Abstract
A direct correlation exists between increased choline kinase (Chk) expression, the resulting increase of phosphocholine levels, and histological tumor grade. To better understand the function of Chk and choline phospholipid metabolism in breast cancer we have stably overexpressed one of the two isoforms of Chk-α, known to be upregulated in malignant cells, in non-invasive MCF-7 human breast cancer cells. Dynamic tracking of cell invasion and cell metabolism was perfomed with a magnetic resonance (MR) compatible cell perfusion assay. The MR based invasion assay demonstrated that MCF-7 cells overexpressing Chk-α (MCF-7-Chk) exhibited an increase of invasion relative to control MCF-7 cells (0.84 vs 0.3). Proton MR spectroscopy studies showed significantly higher phosphocholine and elevated triglyceride signals in Chk overexpressing clones compared to control cells. A test of drug resistance in MCF-7-Chk cells revealed that these cells had an increased resistance to 5-fluorouracil and higher expression of thymidylate synthase compared to control MCF-7 cells. To further characterize increased drug resistance in these cells, we performed rhodamine-123 efflux studies to evaluate drug efflux pumps. MCF-7-Chk cells effluxed twice as much rhodamine-123 compared to MCF-7 cells. Chk-α overexpression resulted in MCF-7 human breast cancer cells acquiring an increasingly aggressive phenotype, supporting the role of Chk-α in mediating invasion and drug resistance, and the use of phosphocholine as a biomarker of aggressive breast cancers.
Keywords: MCF-7 cells, choline kinase, invasion, drug resistance, magnetic resonance spectroscopy
INTRODUCTION
Choline is phosphorylated by choline kinase (Chk) to generate phosphocholine (PC), which represents the first step in the biosynthesis of phosphatidylcholine (1). In mammalian cells, the three known isoforms of Chk (Chk-α1, Chk-α2, and Chk-β) are encoded by two separate genes: Chk-α and Chk-β The two functional isoforms of Chk-α, Chk-α1 and Chk-α2, are the result of alternative splicing of the Chk-α transcript (1). None of the isoforms are active as monomers and the active enzyme consists of homo- or heterodimers (1). PC is a lipid precursor and breakdown product, but some studies have shown that PC can also act as a second messenger in cell growth signaling pathways (2). Thus, an activation of Chk and the resulting increase in PC levels have been proposed as necessary events for the proliferation of certain cell types (3).
Chk activity can be modulated by serum (4) and components of serum such as hormones (4–7), platelet-derived growth factor (8), fibroblast growth factor (8), and epidermal growth factor (5). In addition, it has been found that increased expression of human Chk in fibroblasts increased the mitogenic potential of insulin, insulin-like growth factor-I, fibroblast growth factor, and platelet derived growth factor (9). Recently we observed that hypoxia can induce Chk expression in cancer cells (10). In the same study we also reported a coarse co-localization between total choline (tCho) maps obtained with magnetic resonance spectroscopic imaging (MRSI), and fluorescing hypoxic regions of solid tumors in a human prostate cancer xenograft model that was genetically engineered to express fluorescent protein under the control of a hypoxia response element (10). These data provide evidence that certain microenvironmental conditions within the tumor can regulate Chk levels either by stabilizing the protein, stabilizing its mRNA, or inducing gene expression.
Increased activity of Chk along with the resulting increase in PC levels in malignant cells and tumors have been observed in several studies (3,11,12). For instance, Ramirez De Molina et al. (12) found significant association between Chk overexpression and both histological tumor grade and estrogen receptor status in breast cancers. Chk overexpression and increased activity has also been found in malignant cells and tumors of the prostate, lung, colon, and breast (3).
Over the past decade magnetic resonance spectroscopy (MRS) studies have consistently detected an elevation of PC and tCho in human tumors and cancer cells (13–16). Increased PC is a major contributor to the elevation of the tCho signal detected in preclinical and clinical 1HMRS and magnetic resonance spectroscopic imaging (MRSI) studies of cancer (17). There is increasing evidence to suggest that Chk-α (and not Chk-β) is primarily upregulated in cancers (18–20), and its downregulation has been shown to induce differentiation and apoptosis (21,22). To further understand the role of Chk-α overexpression and choline phospholipid metabolism in breast cancer, we generated MCF-7 breast cancer cell lines that stably overexpress the variant 1 (NM_001277.2) functional isoform of Chk-α. The invasion of MCF-7-Chk cells was compared to MCF-7 cells using an MR-compatible perfused-cell invasion assay, the Metabolic Boyden Chamber (MBC).
Recently we observed that siRNA-mediated downregulation of Chk-α increased the effect of 5-fluorouracil (5-FU) treatment in human breast cancer cells (22). We therefore investigated the relationship between Chk-α overexpression and drug resistance by determining the effect of 5-FU treatment on MCF-7-Chk cells. Consistent with increased resistance to 5-FU treatment, we detected increased thymidylate synthase (TS) expression in MCF-7-Chk cells. To further understand the role of Chk-α in drug resistance, we subjected MCF-7-Chk cells to a rhodamine-123-based efflux assay, which showed that higher efflux occurred in the Chk-α overexpressing cells compared to control MCF-7 cells.
In summary, we found that overexpression of the variant 1 functional isoform of Chk-α in MCF-7 breast cancer cells resulted in the induction of an aggressive phenotype with an increased capacity to invade the extracellular matrix and with increased drug resistance. These results support the role of Chk-α in mediating a drug-resistant and invasive phenotype in breast cancer, and identify it as an important target in breast cancer treatment.
MATERIALS AND METHODS
Cell culture conditions
The MCF-7 human breast cancer cell line was acquired from ATCC and cultured in minimal essential media (MEM) (Sigma, St. Louis, MO) supplemented with 10% (v/v) fetal bovine serum plus 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured in standard cell culture incubator conditions at 37°C in a humidified atmosphere containing 5% CO2.
Cloning the Chk-α coding sequence
The Chk-α variant 1 coding sequence was excised from the pHGCK-1 vector (kindly provided by Hosaka et al. (23)) and cloned into the Not I site of the mammalian expression vector pEF1α-Myc-HisA vector (Invitrogen, Carlsbad, CA), which places the expression of the transgene under the control of the EF-1α promoter. Proper orientation of the cDNA within the context of pEF1α-Myc-HisA was ascertained with an appropriate endonuclease restriction diagnostic of isolated plasmids. Further sequence alignment of the cDNA product confirmed it to be the variant 1 functional isoform of Chk-α (NM_001277.2).
Generation of transgenic MCF-7 cells stably overexpressing Chk-α
Transfections in MCF-7 cells were conducted using the Trans-LT1 transfection reagent (Mirus Biocorp., Madison, WI) according to the manufacturer’s protocol. MCF-7 cells (1 × 106) in a 100 mm dish were transfected with 10 μg pEF1α-Chk-α plasmid. Two days after transfection, cells were dispersed by trypsin/EDTA treatment, diluted (1:10), and seeded into 6 well plates. Cells were maintained in MEM medium supplemented with 10% (v/v) fetal bovine serum plus 100 units/ml penicillin, and 100 μg/ml streptomycin and 800 μg G418/ml for selection. After 4–5 weeks, four large monoclonal colonies were picked and expanded (MCF-7-Chk1 to MCF-7-Chk4). The remaining colonies were maintained and expanded as a polyclonal preparation (MCF-7-ChkP). The empty vector control cells, MCF-7-EV, were produced by transfecting parental MCF-7 cells with the empty vector pEF1α-Myc-HisA following the above protocol.
Immunoblot analyses
Whole-cell extracts were prepared by lysing cells with RIPA lysis buffer supplemented with protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA). Protein concentration was estimated using the Bradford Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Total cellular protein was resolved on SDS-PAGE and specific detection of Chk-α on immunoblots by a custom-designed Chk-α antibody was done as previously described (21). TS expression was scored using a monoclonal antibody (Novus biologicals, NB600-550, Littleton, CO, USA).
Cell proliferation
To measure cell growth, 0.5 × 106 cells were plated on 100-mm dishes. At 24 h, 48 h, and 72 h post-plating, the cells were trypsinized and viable cells counted using trypan blue exclusion.
MR data acquisition with the MBC assay
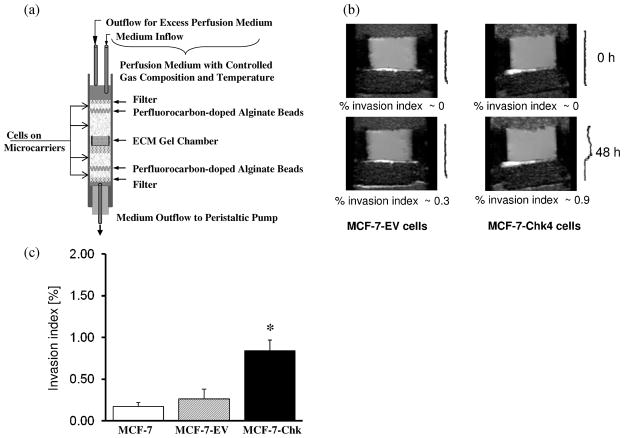
Three days prior to the MR experiments, cells were seeded on Biosilon beads (Nunc, Denmark) at a cell density of 1.5 × 106 cells per 0.5 ml of beads in non–cell culture petridishes (Labtec, Nunc, Denmark) and grown to approximately 70% confluence. A schematic of the MBC is shown in Figure 2a and a detailed description of the MR cell perfusion system can be found in Pilatus et al. (24) and Ackerstaff et al. (25). Briefly, adherently grown cancer cells were layered surrounding an ECM gel chamber. Two layers of perfluorocarbon doped alginate beads were interspersed within the layers of cancer cells grown on Biosilon beads to monitor the oxygen tension in the sample using 19F MR relaxometry.
Figure 2.
(a) Schematic illustration of the Metabolic Boyden Chamber assembly. (b) Representative T1-weighted 1H MR images at the 48 h time point showing degradation of ECM gel by MCF-7 Chk4 cells and no degradation of ECM by MCF-7-EV cells. Corresponding intracellular diffusion-weighted profiles are shown next to the images. (c) Quantitative time-dependent invasion indices I(t) obtained from intracellular diffusion-weighted water profiles at the 36 h time point of MCF-7 cells (open box, n = 2), MCF-7-EV (striped box, n = 4) MCF-7 Chk cells (black box, n = 4). Values are Mean ± SD; *p < 0.05. Data for MCF-7-Chk cells were pooled from MCF-7-Chk4 (n = 3) and MCF-7-ChkP (n = 1) cells.
The following series of MR data were acquired on a 9.4 T MR spectrometer (Bruker, Billerica, MA, USA) every 12 h over a period of 2 days. Proton MR imaging was performed to evaluate the overall sample preparation, to visualize the geometry of the extracellular matrix (ECM) gel, and to detect changes in the integrity of the ECM gel due to invasion and degradation by cancer cells. A one dimensional (1D) 1H profile of intracellular water was acquired along the length of the sample (z-axis), using diffusion-weighted 1D 1H MR imaging with gradient pulses of 3 msec duration, 18 G/cm gradient strength, and diffusion weighting time of 100 msec, to suppress the extracellular water signal. Profiles of intracellular water, acquired with a spatial resolution of 62.5 μm, were used to quantify the number of cancer cells that invaded the ECM gel and to derive an index of invasion. The invasion index I(t) at time t was calculated as follows:
where Ip,7mm(t) and Ip,7mm (t0) is the integral value of the signal at time t and t0 respectively, obtained by integrating the intracellular water signal over a 7 mm region starting at the base of the ECM gel chamber, and Ip(t)and Ip(t0) is the integral of the profile of the entire sample at time t and t0 respectively. The 1st contact of cancer cells with the ECM gel during the loading of the sample was defined as the zero time point, and defined as t0.
Energy metabolites, pH, and the choline phospholipid metabolites, PC, and glycerophosphocholine (GPC), were obtained from unlocalized 1D 31P MR spectra. Intracellular levels of tCho, i.e., signals from PC + GPC + free choline, total creatine (tCr), i.e., signals from creatine + phosphocreatine (PCr), and lactate/triglycerides (LacTG) were derived from unlocalized, diffusion-weighted (DW) 1H MR spectra. DW 1D 1H MR spectra were acquired using lactate-editing to quantify the contribution of Lac and TG to the LacTG signal. Since the slow-diffusing water, which represents intracellular water, is proportional to the number of cells, DW 1D 1H MR water spectra were obtained as an index of cell number to factor in cell proliferation. To quantify the contribution of lactate (Lac) and triglycerides (TGs) to the signal at 1.3 ppm in the unlocalized 1H MR spectra, we acquired diffusion-weighted 1D 1H MR spectra using a spin echo–based pulse sequence with an echo time of 136 ms, 2K data points and 256 scans and lactate editing. Lactate editing was carried out by selective excitation of the lactate methylene resonance (26,27). Spectra acquired with and without selective excitation were used to determine lactate and triglycerides.
Localized DW 1D 1H chemical shift imaging (CSI) spectra with and without water suppression were acquired to obtain metabolic information from 310-μm-thick slices along the z-axis of the sample. In each experiment the metabolites were normalized to intracellular water signal which provided an index of the cell number. Metabolite values normalized to cell number and determined in arbitrary units were averaged from multiple experiments. The oxygen tension was obtained from slice-selective 1D 19F inversion recovery T1 measurements of the perfluorocarbon beads. Localized 1D 1H CSI and 19F MR spectra were acquired every 24 h. The MR data acquisition, processing, and analysis have been described in detail by Ackerstaff et al. (25).
Methylthiazyl blue tetrazolium bromide (MTT) viability assay
The MTT viability assay was carried out using an ATCC kit according to the manufacturer’s protocol (ATCC, Manassas, VA). Briefly, 4–5 × 103 cells were plated in 96 well plates, 24 h later the cells were treated with 2.5 μg/ml 5-FU for 24 h, cultured for another 48 h, then incubated in MTT for 3 h, lysed with detergent, and left overnight. On the following day, absorbance was recorded at 570 nm and percentage survival was determined by comparison with untreated cells.
MR spectroscopy of cell extracts
Cells were harvested and water-soluble cell extracts were obtained using a dual-phase extraction method based on methanol/chloroform/water (1/1/1; v/v/v) as previously described (29). Briefly, approximately 1.5 × 107 cells were harvested by trypsinization, washed twice with saline, and pooled into a glass centrifuge tube. Cell pellets were suspended in ice-cold methanol, vigorously vortexed, and kept on ice for 10 min. Next, 4 ml of chloroform and 4 ml of water were added, the solution was vortexed, and kept at 4°C overnight for phase separation. The samples were centrifuged for 30 min at 7800 g at 4°C and the phases were carefully separated. The water-soluble phase, which contains metabolites such as choline, PC, and GPC, was treated with chelex (Sigma-Aldrich, St. Louis, MO, USA) to remove divalent cations. The chelex beads were removed through filtration. Methanol was removed by rotary evaporation and the remaining water phase was lyophilized. The lipid soluble phase was dried under nitrogen gas. The samples were dissolved in deuterated solvents containing 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid (TSP; Sigma-Aldrich, St. Louis, MO) in the case of water-soluble fractions, or tetramethylsilane (TMS); Cambridge Isotope Laboratories, Inc., Andover, MA, USA) in the case of lipid fractions, to serve as concentration standards and chemical shift reference.
Fully relaxed 1H MR spectra of the extracts were acquired on a Bruker Avance 500 spectrometer operating at 11.7 Tesla (Bruker, Billerica, MA, USA) using a 5-mm inverse probe. MR spectra were analyzed using Bruker XWIN-NMR 3.5 software. Integrals of the N (CH3)3 signals of free Cho at 3.209 ppm, PC at 3.227 ppm, GPC at 3.236 ppm, in the 1H magnetic resonance spectra of water-soluble metabolites, and of phosphatidylcholine at 3.220 ppm and the methylene groups in fatty acids (Fmix) at 1.245 to 1.36 ppm in the 1H magnetic resonance spectra of lipids, were determined and normalized to cell size and number as previously described (21).
Rhodamine-123 efflux assay
For each condition described 1 × 106 cells/ml were used. Rhodamine-123 was added at a final concentration of 0.5 μg/ml for test samples while control cells were left untreated. Cells were gently mixed for 45 min at 37°C, washed twice with cold PBS, and fixed with 0.5% paraformaldehyde. Flow cytometry was performed on a BD FACS Calibur Flow Cytometry System (Becton Dickinson Biosciences, San Jose, CA, USA). Mean rhodamine-123 fluorescence was determined following autofluorescence subtraction. Quantitative analyses were performed using Cellquest software provided by the vendor, for three separate experiments.
Nile-red staining for lipid droplets
Cells were grown on glass chamber slides (Thermo Fisher, Rochester, NY, USA) to 60% to 70% confluence, washed with PBS, and fixed with 3% (w/v) paraformaldehyde. Cells were washed with PBS and incubated with a 1:1,000 dilution in PBS of a 1 mg/ml stock solution of Nile-red (Sigma- Aldrich, St. Louis, MO) in acetone for 10 min at room temperature. Cell nuclei were counterstained with Hoechst H-33342 (Molecular Probes, Eugene, OR). Cells were washed and mounted using Faramount aqueous mounting medium. Fluorescence microscopy was done with a Zeiss LSM 710NLO-Meta confocal laser scanning microscope (Carl Zeiss, Inc., Thornwood, NY, USA) using a C-Apo 40X/1.1W LD water immersion lens. Nile-red stained lipid droplets and Hoechst stained nuclei were excited at 488 and 400 nm, respectively and the fluorescence emission was detected by using 560 nm long pass and 450 to 500-nm band pass filters, respectively. Confocal Z-sections of 1-μm thickness were imaged. The number and size of lipid droplets per cell were quantified using customized software that was developed in-house as previously described (30). Approximately 20–40 cells per field of view from five different fields of view were analyzed for each cell line.
Statistical analyses
Statistical significance was evaluated using the Mann–Whitney U-test. P-values ≤ 0.05 were considered statistically significant unless otherwise stated.
RESULTS
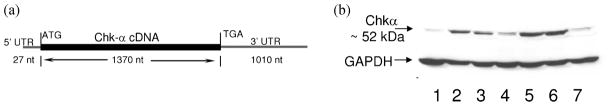
As illustrated in Figure 1a, the 2.4 Kb Chk-α cDNA (Accession number D10704) includes the 1370 nucleotide (nt) coding sequence and some 5′- and 3′-untranslated region (UTR) sequence. MCF-7 clones that stably overexpress variant 1 isoform of functional Chk-α were generated and four monoclonal cell lines (MCF-7-Chk1 to MCF-7-Chk4) along with pooled clones (MCF-7-ChkP) were selected for characterization. The immunoblot assay shown in Figure 1b demonstrates that all Chk-α overexpressing MCF-7 clones (lanes 2–6) exhibit 3–4 fold increased Chk expression levels compared to MCF-7-EV (lane 1) and parental wild-type MCF-7 cells (lane 7). No significant differences in growth rate were observed between MCF-7, MCF-7-EV, MCF-7-Chk4 or MCF-7-ChkP cells.
Figure 1.
(a) Linear representation of the Chk-α cDNA used in the pEF1α-Myc/His vector construct. (b) Representative immunoblot showing relative overexpression levels of Chk in cell lysates of MCF-7-Chk-α clones (Chk1-lane 2, Chk2-lane 3, Chk3-lane 4, Chk4-lane 5, ChkP-lane 6) compared with MCF-7-EV (lane 1) and MCF-7 control cells (lane 7). GAPDH served as a loading control.
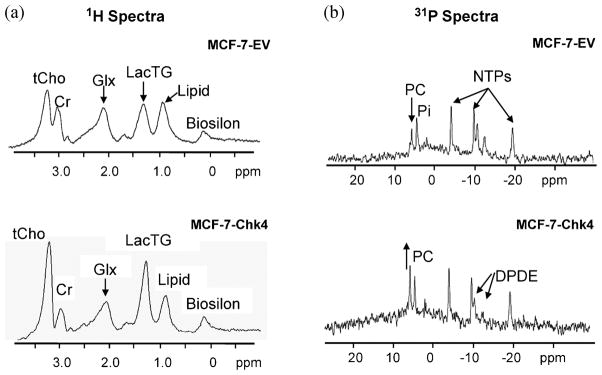
Figure 3.
Representative (a) 1H and (b) 31P MR spectra from perfused cells acquired at 400 MHz, with 128 and 2000 averages respectively, at the 12 h time point. Abbreviations: tCho, total choline; Cr, creatine; Glx, glutamine + glutamate; LacTG, lactate/triglyceride; PC, phosphocholine; DPDE, diphosphodiester, Pi, inorganic phosphate; NTP, nucleoside triphosphate.
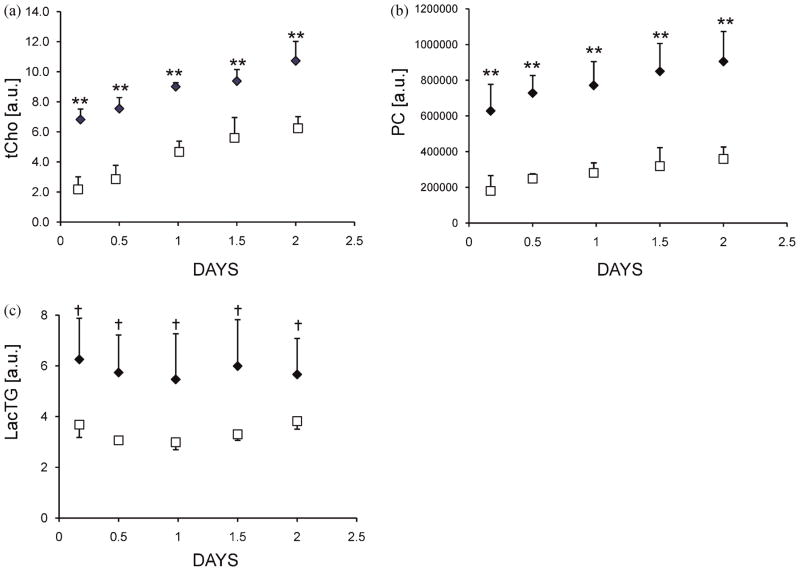
Figure 4.
Quantification of data from 1H and 31P MR spectra in arbitrary units (a.u.) demonstrating differences in (a) tCho, (b) PC and (c) LacTG levels between MCF-7-EV (□; n = 4) and MCF-7-Chk cells (◆; n = 4). Metabolites were integrated and normalized to the intracellular water signal to account for cell proliferation. Values are Mean ± SD; **p < 0.01, +p < 0.1). Data for MCF-7-Chk cells were pooled from MCF-7-Chk4 (n = 3) and MCF-7-ChkP (n = 1) cells.
The non-invasive and longitudinal characterization of the invasion and metabolism of MCF-7-Chk cells or control cells was performed under carefully controlled environmental conditions with the MBC assay (schematic shown in Fig. 2a). Based on the immunoblot characterization of Chk expression levels in the overexpressing clones, MCF-7-Chk4 (lane 5 in Fig. 1b) and MCF-7-ChkP (lane 6 in Fig. 1b) cells were selected for the studies. Representative 1H MR images of the ECM gel region obtained at the start of the MR experiment and 48 h later, are shown in Figure 2b, and demonstrate an increase of invasion into ECM gel by MCF-7-Chk4 cells at 48 h relative to control MCF-7 cells. Quantitative time-dependent invasion indices I(t) obtained from intracellular DW water profiles demonstrated that the invasion of MCF-7-Chk cells (0.84 ± 0.22) was significantly higher than parental wild type MCF-7 (0.21 ±.05) and MCF-7-EV cells (0.305 ± 0.13) (p < 0.04) (Fig. 2c). Overexpression of Chk-α resulted in a small but reproducible increase of invasion.
Representative 1H and 31P spectra obtained from MCF-7-Chk4 and MCF-7-EV cells corresponding to the data shown in Figure 2b at the 12 h time point are shown in Figure 3. The 1H spectra demonstrate that overexpression of Chk-α resulted in an increase of tCho and LacTG in MCF-7-Chk cells relative to MCF-7-EV cells (Fig. 3a). The 31P spectra demonstrated that the observed increase in Chk protein levels on immunoblots was functional since higher levels of PC were observed in MCF-7-Chk4 cells compared to MCF-7-EV cells (Fig. 3b). Quantitative MRS data collected over the course of two days are summarized in Figure 4. Levels of tCho and PC in MCF-7-Chk4 cells were significantly higher than those found in MCF-7-EV cells through the time course of the cell-perfusion experiments (n = 4, p < 0.01). As shown in Figure 4c, the levels of LacTG tended to be higher in MCF-7-Chk cells compared to MCF-7-EV cells (n = 4, p < 0.1). There was no significant difference in lactate levels between MCF-7-EV and MCF-7-Chk4 cells estimated through a spin-echo lactate-edited sequence (data not shown), consistent with the finding that there was no significant difference in pH between these cells. These results also confirm that the increase of the LacTG signal was primarily due to an increase in the triglyceride signal. Our data suggest that the signal from LacTG normalized to cell number remained constant during cell growth. Total choline and PC values normalized to cell number increased during cell growth, consistent with previous observations that tCho and PC increase during the log phase of cell proliferation (31).
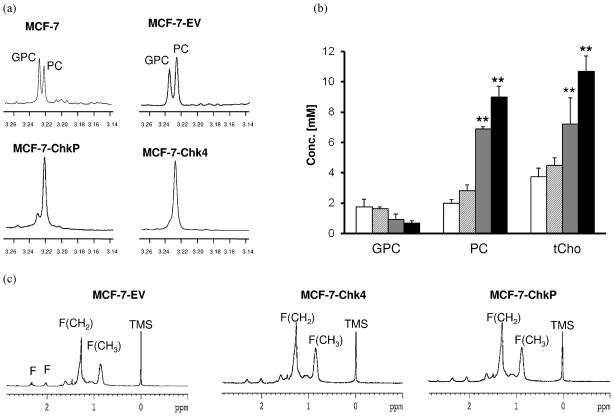
We next studied the contribution of different choline containing compounds to the tCho signal. Representative high-resolution 1H MR spectra from the Cho region obtained from water-soluble tumor extracts of MCF-7, MCF-7-EV, MCF-7-ChkP, and MCF-7-Chk4 cells are presented in Figure 5a. The corresponding PC/GPC ratios for those spectra were 0.90, 1.20, 5.14, and 16.0 respectively. Analyses of the MR spectra from independent experiments demonstrated that the PC/GPC ratios were significantly elevated in MCF-7-ChkP and MCF-7-Chk4 cells relative to MCF-7 control cells ( p < 0.05, n = 3). Levels of PC and GPC along with tCho levels for each cell line are presented in Figure 5b. PC levels were significantly higher in MCF-7-ChkP and MCF-7-Chk4 cells compared to MCF-7 and MCF-7-EV cells ( p < 0.01, n = 3). The increase of tCho in the Chk-α overexpressing cell lines was primarily due to the increase of PC. Significant differences in lactate were not detected in the spectra (data not shown).
Figure 5.
(a) Representative expanded 1H MR spectra showing the choline region from MCF-7, MCF-7-EV, MCF-7-ChkP, and MCF-7-Chk4 cells. (b) Histogram showing levels of GPC, PC and tCho in MCF-7 (open box, n = 3), MCF-7-EV (striped box, n = 3), MCF-7-ChkP (gray box, n = 3), and MCF-7-Chk4 (black box, n = 3) cells. Values are Mean ± SD; **p < 0.01. (c) Representative high resolution 1H MR spectra from lipid extracts of MCF-7-EV cells (n = 2) and Chk-α overexpressing MCF-7 cells (n = 4) demonstrating increase of fatty acid (F)/triglyceride signals (± 1.75 fold for F(CH2), p < 0.03, and ~ 2.0 fold for F(CH3), p < 0.05) in MCF-7-Chk cells when compared to MCF-7-EV cells. Data for MCF-7-Chk cells were pooled from MCF-7-Chk4 (n = 2) and MCF-7-ChkP (n = 2) cells.
Representative 1H MR spectra from the lipid extracts presented in Figure 5c demonstrate the increased fatty acid/triglyceride/phospholipid signal observed in MCF-7-Chk cells compared to MCF-7-EV, consistent with the increase of the LacTG signal in Figures 3a and 4c in the perfused cells. Mean ± SD values in arbitrary units were 7.43 ± 2.43 for MCF-7-Chk (n = 4) vs 4.25 ± 0.35 for MCF-7-EV (n = 2) cells ( p < 0.03) for F(CH2), and 6.25 ± 0.94 for MCF-7-Chk (n = 4) vs 3.25 ± 0.35 for MCF-7-EV (n = 2) cells ( p < 0.05) for F(CH3). There were no significant differences in phospatidylcholine levels between choline kinase overexpressing MCF-7 cells and control MCF-7 cells in the lipid extracts.
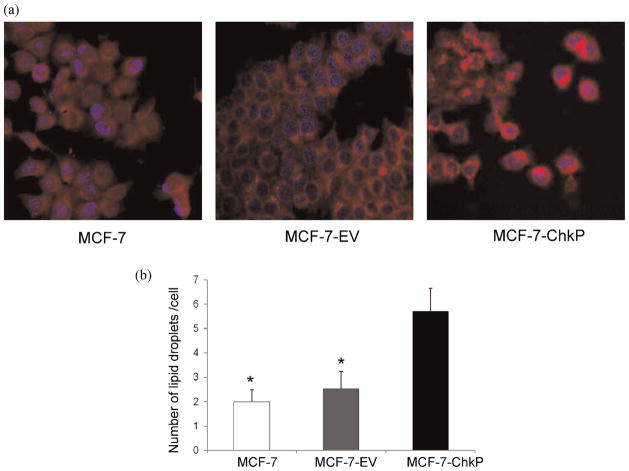
Representative images of Nile red staining for lipid droplets in MCF-7, MCF-7-EV and MCF-7-ChkP cells are shown in Figure 6a, and demonstrate an increase in the number of lipid droplets in MCF-7-ChkP cells. Median values of the number of lipid droplets are presented in Figure 6b that demonstrate a significant increase in the number of lipid droplets in MCF-7-ChkP cells compared to MCF-7 or MCF-7-EV cells ( p < 0.05) (Fig. 6b). There were no significant differences in the size of the lipid droplets.
Figure 6.
(a) Representative confocal images of lipid droplets visualized by Nile red staining (red), with Hoechst-stained nuclei (blue) in MCF-7 control and MCF-7-ChkP breast cancer cells. (b) Quantitation of the number of lipid droplets showing significant differences between MCF-7 or MCF-7-EV and MCF-7-ChkP cells. Values are Median ± SE, *p < 0.05.
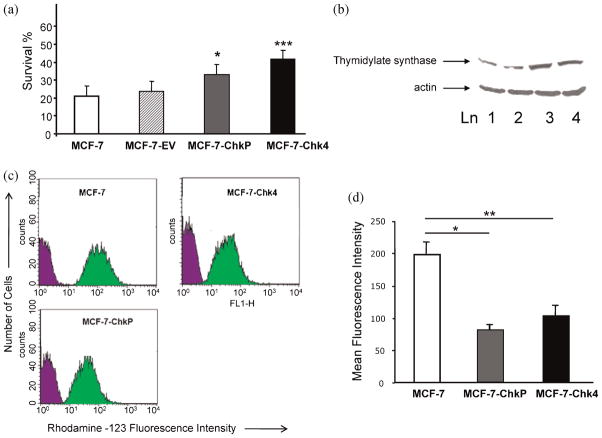
MCF-7-ChkP and MCF-7-Chk4 cells were more resistant to 5-FU treatment than MCF-7-EV and MCF-7 cells, as observed in the MTT viability assay. As shown in Figure 7a, survival of MCF7-Chk4 (41.86 ± 4.90) and MCF-7-ChkP (33.11 ± 5.41) cells was significantly higher than the survival observed in MCF-7 (21.12 ± 5.57) or MCF-7-EV (23.5 ± 5.70) cells ( p < 0.05 n = 4). Since TS is a critical target for fluoropyrimidines such as 5-FU, that are widely used in the treatment of solid tumors, we determined its expression in our cell lines. We found increased expression of TS in MCF-7-Chk cells compared to control cells (Fig. 7b).
Figure 7.
(a) MTT assay of MCF-7 (open box, n = 4), MCF-7-EV (striped box, n = 4), MCF-7-ChkP (gray box, n = 4) and MCF-7-Chk4 (black box, n = 4) cells following treatment with 2.5 μg/ml 5-FU for 24 h. Values are Mean ± SD; *p < 0.05 for MCF-7-ChkP vs MCF-7 and EV; ***p < 0.003 for MCF-7-Chk4 vs MCF-7 and MCF-7-EV). (b) Representative immunoblot from three sets of experiments showing increased levels of TS expression in cell lysates of MCF-7-Chk4 cells (lane 3), MCF-7-ChkP (lane 4) compared with MCF-7 (lane 1) and MCF-7-EV (lane 2) control cells. (c) Representative histograms obtained from rhodamine-123 efflux assay of MCF-7, MCF-7-Chk4, and MCF-7-ChkP cells. Mean fluorescence intensity (MFI) of the cells before (blue) and after (green) incubation with rhodamine-123 is shown on the x-axis. (d) Intracellular accumulation of rhodamine-123 as detected by MFI in MCF-7 (open box, n = 3), MCF-7-ChkP (gray box, n = 3), and MCF-7-Chk4 cells (black box, n = 3). Values are Mean ± SEM, *p < 0.05 and **p < 0.01).
Data from a rhodamine-123 efflux assay of MCF-7, MCF-7-Chk4, and MCF-7-ChkP cell lines presented as flow cytometry histograms of fluorescence intensity vs cell number are shown in Figure 7c. MCF-7-ChkP and MCF-7-Chk4 cells exhibited lower fluorescence intensities than wild-type MCF-7 cells, indicating that Chk-α overexpressing cells effluxed more rhodamine-123. A quantitative summary of multiple repeats of these experiments, displayed as mean fluorescence intensities, is presented in Figure 7d. We observed that the retention of rhodamine-123 in MCF-7 cells was at least two-fold higher than in the Chk overexpressing cells, further confirming the possibility that Chk-α overexpression increases the activity of drug efflux pumps.
DISCUSSION
Chk-α overexpression was found to mediate increased invasion and drug resistance in the ER/PR positive, poorly invasive and non-metastatic MCF-7 breast cancer cell line. As anticipated, a profound increase of PC was observed in these cells, confirming the role of Chk-α in the high PC levels consistently observed in cancer cells and human tumors with MR spectroscopy (32). Additionally we observed a significant increase of fatty acid/triglycerides/phospholipids with Chk-α overexpression in cell extracts.
Chk has previously been linked to cellular processes that result in malignant transformation as recently demonstrated by Ramirez De Molina et al. (33). Their data indicated that elevated Chk levels might modulate a Rho A/ROCK kinase pathway during the oncogenic transformation of human embryo kidney fibroblasts. Our previous studies have also shown that Chk and PC levels are relatively low in non-tumorigenic human breast epithelial cell lines but increase as a function of the aggressiveness of the breast cancer cell lines, with the highest levels found in the most aggressive cell lines (11,21). High grade patient tumor samples also exhibit higher levels of Chk as well as PC levels relative to low grade tumor samples (3,34–36).
In addition to the increase of PC, we also observed a significant increase of fatty acids/triglycerides/phospholipids in cell extracts of Chk-α overexpressing MCF-7 cells. Consistent with the increase of fatty acids/triglyceride signal we also observed a significant increase of lipid droplets in the Chk-α overexpressing MCF-7 cells. Interestingly, we previously observed an increase of fatty acids/triglycerides/phospholipids in the ER/PR negative invasive MDA-MB-231 human breast cancer cells following siRNA-mediated downregulation of Chk-α (21). In that study we also observed an increase of lipid droplets, that is also a marker of cell differentiation, following the downregulation of Chk-α. Others have found higher triglycerides in malignant gliomas (37) and adenocarcinomas (38) compared to benign or normal tissue samples. It is possible that the increase of fatty acids/triglycerides/phospholipids and the formation of lipid droplets following Chk-α overexpression and Chk-α downregulation occurs through entirely independent pathways that require further investigation.
We studied the differential invasiveness of MCF-7 and MCF-7-Chk cells using the MBC assay. This model system has been validated in several of our previous reports (25,39). Those studies have shown consistent and reproducible estimation of cancer cell invasion into the ECM under a variety of well-controlled environmental conditions. The MBC data presented here indicate that MCF-7-Chk cells acquired a modest but significantly increased ability to degrade and invade basement membrane-like ECM gel relative to MCF-7 and MCF-7-EV control cells. This increased invasiveness of MCF-7-Chk cells supports the potential role of Chk-α in mediating tumor progression and metastasis.
There is a direct correlation between Chk levels and the aggressiveness of various cancers (3,19), and the latter has been associated with drug resistance. We previously found that siRNA mediated downregulation of Chk-α increased cell kill following 5-FU treatment in human breast cancer cells (22). Here we found that Chk-α overexpression imparted resistance to 5-FU in MCF-7 cells. Inhibition of TS by 5-fluoro-dUMP (FdUMP), the active metabolite of 5-FU, is considered to be the main mechanism for the action of 5-FU, and a high enzyme level before treatment has been related to intrinsic resistance (40). We examined TS levels in MCF-7 cells overexpressing Chk-α and observed an increase in TS expression in MCF7-Chk cells. Mechanisms resulting in the increase of TS with Chk-α overexpression, especially in the absence of any change in cell proliferation, require further investigation.
To further understand the role of Chk-α in drug resistance we characterized rhodamine-123 efflux in Chk overexpressing and control cells. Three ATP-binding cassette (ABC) drug efflux pumps account for most of MDR in both human and rodent cells: P-glycoprotein (Pgp), multidrug resistance associated protein 1 (MRP1), and breast cancer resistance protein (BCRP) (41). These pumps provide mechanisms for cellular detoxification, and provide protection against xenobiotic substances (41). In many cases the MDR of cancer cell lines has been shown to be mediated by overexpression of Pgp (41). This overexpression results in the enhanced elimination of a variety of drugs from the cells with concomitant lower cytotoxicity. When added to cultured cells, the fluorescent dye rhodamine-123 normally accumulates in the mitochondria (42). It is a substrate for Pgp and is used as a molecular probe in studies of the MDR phenotype (42–44). An aberrant increase in Pgp activity results in the efflux of rhodamine-123 from cells, which can be monitored with FACS studies. Our data indicate that increased Chk-α expression results in the increased efflux of rhodamine-123 suggesting that Chk-α can modulate Pgp or other ABC transporters that result in increased efflux of a typical substrate. Consistent with these data, Comerford et al. (45) observed that hypoxia enhances Pgp function by 7-fold over normoxia and increased the MDR of the cells. Similarly, Krishnamurthy et al. (46) showed that increased BCRP expression during hypoxia provided an important cell survival advantage, which enhanced cell survival by reducing the accumulation of toxic heme metabolites. We have also shown an induction of Chk expression in cancer cells subjected to hypoxia and found that this can be modulated by hypoxia inducible factor-1 via hypoxia response elements within the Chk-α1 promoter region (10). Hypoxia-induced overexpression of these ABC transporters along with Chk may thus represent a pathway for cell survival under hypoxia and resistance to chemotherapeutic treatment of solid tumors.
In summary, Chk-α overexpression in breast cancer cells may represent a pathway for cell survival and resistance to chemotherapeutic treatment of solid tumors, as well as increased invasion. As Chk-α is increased in a number of other cancers, its contribution to drug resistance associated with increased expression of TS and increased cell invasion might be a general phenomenon. Our data suggest Chk-α may represent a novel and critical target to reduce cancer cell invasion and drug resistance, and support further preclinical and clinical studies of this target with a wide spectrum of cell lines and tumors.
Acknowledgments
Contract/grant sponsor: NIH; contract/grant numbers: P50 CA103175, R01 CA73850, RO1 CA82337.
This work was supported by NIH P50 CA103175, R01 CA73850 and RO1 CA82337. We gratefully acknowledge and thank Dr K. Hosaka for providing the variant 1 functional isoform of Chk-α cDNA. We thank Drs C. Aoyama and K. Ishidate for their assistance. We thank Drs D. C. Shungu and X. Mao for generously providing the XsOsNMR software used for MR data analyses, and Mr M. Solaiyappan for providing the software used for the lipid droplet analysis.
Abbreviations used
- Chk
choline kinase
- Chk P
choline kinase overexpressing pooled clones
- Chk-4
choline kinase overexpressing clone 4
- ECM
extracellular matrix
- FdUMP
5-fluoro-dUMP
- GPC
glycerophosphocholine
- MBC
Metabolic Boyden Chamber
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance, spectroscopic imaging
- PC
phosphocholine
- Pgp
P-glycoprotein
- tCho
total choline-containing metabolites (GPC + PC + free choline)
- TMS
tetramethylsilane
- TS
thymidylate synthase
- TSP
3-(trimethylsilyl) propionic-2,2,3,3,-d4 acid
References
- 1.Aoyama C, Liao H, Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog Lipid Res. 2004;43:266–281. doi: 10.1016/j.plipres.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Kent C. Regulation of phosphatidylcholine biosynthesis. Prog Lipid Res. 1990;29:87–105. doi: 10.1016/0163-7827(90)90010-i. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez J, Bonilla F, Rosell R, Lacal J. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 4.Warden CH, Friedkin M. Regulation of choline kinase activity and phosphatidylcholine biosynthesis by mitogenic growth factors in 3T3 fibroblasts. J Biol Chem. 1985;260:6006–6011. [PubMed] [Google Scholar]
- 5.Uchida T. Stimulation of phospholipid synthesis in HeLa cells by epidermal growth factor and insulin: activation of choline kinase and glycerophosphate acyltransferase. Biochim Biophys Acta. 1996;1304:89–104. doi: 10.1016/s0005-2760(96)00109-9. [DOI] [PubMed] [Google Scholar]
- 6.Pelech SL, Vance DE. Regulation of phosphatidylcholine biosynthesis. Biochim Biophys Acta. 1984;779:217–251. doi: 10.1016/0304-4157(84)90010-8. [DOI] [PubMed] [Google Scholar]
- 7.Vigo C, Vance DE. Effect of diethylstilboestrol on phosphatidylcholine biosynthesis and choline metabolism in the liver of roosters. Biochem J. 1981;200:321–326. doi: 10.1042/bj2000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jimenez B, del Peso L, Montaner S, Esteve P, Lacal JC. Generation of phosphorylcholine as an essential event in the activation of Raf-1 and MAP-kinases in growth factors-induced mitogenic stimulation. J Cell Biochem. 1995;57:141–149. doi: 10.1002/jcb.240570114. [DOI] [PubMed] [Google Scholar]
- 9.Chung T, Huang JS, Mukherjee JJ, Crilly KS, Kiss Z. Expression of human choline kinase in NIH 3T3 fibroblasts increases the mitogenic potential of insulin and insulin-like growth factor I. Cell Signal. 2000;12:279–288. doi: 10.1016/s0898-6568(00)00065-6. [DOI] [PubMed] [Google Scholar]
- 10.Glunde K, Shah T, Winnard PT, Jr, Raman V, Takagi T, Vesuna F, Artemov D, Bhujwalla ZM. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 alpha signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 12.Ramirez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J, Bonilla F, Sanchez JJ, Lacal JC. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–4322. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 13.Hourani R, Horska A, Albayram S, Brant LJ, Melhem E, Cohen KJ, Burger PC, Weingart JD, Carson B, Wharam MD, Barker PB. Proton magnetic resonance spectroscopic imaging to differentiate between nonneoplastic lesions and brain tumors in children. J Magn Reson Imaging. 2006;23:99–107. doi: 10.1002/jmri.20480. [DOI] [PubMed] [Google Scholar]
- 14.Casciani E, Polettini E, Bertini L, Emiliozzi P, Amini M, Pansadoro V, Gualdi GF. Prostate cancer: evaluation with endorectal MR imaging and three-dimensional proton MR spectroscopic imaging. Radiol Med (Torino) 2004;108:530–541. [PubMed] [Google Scholar]
- 15.Kim JK, Park SH, Lee HM, Lee YH, Sung NK, Chung DS, Kim OD. In vivo 1H-MRS evaluation of malignant and benign breast diseases. Breast. 2003;12:179–182. doi: 10.1016/s0960-9776(03)00012-2. [DOI] [PubMed] [Google Scholar]
- 16.Bhakoo KK, Williams SR, Florian CL, Land H, Noble MD. Immortalization and transformation are associated with specific alterations in choline metabolism. Cancer Res. 1996;56:4630–4635. [PubMed] [Google Scholar]
- 17.Glunde K, Ackerstaff E, Mori N, Jacobs MA, Bhujwalla ZM. Choline phospholipid metabolism in cancer: consequences for molecular pharmaceutical interventions. Mol Pharm. 2006;3:496–506. doi: 10.1021/mp060067e. [DOI] [PubMed] [Google Scholar]
- 18.Banez-Coronel M, de Molina AR, Rodriguez-Gonzalez A, Sarmentero J, Ramos MA, Garcia-Cabezas MA, Garcia-Oroz L, Lacal JC. Choline kinase alpha depletion selectively kills tumoral cells. Curr Cancer Drug Targets. 2008;8:709–719. doi: 10.2174/156800908786733432. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez deMolina, Sarmentero-Estrada A, Belda-Iniesta J, Taron C, Ramirez M, de Molina V, Cejas P, Skrzypski M, Gallego-Ortega D, de Castro J, Casado E, Garcia-Cabezas MA, Sanchez JJ, Nistal M, Rosell R, Gonzalez-Baron M, Lacal JC. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 20.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 21.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 22.Mori N, Glunde K, Takagi T, Raman V, Bhujwalla ZM. Choline kinase down-regulation increases the effect of 5-fluorouracil in breast cancer cells. Cancer Res. 2007;67:11284–11290. doi: 10.1158/0008-5472.CAN-07-2728. [DOI] [PubMed] [Google Scholar]
- 23.Hosaka K, Tanaka S, Nikawa J, Yamashita S. Cloning of a human choline kinase cDNA by complementation of the yeast cki mutation. FEBS Lett. 1992;304:229–232. doi: 10.1016/0014-5793(92)80625-q. [DOI] [PubMed] [Google Scholar]
- 24.Pilatus U, Aboagye E, Artemov D, Mori N, Ackerstaff E, Bhujwalla ZM. Real-time measurements of cellular oxygen consumption, pH, and energy metabolism using nuclear magnetic resonance spectroscopy. Magn Reson Med. 2001;45:749–755. doi: 10.1002/mrm.1102. [DOI] [PubMed] [Google Scholar]
- 25.Ackerstaff E, Gimi B, Artemov D, Bhujwalla ZM. Anti-inflammatory agent indomethacin reduces invasion and alters metabolism in a human breast cancer cell line. Neoplasia. 2007;9:222–235. doi: 10.1593/neo.06673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hetherington HP, Avison MJ, Shulman RG. 1H homonuclear editing of rat brain using semiselective pulses. Proc Natl Acad Sci USA. 1985;82:3115–3118. doi: 10.1073/pnas.82.10.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hetherington HPHJ, Pan JW, Rothman DL, Shulman RG. A fully localized 1H homonuclear editing sequence to observe lactate in human skeletal muscle after exercise. J Magn Reson. 1989;82:86–96. [Google Scholar]
- 28.Pilatus U, Ackerstaff E, Artemov D, Mori N, Gillies RJ, Bhujwalla ZM. Imaging prostate cancer invasion with multi-nuclear magnetic resonance methods: the Metabolic Boyden Chamber. Neoplasia. 2000;2:273–279. doi: 10.1038/sj.neo.7900089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn Reson Med. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- 30.Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–545. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aiken NR, Gillies RJ. Phosphomonoester metabolism as a function of cell proliferative status and exogenous precursors. Anticancer Res. 1996;16:1393–1397. [PubMed] [Google Scholar]
- 32.Jagannathan NR, Kumar M, Seenu V, Coshic O, Dwivedi SN, Julka PK, Srivastava A, Rath GK. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Cancer. 2001;84:1016–1022. doi: 10.1054/bjoc.2000.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramirez de Molina A, Gallego-Ortega D, Sarmentero J, Banez-Coronel M, Martin-Cantalejo Y, Lacal JC. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer Res. 2005;65:5647–5653. doi: 10.1158/0008-5472.CAN-04-4416. [DOI] [PubMed] [Google Scholar]
- 34.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001;61:3599–3603. [PubMed] [Google Scholar]
- 35.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 36.Morse DL, Carroll D, Day S, Gray H, Sadarangani P, Murthi S, Job C, Baggett B, Raghunand N, Gillies RJ. Characterization of breast cancers and therapy response by MRS and quantitative gene expression profiling in the choline pathway. NMR Biomed. 2009;22:114–127. doi: 10.1002/nbm.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tugnoli V, Tosi MR, Barbarella G, Bertoluzza A, Ricci R, Trevisan C. In vivo 1H MRS and in vitro multinuclear MR study of human brain tumors. Anticancer Res. 1996;16:2891–2899. [PubMed] [Google Scholar]
- 38.Calabrese C, Pisi A, Di Febo G, Liguori G, Filippini G, Cervellera M, Righi V, Lucchi P, Mucci A, Schenetti L, Tonini V, Tosi MR, Tugnoli V. Biochemical alterations from normal mucosa to gastric cancer by ex vivo magnetic resonance spectroscopy. Cancer Epidemiol Bio-markers Prev. 2008;17:1386–1395. doi: 10.1158/1055-9965.EPI-07-2676. [DOI] [PubMed] [Google Scholar]
- 39.Ackerstaff E, Artemov D, Gillies RJ, Bhujwalla ZM. Hypoxia and the presence of human vascular endothelial cells affect prostate cancer cell invasion and metabolism. Neoplasia. 2007;9:1138–1151. doi: 10.1593/neo.07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestalozzi BC, Peterson HF, Gelber RD, Goldhirsch A, Gusterson BA, Trihia H, Lindtner J, Cortes-Funes H, Simmoncini E, Byrne MJ, Golouh R, Rudenstam CM, Castiglione-Gertsch M, Allegra CJ, Johnston PG. Prognostic importance of thymidylate synthase expression in early breast cancer. J Clin Oncol. 1997;15:1923–1931. doi: 10.1200/JCO.1997.15.5.1923. [DOI] [PubMed] [Google Scholar]
- 41.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 42.Johnson LV, Walsh ML, Bockus BJ, Chen LB. Monitoring of relative mitochondrial membrane potential in living cells by fluorescence microscopy. J Cell Biol. 1981;88:526–535. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neyfakh AA. Use of fluorescent dyes as molecular probes for the study of multidrug resistance. Exp Cell Res. 1988;174:168–176. doi: 10.1016/0014-4827(88)90152-8. [DOI] [PubMed] [Google Scholar]
- 44.Tapiero H, Munck JN, Fourcade A, Lampidis TJ. Cross-resistance to rhodamine 123 in Adriamycin- and daunorubicin-resistant Friend leukemia cell variants. Cancer Res. 1984;44:5544–5549. [PubMed] [Google Scholar]
- 45.Comerford KM, Wallace TJ, Karhausen J, Louis NA, Montalto MC, Colgan SP. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62:3387–3394. [PubMed] [Google Scholar]
- 46.Krishnamurthy P, Ross DD, Nakanishi T, Bailey-Dell K, Zhou S, Mercer KE, Sarkadi B, Sorrentino BP, Schuetz JD. The stem cell marker Bcrp/ABCG2 enhances hypoxic cell survival through interactions with heme. J Biol Chem. 2004;279:24218–24225. doi: 10.1074/jbc.M313599200. [DOI] [PubMed] [Google Scholar]