratio of the size of the cheek marking to flank (shoulder + trunk) marking shows distinct widely separated modes with almost no overlap. The cheek (B), shoulder (C), and trunk (D) frequency against size in number of scales covered.
It is suggested (18) that these heliotherms warm rapidly in the hot southern biotope, which allows them to avoid predation, whereas in the cloudy north, longer periods are required for thermoregulation, which renders the lizard vulnerable to predation. Consequently, the greater need for crypsis in the northern biotope switches the balance between sexual selection and crypsis such that the northern specimens have disruptive dorsal/dorso-lateral cross-bars, which would interfere with lateral trunk sexual selection markings. Therefore these markings are based on the cheek, rather than the trunk, in the north (18).
The island of Tenerife has a geological history that allows for distinct phylogenetic lineages (19, 36–38). The western precursor islands (Adeje/Teno) joined the eastern precursor island of Anaga to form Tenerife after the emergence of the Canadas/Teide edifice. Molecular studies using mtDNA show that both the gecko, Tarentola delalandii (37), and the lacertid (19) have distinct lineages on Tenerife, with numerical hypothesis testing suggesting secondary contact of separate lineages from these eastern and western precursor islands. In G. galloti this results in an approximate east–west distribution of two lineages with some interdigitation. Hence, one has approximately east–west mitochondrial lineages and north–south biotopes (Fig. 1). However, it is not as simple as this, because in both geckos and lacertids the eastern lineage has spread further along the north coast than the western lineage (37). Consequently, the two patterns are not entirely independent, and appropriate procedures are required to disentangle lineage and biotope effects. When this is done, both by partialling out effects and by analyzing separately within eastern and western lineages, the color pattern is clearly associated with biotope (19).
Gene Flow Patterns.
Highly polymorphic, codominant, tandemly repeated, nuclear markers such as microsatellites may be used to estimate gene flow among largely contiguous populations distributed over geographic space (25, 39) by using metrics such as FST. However, this involves several assumptions. Aside from the assumptions of mutation model of the specific metric used (25) there are general assumptions. For example, it is assumed that there is no generation overlap. In this model, sample localities could in theory include both offspring and parents, but this is unlikely to be a major perturbation. More importantly, there is an assumption of genetic equilibrium that may be unfounded if there have been founder effects in recent bottlenecks. Such founder effects should result in a reduction in allele size range, departure from Hardy–Weinberg equilibrium, and linkage disequilibrium between loci. Allele size range, observed (HO) and expected (HE) heterozygosity, departures from Hardy–Weinberg equilibrium (24), and linkage disequilibrium are given in Table 1. There is high gene diversity (0.525–0.925), no significant evidence of null alleles from heterozygote deficit, no notable reduction in allele range, no consistent departure from Hardy–Weinberg equilibrium, and no consistent departure from linkage expectations. Consequently, the populations studied largely appear to be in genetic equilibrium. Moreover, the lack of consistent departure from Hardy–Weinberg indicates that the assumptions of no population substructuring or selection are broadly valid. Finally, FSTwill measure both past and current gene flow. Here we specifically test for, and regress out, historical factors represented by the phylogeny.
Table 1.
Observed (HO) and expected (HE) heterozygosity and allele range for microsatellite loci (A348, A49, B81, B821, and B967) and Tenerife localities (T1–T63)
| A348 | A49 | B81 | B821 | B967 | All loci | ||
|---|---|---|---|---|---|---|---|
| T1 | HO | 0.686 | 0.943 | 0.857 | 0.735 | 0.886 | 0.821 |
| HE | 0.767 | 0.878 | 0.841 | 0.861 | 0.892 | 0.848 | |
| Alleles | 10–19 | 8–20 | 7–19 | 11–22 | 8–23 | ||
| T6 | HO | 0.733 | 0.833 | 0.867 | 0.867 | 0.900 | 0.840 |
| HE | 0.757 | 0.893 | 0.895 | 0.827 | 0.901 | 0.855 | |
| Alleles | 10–20 | 7–20 | 6–23 | 10–22 | 8–21 | ||
| T9 | HO | 0.667 | 0.767a | 0.833 | 0.700b | 0.867a,b | 0.767 |
| HE | 0.749 | 0.884 | 0.836 | 0.819 | 0.888 | 0.835 | |
| Alleles | 10–20 | 8–20 | 6–19 | 11–22 | 8–21 | ||
| T10 | HO | 0.519 | 0.964 | 0.821 | 0.750 | 0.643* | 0.739 |
| HE | 0.667 | 0.895 | 0.848 | 0.742 | 0.858 | 0.802 | |
| Alleles | 10–19 | 7–20 | 7–21 | 11–23 | 9–22 | ||
| T12 | HO | 0.607a | 0.929 | 0.929 | 0.786 | 0.857a | 0.821 |
| HE | 0.757 | 0.868 | 0.879 | 0.812 | 0.892 | 0.841 | |
| Alleles | 9–18 | 8–19 | 6–20 | 12–22 | 7–22 | ||
| T17 | HO | 0.786 | 0.929 | 0.679 | 0.786 | 0.929 | 0.821 |
| HE | 0.838 | 0.892 | 0.797 | 0.883 | 0.925 | 0.867 | |
| Alleles | 10–20 | 7–20 | 6–21 | 11–23 | 9–22 | ||
| T19 | HO | 0.467 | 1.000 | 0.900 | 0.833 | 0.867 | 0.813 |
| HE | 0.525 | 0.902 | 0.814 | 0.861 | 0.916 | 0.804 | |
| Alleles | 10–19 | 8–21 | 6–22 | 11–22 | 8–22 | ||
| T21 | HO | 0.556a | 0.704 | 0.704 | 0.815 | 0.889a | 0.733 |
| HE | 0.766 | 0.887 | 0.861 | 0.875 | 0.887 | 0.855 | |
| Alleles | 10–19 | 7–18 | 7–19 | 11–22 | 9–21 | ||
| T24 | HO | 0.577 | 0.692 | 0.808 | 0.769 | 0.885 | 0.746 |
| HE | 0.554 | 0.870 | 0.903 | 0.869 | 0.851 | 0.809 | |
| Alleles | 10–18 | 8–20 | 6–21 | 11–22 | 11–23 | ||
| T26 | HO | 0.607 | 0.964 | 0.857a | 0.857a,b | 0.893b | 0.836 |
| HE | 0.768 | 0.863 | 0.884 | 0.852 | 0.925 | 0.859 | |
| Alleles | 9–19 | 7–20 | 6–17 | 8–22 | 11–23 | ||
| T29 | HO | 0.781 | 0.906 | 0.813 | 0.844 | 0.969 | 0.863 |
| HE | 0.743 | 0.894 | 0.852 | 0.809 | 0.921 | 0.844 | |
| Alleles | 3,10–20 | 8–21 | 7–23 | 11–20 | 7–22 | ||
| T39 | HO | 0.806 | 0.935 | 0.871 | 0.871 | 1.000 | 0.897 |
| HE | 0.785 | 0.891 | 0.856 | 0.889 | 0.909 | 0.866 | |
| Alleles | 10–20 | 8–20 | 6–21 | 11–23 | 9–23 | ||
| T48 | HO | 0.590a | 0.795 | 0.872a | 0.769 | 0.795 | 0.764 |
| HE | 0.679 | 0.851 | 0.845 | 0.826 | 0.882 | 0.817 | |
| Alleles | 10–20 | 8–19 | 6–21 | 11–21 | 8–22 | ||
| T51 | HO | 0.763a | 0.763b | 0.658a,b | 0.816 | 0.753 | |
| HE | 0.740 | 0.885 | 0.836 | 0.750 | 0.887 | 0.820 | |
| Alleles | 10–21 | 8–20 | 7–22 | 11–21 | 11–24 | ||
| T53 | HO | 0.741a,b | 0.926 | 0.778a,c | 0.778c,d | 0.926b,d | 0.830 |
| HE | 0.760 | 0.880 | 0.869 | 0.869 | 0.915 | 0.858 | |
| Alleles | 3,10–19 | 8–19 | 7–21 | 10–22 | 6–24 | ||
| T61 | HO | 0.760 | 0.760* | 0.840 | 0.840a | 0.840a | 0.808 |
| HE | 0.826 | 0.879 | 0.897 | 0.839 | 0.896 | 0.867 | |
| Alleles | 10–20 | 8–22 | 6–21 | 10–22 | 6–22 | ||
| T63 | HO | 0.622 | 0.944 | 0.838 | 0.838 | 0.919 | 0.932 |
| HE | 0.722 | 0.893 | 0.877 | 0.876 | 0.909 | 0.850 | |
| Alleles | 10–21 | 8–23 | 7–22 | 11–22 | 8–23 |
Asterisk indicates a departure from Hardy–Weinberg equilibrium, and shared alphabetic superscript indicates pair-wise linkage disequilibrium between those loci, at that locality, when departure from Hardy–Weinberg equilibrium is not involved.
Bearing in mind the above assumptions, gene flow among populations may be proportional to the geographic distance between them (isolation-by-distance). A tendency for reproductive isolation between mtDNA lineages would result in a departure from this and be reflected in pattern of gene flow associated with the east–west distribution of the lineages. Moreover, assortative mating among populations would also predict departure from isolation-by-distance. Previous behavioral experiments on birds suggest a role for UV markings in sexual selection. These lizards have a visually dominated sexual selection system (16, 17) with sexually mature males in breeding condition having bright lateral UV markings. If there is less interbreeding between populations with different types of UV marks than between populations with the same type of marks because of assortative mating, then the pattern of gene flow should be associated with the type of UV mark.
This is tested by matrix correspondence between gene flow estimated across five microsatellites and the three alternative hypotheses. In pair-wise tests across all 17 localities (Table 2), isolation-by-distance can be rejected, and in subsequent partial tests both isolation-by-distance and reduced gene flow between mtDNA lineages can be rejected. However, gene flow and the type of UV marking are significantly and highly associated in the pair-wise test, and in the partial test when isolation-by-distance and lineage are taken into account.
Table 2.
Matrix correspondence (Mantel) tests across all 17 localities
| Pair-wise/Partial | mtDNA | Prox | UV |
|---|---|---|---|
| Pair-wise | |||
| r | 0.28 | 0.14 | 0.52 |
| P | 0.0005 | 0.1400 | 0.0000 |
| Partial | |||
| g | 0.02 | 0.09 | 0.52 |
| P | 0.8400 | 0.4469 | 0.0005 |
Correspondence is measured as absolute correlation (r) in the pair-wise tests, and absolute standardized partial regression (g) in the simultaneous tests. The probability (P), of the null hypothesis of no association, between gene flow (dependent variable) and mtDNA lineage (mtDNA), isolation-by-distance (Prox), and type of UV pattern (UV) (independent variables) is based on 10,000 randomizations. Significant correspondence in bold. The UV pattern matrix is based on taxonomic distance. The same results are found when the UV pattern matrix is based on Mahalonobis distance (UV-FSTassociation: pairwise r = 0.52, P = 0.0000; partial g = 0.49, P = 0.0005).
Hence, the pattern of gene flow is strongly structured such that there is less gene flow between populations with different types of UV marks than between populations with similar UV marks. There has been no previous empirical molecular genetic evidence that UV markings determine patterns of evolution in natural populations. However, this data associates UV markings with molecular gene flow in natural populations. The results suggest that irrespective of the influence of phylogeny or isolation-by-distance, there is less interbreeding between biotope types with different UV marks than the same UV marks. The mechanism for this may be assortative mating via female choice and this study points to the need for experimental investigation of this area to test this possibility. The behavioral experiments on the role of UV marks in assortative mating in birds (10–15) suggest this is feasible, and open the way for such experiments in these and other lizards. For these hypotheses to be acceptable, G. galloti would have to be able to perceive in the UV spectrum. Though there have been no specific studies of vision in this species, UV vision is becoming increasingly apparent in many vertebrate groups (1–3, 9, 10) including lizards (4–8) and it may be that UV vision is conservative in some lizards and occurs even when UV markings do not (6). Indeed, recent phylogenetic analysis of the genetic basis of UV vision suggests that it is the ancestral state for all vertebrates and secondarily lost in only a few groups—e.g., primates (40). The peak reflectance of these UV markings in G. galloti coincides with the known peak absorption of UV cones in lizards (4–8). This, and the orientation of these marks so that they are seen by lateral conspecifers rather than overhead predators, predicts the existence of UV vision in this group and suggests that this should be tested.
It is also possible that it is characters, other than these UV markings, that are primarily adapted to these specific latitudinal biotopes, which in turn renders biotope genotypes less compatible, and hence limits gene flow between them (41, 42). However, this is less likely because patterns of geographic variation in various character systems have been studied in this lizard and only the color pattern of sexually mature males shows such a distinct categorical latitudinal biotope pattern Fig. 1 (18). There is no evidence of such a geographic pattern in the color pattern of juveniles or females, or in the size, shape, and scalation of these lizards (20, 43). Size variation is primarily smoothly altitudinal (20), shape variation is mosaic (43), and the pattern of generalized scalation (43) appears, in retrospect, to closely reflect mtDNA lineage (19). Consequently, while behavioral experiments are called for, current data suggests that the most likely explanation is that sexual selection based on UV markings plays a central role in influencing genetic structure.
There is much debate on the relative roles of selection (both natural and sexual) and vicariance in speciation (41, 42, 44). Initially, we expected the nuclear gene flow to be primarily structured according to the distribution of the mtDNA lineages (reflecting historical vicariance), but this can clearly be rejected. This study suggests that, in this particular case, sexual selection linked to biotopes may be more influential than past vicariance in generating reproductive isolation, and therefore supports the role of biotopes in generating biodiversity (42). In visually orientated vertebrates, sexual selection via UV markings may be an important component in shaping the evolution of natural populations.
Acknowledgments
We thank A. Wootton for his contribution to character recording and two anonymous referees for their comments. The Leverhulme Trust (RF&G/2/9900012) supported the UV work, The Wellcome Trust (057257/Z/99/Z) supplied equipment, and a joint European Community grant (EU-ERBCHRXCT-940585) supported the molecular work (we thank the coholder of the award, M. Baez, University of La Laguna, for obtaining permissions).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Douglas R H, Djamgoz M B A. The Visual System of Fish. London: Chapman & Hall; 1990. [Google Scholar]
- 2.McFarland W N, Loew E R. Vision Res. 1994;34:1393–1396. doi: 10.1016/0042-6989(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 3.Perry R J, McNaughton P A. J Physiol. 1991;433:561–587. doi: 10.1113/jphysiol.1991.sp018444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleishman L J, Bowman M, Saunders D, Miller W E, Rury M J, Loew E R. J Comp Physiol A. 1997;181:446–460. [Google Scholar]
- 5.Fleishman L J, Loew E R, Leal M. Nature (London) 1993;365:397. [Google Scholar]
- 6.Macedonia J M. Anolis Newsletter. 1999;1999:67–80. [Google Scholar]
- 7.Loew E R. Vision Res. 1994;26:291–298. [Google Scholar]
- 8.Ellingson J M, Fleishman L J, Loew E R. J Comp Physiol A. 1995;177:559–567. doi: 10.1007/BF00207185. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs G H. Am Zool. 1992;32:544–554. [Google Scholar]
- 10.Bennett A T D, Cuthill I C. Vision Res. 1994;34:1471–1478. doi: 10.1016/0042-6989(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 11.Andersson S, Amundsen T. Proc R Soc London Ser B. 1997;264:1587–1591. [Google Scholar]
- 12.Andersson S, Ornborg J, Anderson M. Proc R Soc London Ser B. 1998;265:445–450. [Google Scholar]
- 13.Bennett A T D, Cuthill I C, Partridge J C, Maier E J. Nature (London) 1996;380:433–435. [Google Scholar]
- 14.Bennett A T D, Cuthill I C, Partridge J C, Lunau K. Proc Natl Acad Sci USA. 1997;94:8618–8621. doi: 10.1073/pnas.94.16.8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt S, Bennett A T D, Cuthill I C, Griffiths R. Proc R Soc London Ser B. 1998;265:451–455. [Google Scholar]
- 16.Molina-Borja M. Ethology. 1987;5:11–15. [Google Scholar]
- 17.Molina-Borja M, Padron-Fumero M, Alfonso-Martin T. Ethology. 1988;104:314–322. [Google Scholar]
- 18.Thorpe R S, Brown R P. Biol J Linn Soc. 1989;38:303–322. [Google Scholar]
- 19.Thorpe R S, Black H, Malhotra A. Syst Biol. 1996;45:335–343. [Google Scholar]
- 20.Thorpe R S, Brown R P. Herpetologica. 1991;47:28–37. [Google Scholar]
- 21.Richard M, Thorpe R S. Mol Ecol. 2000;9:1919–1920. doi: 10.1046/j.1365-294x.2000.01052.x. [DOI] [PubMed] [Google Scholar]
- 22.Slatkin M, Excoffier L. Heredity. 1996;76:377–383. doi: 10.1038/hdy.1996.55. [DOI] [PubMed] [Google Scholar]
- 23.Schneider S, Kueffer J-M, Roessli D, Excoffier L. arlequin. University of Geneva: Genetics and Biometry Lab; 1996. , Version 1.1. [Google Scholar]
- 24.Raymond M, Rousset F. J Hered. 1995;83:239. [Google Scholar]
- 25.Gaggiotti O E, Lange O, Rassmann K, Gliddon C J. Mol Ecol. 1999;8:1513–1520. doi: 10.1046/j.1365-294x.1999.00730.x. [DOI] [PubMed] [Google Scholar]
- 26.Slatkin M. Annu Rev Ecol Syst. 1985;16:393–430. [Google Scholar]
- 27.Manly B J F. Multivariate Statistical Methods: a primer. London: Chapman & Hall; 1986. [Google Scholar]
- 28.Manly B J F. Res Popul Ecol. 1986;28:201–218. [Google Scholar]
- 29.Smouse P E, Long E, Sokal R R. Syst Zool. 1986;35:627–632. [Google Scholar]
- 30.Daltry J, Wüster W, Thorpe R S. Nature (London) 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 31.Sheldon B C, Andersson S, Griffith S C, Ornborg J, Sendecka J. Nature (London) 1999;402:874–876. [Google Scholar]
- 32.Finger E, Burkhadt D. Vision Res. 1993;34:1509–1514. doi: 10.1016/0042-6989(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 33.Burkhadt D. J Comp Physiol A. 1989;164:787–796. [Google Scholar]
- 34.Cuthill I C, Bennett A T D, Partridge J C, Maier E J. Amer Nat. 1999;160:183–200. doi: 10.1086/303160. [DOI] [PubMed] [Google Scholar]
- 35.Thorpe R S, Brown R P, Day M, Malhotra A, McGregor D P, Wuster W. In: Phylogenetics and Ecology. Eggleton P, Vane-Wright R, editors. London: Academic; 1994. pp. 189–206. [Google Scholar]
- 36.Juan C, Ibrahim K M, Oromi P, Hewitt G M. Heredity. 1996;77:589–598. doi: 10.1038/hdy.1996.186. [DOI] [PubMed] [Google Scholar]
- 37.Gübitz T, Thorpe R S, Malhotra A. Mol Ecol. 2000;9:1213–1221. doi: 10.1046/j.1365-294x.2000.00997.x. [DOI] [PubMed] [Google Scholar]
- 38.Brown R P, Campos-Delgado R, Pestano J. Mol Ecol. 2000;9:1061–1069. doi: 10.1046/j.1365-294x.2000.00962.x. [DOI] [PubMed] [Google Scholar]
- 39.Jarne P, Lagoda P J L. Trends Ecol Evol. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama S, Shi Y. FEBS Lett. 2000;486:167–172. doi: 10.1016/s0014-5793(00)02269-9. [DOI] [PubMed] [Google Scholar]
- 41.Orr M R, Smith T B. Trends Ecol Evol. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- 42.Smith T B, Wayne R K, Girman D J, Bruford M W. Science. 1997;276:1855–1857. [Google Scholar]
- 43.Thorpe R S, Baez M. Evolution. 1987;41:256–268. doi: 10.1111/j.1558-5646.1987.tb05795.x. [DOI] [PubMed] [Google Scholar]
- 44.Foster S A, Scott R J, Cresko W A. Proc R Soc London Ser B. 1998;353:207–218. [Google Scholar]