Abstract
Staphylococcus aureus is a common cause of infections that has undergone rapid global spread over recent decades. Formal phylogeographic methods have not yet been applied to the molecular epidemiology of bacterial pathogens because the limited genetic diversity of data sets based on individual genes usually results in poor phylogenetic resolution. Here, we investigated a whole-genome single nucleotide polymorphism (SNP) data set of health care-associated Methicillin-resistant S. aureus sequence type 239 (HA-MRSA ST239) strains, which we analyzed using Markov spatial models that incorporate geographical sampling distributions. The reconstructed timescale indicated a temporal origin of this strain shortly after the introduction of Methicillin, followed by global pandemic spread. The estimate of the temporal origin was robust to the molecular clock, coalescent prior, full/intergenic/synonymous SNP inclusion, and correction for excluded invariant site patterns. Finally, phylogeographic analyses statistically supported the role of human movement in the global dissemination of HA-MRSA ST239, although it was unable to conclusively resolve the location of the root. This study demonstrates that bacterial genomes can indeed contain sufficient evolutionary information to elucidate the temporal and spatial dynamics of transmission. Future applications of this approach to other bacterial strains may provide valuable epidemiological insights that may justify the cost of genome-wide typing.
Keywords: Bayesian inférence, phylogeography, phylogenetics, measurably evolving population
Introduction
High-resolution Bayesian phylogenetic methods are powerful tools, capable of elucidating the temporal and spatial dynamics of viral epidemics (Pybus and Rambaut 2009), including those of HIV-1 (e.g., Gray et al. 2009), HCV (i.e., Markov et al. 2009), and influenza (e.g., Rambaut et al. 2008). Because these methods assume a molecular clock, the data set under study must either represent sequences sampled at different times from a measurably evolving population (MEP; Drummond, Pybus, and Rambaut 2003) or the study must incorporate an independent calibration date or evolutionary rate (Pybus 2006). A population can be considered to be measurably evolving only if a significant number of mutations accumulate between sampling times (Drummond et al. 2002), which can be evaluated using a likelihood ratio test (Rambaut 2000). Until now, these methods have been typically applied to RNA viruses, which accumulate mutations at rates around 10−3–10−4 nucleotide substitutions/site/year, thereby generating sufficient genetic diversity within their relatively short genomes (∼103 nucleotides) over limited periods of time (months, years, or decades; Drummond, Pybus, Rambaut, Forsberg, and Rodrigo 2003; Belshaw et al. 2008). A recent study has evaluated the reliability of using MEP methods when applied to DNA viruses, whose evolutionary rates are thought to be orders of magnitude lower than RNA viruses (Firth et al. 2010). Based on extensive simulations, the authors concluded that, given enough variable sites, accurate estimation of divergence times was possible even when the underlying evolutionary rate was 10−7 substitutions/site/year (Firth et al. 2010).
Bacterial populations, on the other hand, have typically not been considered as MEPs because the limited genetic diversity of individual genes, or of multi-locus sequence typing (MLST) loci, contain insufficient phylogenetic signal (Achtman 2008) to exhibit measurable evolution over the timeframes of typical epidemiological studies. Only three studies to date have used serial-sample coalescent methods to reconstruct evolutionary patterns in bacteria: Staphylococcus aureus sequence type (ST)-5 (Lowder et al. 2009) and ST-225 (Nubel et al. 2010) and Neisseria gonorrhoeae (Tazi et al. 2010), although none of these studies employed Bayesian phylogeography. The evolutionary rates estimated for S. aureus when the sampling dates of serially sampled sequences were taken into account were on the order of 10−6 substitutions/site/year (Harris et al. 2010; Nubel et al. 2010). In contrast, older estimates of bacterial evolutionary rates, based on external calibration dates, were on the order of 10−9 substitutions/site/year (Ochman et al. 1999). It is known that using an independent calibration date or evolutionary rate can often lead to erroneous timescales if chosen without diligence (Shapiro et al. 2006), as rates of nucleotide change can differ by several orders of magnitude depending on the amount of time separating two sequences (Ho et al. 2005). Indeed, the slower rate estimate for bacteria has been acknowledged to suspect by the original author (Achtman 2008). On the other hand, is unclear whether heterochronous methods can be applied to bacteria, that is, whether they represent an MEP. If Bayesian methods are applied inappropriately to serially sampled sequences then inappropriate conclusions may be drawn (Firth et al. 2010).
Recent advances in sequencing technologies mean that the investigation of bacterial genetic variation at the whole-genome level is now experimentally and economically feasible, as recently demonstrated by Harris et al. (2010). This gives rise to the possibility that enough bacterial genetic diversity can be observed over short timeframes to consider such populations as measurably evolving, thereby allowing the estimation of divergence times and the reconstruction of dispersal patterns without the need for an independent temporal calibration, although this was not specifically tested in the previous publication. In this study, we investigated a data set of 4,310 genome-wide single nucleotide polymorphisms (SNPs) of health care-associated Methicillin-resistant S. aureus (HA-MRSA), present within an alignment of 63 geographically diverse sequences sampled over a 24 year period (Harris et al. 2010; for strains, see supplementary table S1, Supplementary Material online). We aimed to determine whether this genome-wide data set met the requirements for Bayesian coalescent analysis. First, a significant number of nucleotide changes must be observed, thus suggesting strong phylogenetic signal. Second, the data set must demonstrate low signal for recombination. Third, the amount of time between sampling times must provide enough time for a significant number of mutations to have arisen. These conditions were tested on 1) the full data set, comprising 4,310 SNPs among 63 taxa, 2) a subset that included only synonymous changes (1,055 SNPs), and 3) a subset that included only intergenic changes (962 SNPs). Further Bayesian phylogeographic analyses were then performed to investigate the temporal and spatial spread of HA-MRSA ST-239.
Materials and Methods
Data sets
The alignment of 4,310 SNPs among 63 isolates was downloaded from the Supplementary files of Harris et al. (2010). Subsampled data sets were constructed as described above. Sequences were named following the convention of Harris et al. (2010) including the year of sampling (supplementary table S1, Supplementary Material online).
Likelihood Mapping
To investigate the phylogenetic signal of each data set, likelihood mapping was performed using the Tree-Puzzle program by analyzing 10,000 random quartets (Schmidt et al. 2002). This method proceeds by evaluating, using maximum likelihood, groups of four randomly chosen sequences (quartets). For each quartet, the three possible unrooted tree topologies are weighted. The posterior weights are then plotted using triangular coordinates, such that each corner represents a fully resolved tree topology. Hence, dots localized close to the triangle vertices represent tree-like phylogenetic signal, whereas those close to the center and on the sides represent star-like (completely unresolved) and network-like (partially unresolved) signal, respectively (Strimmer and von Haeseler 1997).
Recombination
Recombination creates mosaic genomes, which violate the assumption of tree-like evolution. Therefore, a network was inferred for each data set using SplitsTree (Huson and Bryant 2006). Each data set was analyzed for the presence of recombinant sequences using the PHI test (Bruen et al. 2006) with alpha = 0.001 (Salemi et al. 2008).
Bayesian Phylogenetic Inference
To estimate the phylogeny for each alignment, we used the Bayesian framework implemented in BEAST software package version 1.5.4 (Drummond and Rambaut 2007) under the general time reversible nucleotide substitution model. The molecular clock was calibrated under either a strict molecular clock (which assumes the same evolutionary rates for all branches in the tree) or a relaxed clock (which allows different rates on different branches, drawn from a specified distribution; Drummond et al. 2006). Both the constant population size coalescent prior or the extended Bayesian skyline plot model (allowing change in effective population size over time; Heled and Drummond 2010) were tested. The Markov chain Monte Carlo (MCMC) analysis was run until evidence of proper mixing was obtained (up to 108 generations, see below); the chain was sampled every 10,000th generation. For analyses assuming the relaxed clock, three independent runs were combined to achieve proper mixing of the chain. Results were visualized in Tracer v.1.5, and proper mixing of the MCMC was assessed by calculating the effective sampling size (ESS) for each parameter (Drummond and Rambaut 2007). All ESS values were >200. For each data set, the maximum clade credibility (MCC) tree, which is the tree with the largest product of posterior clade probabilities, was selected from the posterior tree distribution (after removal of 50% burn-in) using the program TreeAnnotator version 1.5.4 (available as part of the BEAST package). Final trees were annotated with FigTree version 1.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Because only variable sites were used in the analysis, an ascertainment bias correction (ABC) model was implemented in a subset of analyses. For a nucleotide model with four states and a N-taxon tree, there are 4N possible site patterns. Without an ascertainment bias, the sum of the probabilities of all 4N patterns = 1. If specific types of patterns (such as all invariant sites) are deliberately excluded, however, then the probabilities of site patterns no longer sum to 1. To correct for this, the computed probabilities for the observed site patterns can be renormalized, which is implemented in the trunk source code of BEAST available from http://code.google.com/p/beast-mcmc/source/checkout.
Evaluation of Competing Models
Models were compared by calculating the Bayes factor (BF), which is the ratio of the marginal likelihoods (marginal with respect to the prior) of the two models being compared (Kass and Raftey 1995; Suchard et al. 2001). We calculated approximate marginal likelihoods for each coalescent model via importance sampling (1,000 bootstraps) using the harmonic mean of the sampled likelihoods (with the posterior as the importance distribution). The ratio of the marginal likelihoods between any two models is the BF. Evidence against the null model (i.e., the one with lower marginal likelihood) is indicated by 2 ln(BF) > 3 (positive) and > 10 (strong). The calculations were performed with BEAST version 1.5.4 and Tracer v.1.5.
Bayesian Phylogeographic Analysis
The full data set was used in this analysis, although the DEN907 strain was excluded because of uncertainly in its location of origin (n = 62). Ancestral reconstruction of discrete states was tested in a Bayesian statistical framework implemented in BEAST 1.5.4 (Lemey et al. 2009). A matrix of geographic locations was constructed based on the city of sampling for each sequence. Using the full city data set resulted in poor mixing of the MCMC chain because the model was overparameterized; that is, too little information in the sequence data to accurately estimate the migration matrix among locations. Thus, for sequences that were sampled from multiple cities in the same country (e.g., Hungary, Portugal, Brazil, and Czech Republic), the centroid distance between cities was calculated and used as the sampling location in order to reduce the number of locations. This approach retained the global distribution of the sampling locations whereas only minimally sacrificing local resolution within a country. This is true even for Brazil, where the two sampled cities (Sao Paulo and Rio de Janeiro) are only ∼200 miles apart. A total of 13 sampling locations were used: Capital Federal/Buenos Aires (Argentina), Melbourne (Australia), Brazil, Santiago (Chile), Nanjing (China), Czech Republic, Patras (Greece), Hungary, Portugal, Udon (Thailand), Ankara (Turkey), Montevideo (Uruguay), and New York (United States). Strain TW20 was coded as Udon because of its probable epidemiological link with the Thailand outbreak (Harris et al. 2010). A full model was used in which all 78 possible reversible exchange rates between locations were positive. A mixed model was also tested which uses the Bayesian stochastic search variable selection (BSSVS) procedure in which exchange rates are allowed to be zero with some probability. Specifically, a truncated Poisson prior that assigns 50% prior probability on the minimal rate configuration, comprising 12 nonzero rates connecting the 13 locations, was assumed (Lemey et al. 2009). Three different priors on the rate matrix were used under both the full and the BSSVS model: a flat prior (equal probability for all rates); a distance informed prior which was proportional to the Euclidean distance between cities/centroids; and a migration-informed prior, proportional to the number of migrants from each sampled county living in any other sampled county normalized by the total population size.
Files for viewing in Google Earth were created using the MCC treefile, geographic coordinates for each of the sampled cities/centroids, and a script available at http://beast.bio.ed.ac.uk. The xml files for all BEAST analyses are available from http://datadryad.org/.
Summarizing Posterior Location Uncertainty
A model of low statistical power makes poor use of the information in the data, whereas a successful model exploits this information to generate posterior distributions that are maximally different from prior beliefs. As in indication of the amount of information extracted under each mode/prior, the Kullback–Leibler (KL) divergence (Kullback and Leibler 1951) was calculated for the uncertainty in the ancestral reconstruction of the root location, using a uniform discrete distribution as reference distribution, as described by Lemey et al. (2009). KL divergence is a nonsymmetric measure of the difference between the prior and the posterior distribution, where larger values signify that a given Bayesian phylogeographic model extracts more information from the data. KL divergence values for the root location were compared for analyses using different prior distributions on the migration matrix (uniform, human migration, and inverse distance).
Geographic Information System Data
The great-circle distances (i.e., the shortest distance between any two points on the surface of a sphere or orthodromic distance) between each pair of sample locations were calculated using ArcGIS 9.3. To obtain rates of global human migration, we initially sought data at a subnational, regional level, in order to capture the full range of relevant movements. However, such data are nonexistent for the vast majority of countries and are patchy, unstandardized, and variable for the remainder. Foreign-born and foreign-national population data derived from recent censuses represent the most complete and comparable data sets for global and regional analyses that most readily accord with actual population movements (Parsons et al. 2007), and these were used here as a measure of the relative levels of movement between countries.
Data on international bilateral migrant stocks for 226 countries and territories in 2000–2002 were obtained (Parsons et al. 2007). Wherever possible, these data were derived from the latest round of censuses, as these were considered most comparable at the global level. Where unavailable, population registers were drawn upon, and in the cases of missing data, a variety of techniques and tests were employed to create and validate a complete matrix of international bilateral migrant stocks (Parsons et al. 2007). Finally, all data prior to 2000 were scaled to the United Nations midyear totals of migrant stocks for 2000 (United Nations 2004). For each country or territory, the completed data set represented the number of foreign-born and foreign-nationality people in residence in 2000–2002 and which country/territory in which they were born or from which they had traveled. It should of course be noted that these data do not capture either very short-term or illegal movements, which themselves can be substantially larger than those in official records. The migrant data set was rescaled to account for the population size of each country of origin, providing a measure of the strength of migration between countries. For instance, a country containing 100,000 residents born in country A (population 1 million) and 100,000 residents born in country B (population 10 million) has a stronger migratory pull to country A than B. National population totals of each country were obtained (United Nations Population Division 2008) and used to convert the migrant stock numbers into percentages of national origin country population.
Results
Lack of Recombination and Strong Phylogenetic Signal
Likelihood mapping analyses were performed to quantify the nature of the phylogenetic information in the data. These indicated high phylogenetic signal (>85%) for all three data sets (supplementary fig. S1, Supplementary Material online). In addition, the Thailand data set, which includes 20 strains sampled during a 2006–2007 single hospital outbreak, was analyzed independently. Interestingly, the signal was the highest (98.8%) for the Thailand subset even though the sequences were only sampled over an interval of 154 days (Nickerson et al. 2009). The site frequency spectrum was calculated for the full (4,310 SNP) data set (supplementary fig. S2, Supplementary Material online). Although about 50% of the observed changes were singletons, about 15% of the changes were polymorphic at a frequency of >10%. This result is consistent with the strong phylogenetic signal revealed in the likelihood mapping analysis. Neighbor-nets were inferred for the full, synonymous, and intergenic data sets (supplementary fig. S3, Supplementary Material online). In all cases, no major splits were found, and the PHI test gave P values greater than 0.001, suggesting low signal for recombination. This is consistent with a previous in vitro study that found limited uptake of foreign genetic material in S. aureus (Waldron and Lindsay 2006).
HA-MRSA ST239 Is an MEP
Maximum likelihood trees were inferred for each of the three data sets using either a global clock model (“single rate”) or a clock model that incorporated times of sequence sampling (“single rate dated tips”). The two models were compared by taking twice the difference of the natural log of the likelihoods, which is assumed to follow a chi-squared distribution. In all three cases, the single rate dated tips model was a significantly better fit to the data than the single rates model (table 1). This result indicates that ST239 HA-MRSA is a MEP.
Table 1.
Phylogenetic Test for MEPs.
| Data set | Model | LnL |
| Full | SR | −50,848.15 |
| SRDT | −50,710.25* | |
| Intergenic | SR | −7,410.00 |
| SRDT | −7,365.51* | |
| Synonymous | SR | −7,718.34 |
| SRDT | −7,691.08* |
NOTE.—SR, single rate; SRDT, single rate dated tips; LnL, Ln likelihood for the specified model. Values with an asterisk indicate significance for the SRDT over the SR, P < 0.01.
Bayesian Molecular Clock Analysis
The above results indicated that the HA-MRSA ST239 data set is suitable for Bayesian phylogenetic analysis. Thus, Bayesian coalescent molecular clock analyses (Drummond and Rambaut 2007) were performed to reconstruct the temporal history of HA-MRSA ST239. Different molecular clock (strict vs. relaxed) (Drummond et al. 2006) and coalescent (constant vs. nonparametric growth; Heled and Drummond 2010) models were tested for the full, intergenic and synonymous data sets.
In any given alignment of closely related bacterial sequences, the vast majority of sites will be invariant and, thus, phylogenetically uninformative. These sites can be excluded from the analysis as long as the site probabilities are renormalized using an ABC to account for the difference between unobserved and excluded site patterns. We report results with and without this correction (table 2). Evidence against the null model of constant effective population size over time was only weak or moderate (BF < 10) (Suchard et al. 2001). Evidence against the simpler strict clock model was very strong (BF > 200) in all cases. Median estimates of the time to the most recent ancestor (TMRCA) of ST239 were similar across models, data sets, and with and without the bias correction, ranging from 1957 to 1972 with largely overlapping 95% highest posterior density (HPD) intervals (fig. 1), considerably earlier than the earliest identified ST239 strain from 1985 and in agreement with the estimate of Harris et al. (2010). The consistency of timing among models reflected the strong temporal signal in the genome-wide SNPs data. The nucleotide divergence rates are given for each model in supplementary table S2 (Supplementary Material online). In general, they are on the order of 10−3/site/year for the uncorrected estimates and 10−4 for the ABC. For the full data set, these rates can be transformed to reflect the divergence rate “per nucleotide site” (i.e., [10−3 × 4,310]/[2.5 × 106]), resulting in rates of 10−6 to 10−7, consistent with recent estimates using related approaches (Harris et al. 2010; Nubel et al. 2010). However, the appropriate number of total sites in the denominator for the synonymous and intergenic sites is less obvious. It is clear that tight HPDs accompany these estimates, even under the relaxed molecular clock, suggesting that strong temporal information is present in the data set. This result is in contrast to the predicted distribution of rates when little signal is present, that is, wide confidence intervals that tend toward zero (Firth et al. 2010). Great caution should be used in interpreting these rates as long-term evolutionary or substitution rates, as it is likely they are closer to the mutation rate of the organism. Given that the estimates of the TMRCAs were very similar among data sets with different rates, the information contained in the data set is sufficient for inference of divergence times.
Table 2.
Bayesian Estimates of the Evolutionary Rate and Median Root for Major Clades.
| Data seta | Clock Model | Coalescent Prior | Marginal Likelihood | BF (CP)b | BF (clock)c | Mean and 95% HPDs for the TMRCA of ST239 | |
| F | Strict | Constant | −32,217.669 | 1958 | 1953–1962 | ||
| F | Strict | BSP | −32,216.604 | 2.13 | 1958 | 1953–1962 | |
| F | Relaxed | Constant | −32,094.852 | 245.634 | 1964 | 1949–1975 | |
| F | Relaxed | BSP | −32,093.590 | 2.524 | 246.028 | 1968 | 1960–1975 |
| I | Strict | Constant | −7,277.183 | 1970 | 1965–1974 | ||
| I | Strict | BSP | −7,273.335 | 7.696 | 1969 | 1964–1974 | |
| I | Relaxed | Constant | −7,262.205 | 29.956 | 1971 | 1965–1977 | |
| I | Relaxed | BSP | −7,264.389 | 4.368 | 17.892 | 1970 | 1964–1976 |
| S | Strict | Constant | −7,530.590 | 1950 | 1940–1959 | ||
| S | Strict | BSP | −7,525.234 | 10.712 | 1948 | 1937–1958 | |
| S | Relaxed | Constant | −7,504.137 | 52.906 | 1954 | 1933–1974 | |
| S | Relaxed | BSP | −7,499.264 | 9.746 | 51.94 | 1970 | 1953–1979 |
| F | Strict | Constant | −28,238.886 | 1959 | 1955–1964 | ||
| F | Strict | BSP | −28,236.476 | 4.82 | 1959 | 1955–1964 | |
| F | Relaxed | Constant | −28,119.184 | 239.404 | 1964 | 1950–1975 | |
| F | Relaxed | BSP | −28,118.495 | 1.378 | 235.962 | 1968 | 1961–1975 |
| I | Strict | Constant | −6,429.779 | 1970 | 1966–1975 | ||
| I | Strict | BSP | −6,426.129 | 7.3 | 1970 | 1965–1974 | |
| I | Relaxed | Constant | −6,413.103 | 33.352 | 1972 | 1965–1978 | |
| I | Relaxed | BSP | −6,412.846 | 0.514 | 26.566 | 1972 | 1966–1978 |
| S | Strict | Constant | −6,538.188 | 1953 | 1944–1962 | ||
| S | Strict | BSP | −6,532.953 | 10.47 | 1951 | 1942–1961 | |
| S | Relaxed | Constant | −6,513.408 | 49.56 | 1957 | 1938–1975 | |
| S | Relaxed | BSP | −6,509.456 | 7.904 | 46.994 | 1969 | 1951–1979 |
Note.—The upper half of the table gives values without the ABC, and the lower half gives estimates incorporating the correction.
Data sets are coded as F, full; I, intergenic; S, synonymous.
BFs are given for the coalescent prior (CP): constant population size and the nonparametric Bayesian skyline plot (BSP) model (under the same clock model).
BFs are given for the constant and relaxed molecular clock (under the same coalescent model).
FIG. 1.
Posterior estimates of the TMRCA for the ST239 phylogeny. Posterior estimates for 3 data sets (full, intergenic, and synonymous), two clock models (strict and relaxed), two coalescent priors (constant and Bayesian skyline plot), and with and without the ABC. The distributions are colored as follows: red = strict clock, constant; blue = strict clock, BSP; black = relaxed clock, constant; green = relaxed clock, BSP. Note that the earliest time is on the right of the x axis, and the most recent times are on the left.
Bayesian Phylogeography of HA-MRSA ST239
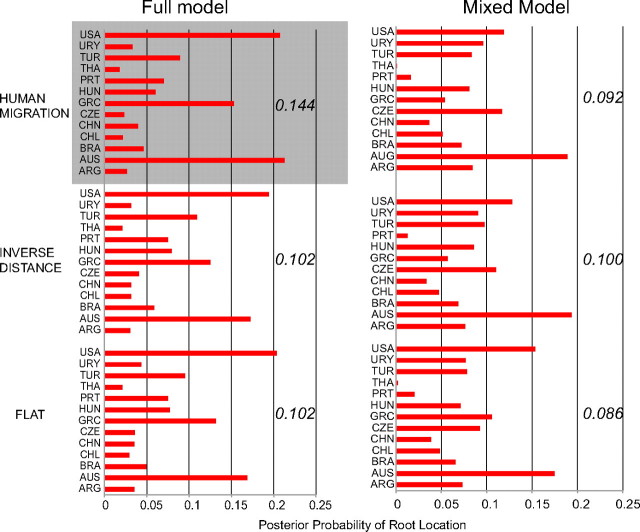
The spatiotemporal dynamics of HA-MRSA ST239 was reconstructed using a Bayesian phylogeographic model that assumed a relaxed molecular clock, constant population size, and included the ABC. “Geographic coordinates” were assigned to each sequence based on the cities from which the strains were sampled (see Materials and Methods). The full model, in which all 78 exchange rates are positive, was compared with a mixed model, in which a subset of the exchange rates is allowed to be zero through the use of a BSSVS (Lemey et al. 2009). For all three priors (see Materials and Methods), under both the full and mixed models, the United States or Australia showed approximately equal probability for being the location for the origin of the sampled HA-MRSA ST239 strains, although their probabilities relative to the other locations were low (fig. 2). Given the small number of strains sampled in each location, and the absence of any strain phylogenetically close to the root, low confidence in assigning the location of the root is perhaps to be expected. Additional sequences would need to be included from each of the locations to ensure that sampled strains are representative. In the present study, the statistical power of each model to extract information from the data was assessed using the KL divergence. In general, full models resulted in higher KL values than BSSVS models (fig. 2). This indicated that the posterior distributions of such models were in fact further away from the prior distributions (equal probability for all locations). The minimal rate configuration (see Materials and Methods) strongly favored by the mixed model is analogous to a parsimony model, which is often the default assumption used in many phylogeographic studies but rarely tested against other possibilities (Lemey et al. 2009). KL divergence was higher for the migration-informed prior (0.144) than the distance-informed (0.102) or flat (0.102) priors, under the full model, suggesting that human migration patterns are associated with the global migration of HA-MRSA ST239.
FIG. 2.
Bayesian posterior estimates of root location. Posterior probability estimates (y axis) for the root location (x axis) under the full and BSSVS models with three different priors (migration-informed, distance-informed, and flat). The KL divergence measure is reported for each model/prior. The model with the largest KL is highlighted in bold. All analyses assumed a relaxed clock, general time reversible model of nucleotide substitution, and constant coalescent prior.
The MCC tree was obtained for the model with the highest KL divergence (full model with human migration prior, fig. 3). The geographic location of the internal nodes closest to the root had low posterior support (<0.5), suggesting that the early geographic spread of the epidemic, including the epidemic origin and initial routes of HA-MRSA ST239 spread, should be interpreted with caution. However, stronger support was found for the geographic reconstruction of the internal nodes in the more derived clades, particularly those form South America + Europe and Asia (fig. 3), which led to a robust reconstruction of bacterial gene flow during the 1990s (fig. 4). Finally, the MCC tree was converted into a Google Earth compatible movie (see Supplementary movie file, Supplementary Material online) to visualize a possible scenario of ST239 global dissemination.
FIG. 3.
Bayesian inference of ST239 evolutionary history. MCC tree selected from the posterior distribution for the model/prior with the highest KL divergence. Branches are scaled by time according to the scale at the bottom. Terminal branches are colored by location of sampling according to the legend on the left. Internal branches are colored according to the most probable location. Posterior support for the geographic location for each internal node is given along the subtending branch. All nodes that are unlabeled have a posterior >0.90. Nodes with low posterior probabilities are labeled A–F. Piecharts are shown for these nodes at the bottom of the figure, with the support for each geographic location shown as a proportion of the circle, colored according to the same color scheme as the tree. All analyses assumed a relaxed clock, general time reversible model of nucleotide substitution, and constant coalescent prior.
FIG. 4.

Spatial spread of HA-MRSA ST239 during the 1990s. Major events in the global dissemination of HA-MRSA are shown for (A) Europe and South America and (B) Asia corresponding to the South America + Europe and Asia clades in figure 3. Countries are colored according the legend. Lines denote migration routes between countries with the arrow indicating the direction of the migration.
Discussion
For the first time, we successfully employed Bayesian phylogeographic methods to a bacterial pathogen. Staphylococcus aureus is a common cause of infections and therefore has not generally been thought of as a pandemic disease. However, with the emergence of specific antibiotic-resistant strains, it has become clear that S. aureus can spread in epidemic waves across the globe. In this study, we demonstrate that the TMRCA of the ST239 lineage occurred shortly after the introduction of Methicillin in 1959 (Batchelor et al. 1959), in agreement with the recent findings of Harris et al. (2010). The type III SCCmec cassette also encodes resistance to penicillin, tetracycline, and erythromycin (Deurenberg and Stobberingh 2008), all of which were in use before 1960 (Khardori 2006). Multiple drug resistance could have provided this strain with a strong fitness advantage in the hospital setting. We estimate that ST239 was spreading worldwide for almost 20 years before the initial identification of an isolate, in 1985, carrying the type III pathogenic cassette (Deurenberg and Stobberingh 2008). Such a discrepancy is alarming because it suggests that antibiotic-resistant bacteria can circulate globally undetected for a relatively long time. Except for a single introduction into Asia, ST239 lineages were not geographically restricted but rather regularly moved among continents, indicating that HA-MRSA diffusion has the characteristics of a pandemic rather than regionally restricted outbreaks. Such results are in contrast with previous reports suggesting that MRSA is characterized by limited geographic dispersal in Europe (Grundmann et al. 2010) and worldwide (Nübel et al. 2008), but consistent with the qualitative observations of Harris et al. (2010) that were based on a single ML tree reconstruction. However, these studies used limited genetic data, which may have contained too little resolution to infer detailed geographic patterns. Furthermore, because the genome-wide SNPs data set used in the present analysis only included the core genome (excluding the SCCmec cassette; Harris et al. 2010), the inferred evolutionary history is representative of the population history rather than the recombinant events within mobile elements.
The rate of evolution of bacteria species is uncertain (Achtman 2008). In this study, the use of the ascertainment correction bias provided estimates of evolutionary rates that were intermediate between those estimated using tip-dated sequences without the bias correction reported here and elsewhere (Harris et al. 2010; Nubel et al. 2010) and those inferred using species divergence dating techniques (Ochman et al. 1999). It is likely that previously estimated rates without the bias correction were artificially high, as the lack of invariant site patterns are not taken into account, in which case the sum of all possible site pattern probabilities is <1. Correcting this bias allowed the invariant sites to be properly incorporated into the probability summation. Evolutionary rates estimates were still 100 times faster than rate estimates based on species divergence times (external calibrations). Reasons for this include the fact that transient polymorphisms exist within a population that will eventually be selected against over time, thereby driving up the estimated evolutionary rate when inferences are drawn from intrapopulation samples. Therefore, this rate reflects a different population process to the long-term fixation process between species. However, the important result from the present analysis is that estimates of bacterial evolutionary timescales are feasible using serially sampled sequence data alone and do not always require a calibration date or external rate.
Recent theoretical advances in Bayesian phylogeography have made it possible to include the geographic location of the samples into the analysis, thus providing a formal statistical framework in which hypotheses concerning the spatial origin and dissemination of epidemics can be investigated. Bayesian methods account for phylogenetic uncertainty inherent in any reconstruction of the evolutionary history of a group of organisms, both in the tree topology and the assignment of geographic states to ancestral nodes, by estimating a probability distribution for parameters of interest (Lemey et al. 2009). This approach constitutes a significant improvement (Sanmartín et al. 2008) upon traditional parsimony-based models (Slatkin and Maddison 1989) that only consider one reconstruction of migration on a fixed tree. Furthermore, although maximum parsimony can reliably be used in simple dissemination scenarios, minimizing the number of migrations is inappropriate in more complex situations, for example, in cases of continuous multidirectional gene flow (Cunningham et al. 1998). The Bayesian framework, on the other hand, can account for more complex models by allowing the calculation of probabilities for the ancestral state (nucleotide and geographic location) reconstruction. This framework also allows testing of specific hypothesis of the driving factors of the migration (Lemey et al. 2009). Here, we show that human migration appears to be significantly associated with the current HA-MRSA pandemic spread. Traveling and migration have been linked anecdotally with community-acquired MRSA (CA-MRSA) (Ellington et al. 2010) and Methicillin-sensitive S. aureus (MSSA) (Schleucher et al. 2008). Although CA-MRSA has traditionally been viewed as demographically and clinically distinct from HA-MRSA and more similar to MSSA (Groom et al. 2001; Naimi et al. 2003), recent evidence suggests that the epidemiological behavior of both CA- and HA-MRSA have begun to overlap (Seybold et al. 2006; Klevens et al. 2007). Furthermore, asymptomatic carriers of HA-MRSA could act as vectors in transmitting the pathogen (Zanger 2010); in fact, the majority of HA-MRSA cases in the United States from 2007 occurred outside of the hospital (Klevens et al. 2007), providing ample opportunity for transmission in the community setting. Given the rapid increase of international air travel during the past several decades, emergence of drug-resistant pathogens in a specific locale should not be considered as an isolated event but rather within a larger global context. Specific migration patterns (such as those reported here) can then be incorporated into monitoring and intervention strategies.
To strengthen the phylogeographic analysis of viral gene flow, future studies should include multiple strains from each sampled location to avoid uncertainty in the reconstruction of ancestral locations. The cost of genome-wide typing is justified by the potential public health utility of such data (Harris et al. 2010). Future applications of this approach may aid the control and prediction of newly emergent drug-resistant pathogens.
Supplementary Material
Supplementary movie file, tables S1–S2, and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was funded in part through the UF Emerging Pathogens Institute Seed Funding, the RAPIDD programme of the Science and Technology Directorate, Department of Homeland Security, and the Fogarty International Center, US National Institutes of Health. RRG was funded through the National Cancer Institute, National Institutes of Health (CA09126). AJT was supported by grants from the Bill & Melinda Gates Foundation (#49446) and Transportation Research Board of the National Academies (ACRP 02-20). OGP was funded through the Royal Society. MS was funded through the National Institute of Allergy and Infectious Diseases, National Institutes of Health (R01 NS063897).
References
- Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Batchelor F, Doyle F, Naylor J, Rolinson G. Synthesis of penicillin: 6-aminopenicillanic acid in penicillin fermentations. Nature. 1959;183:257–258. doi: 10.1038/183257b0. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Gardner A, Rambaut A, Pybus O. Pacing a small cage: mutation and RNA viruses. Trends Ecol Evol. 2008;23:188–193. doi: 10.1016/j.tree.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Omland K, Oakley T. Reconstructing ancestral character states: a critical reappraisal. Trends Ecol Evol. 1998;13:361–366. doi: 10.1016/s0169-5347(98)01382-2. [DOI] [PubMed] [Google Scholar]
- Deurenberg R, Stobberingh E. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8:747–763. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Pybus OG, Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv Parasitol. 2003;54:331–358. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. Measurably evolving populations. Trends Ecol Evol. 2003;18:481–488. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington M, Ganner M, Warner M, Boakes E, Cookson B, Hill R, Kearns A. First international spread and dissemination of the virulent Queensland community-associated methicillin-resistant Staphylococcus aureus strain. Clin Microbiol Infect. 2010;16:1009–1012. doi: 10.1111/j.1469-0691.2009.02994.x. [DOI] [PubMed] [Google Scholar]
- Firth C, Kitchen A, Shapiro B, Suchard M, Holmes E, Rambaut A. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol. 2010;27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R, Tatem A, Lamers S, et al. (12 co-authors) Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009;10:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom A, Wolsey D, Naimi T, Smith K, Johnson S, Boxrud D, Moore K, Cheek J. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA. 2001;286:1201–1205. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aanensen D, van den Wijngaard C, Spratt B, Harmsen D, Friedrich A. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Feil E, Holden M, et al. (15 co-authors) Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond A. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S, Phillips M, Cooper A, Drummond A. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kass R, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Khardori N. Antibiotics—past, present, and future. Med Clin North Am. 2006;90:1049–1076. doi: 10.1016/j.mcna.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Klevens R, Morrison M, Nadle J, et al. (16 co-authors) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Kullback S, Leibler R. On information and sufficiency. Ann Math Statist. 1951;22:290–295. [Google Scholar]
- Lemey P, Rambaut A, Drummond A, Suchard M. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nübel U, Fitzgerald JR. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov P, Pepin J, Frost E, Deslandes S, Labbé A, Pybus O. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol. 2009;90:2086–2096. doi: 10.1099/vir.0.011569-0. [DOI] [PubMed] [Google Scholar]
- Naimi T, LeDell K, Como-Sabetti K, et al. (12 co-authors) Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- Nickerson E, Wuthiekanun V, Wongsuvan G, et al. (13 co-authors) Factors predicting and reducing mortality in patients with invasive Staphylococcus aureus disease in a developing country. PLoS One. 2009;4:e6512. doi: 10.1371/journal.pone.0006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nubel U, Dordel J, Kurt K, et al. (12 co-authors) A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-Resistant Staphylococcus aureus. PLos Pathog. 2010;6:e1000855. doi: 10.1371/journal.ppat.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Roumagnac P, Feldkamp M, et al. (16 co-authors) Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008;105:14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran N. Calibrating bacterial evolution. Proc Natl Acad Sci U S A. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons CR, Skeldon R, Walmsley TL, Winters LA. Quantifying international migration: a database of bilateral migrant stocks. 2007. World Bank Policy Research Working Paper 4165. [Google Scholar]
- Pybus O. Model selection and the molecular clock. PLoS Biol. 2006;4:e151. doi: 10.1371/journal.pbio.0040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus O, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540–550. doi: 10.1038/nrg2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395–399. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Pybus O, Nelson M, Viboud C, Taubenberger J, Holmes E. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salemi M, Gray R, Goodenow M. An exploratory algorithm to identify intra-host recombinant viral sequences. Mol Phylogenet Evol. 2008;49:618–628. doi: 10.1016/j.ympev.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín I, van der Mark P, Ronquist F. Inferring dispersal: a Bayesian, phylogeny-based approach to island biogeography, with special reference to the Canary Islands. J Biogeo. 2008;35:428–449. [Google Scholar]
- Schleucher R, Gaessler M, Knobloch J. Panton-Valentine leukocidin-producing methicillin-sensitive Staphylococcus aureus as a cause for recurrent, contagious skin infections in young, healthy travelers returned from a tropical country: a new worldwide public health problem? J Travel Med. 2008;15:137–139. doi: 10.1111/j.1708-8305.2008.00194.x. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Seybold U, Kourbatova E, Johnson J, Halvosa S, Wang Y, King M, Ray S, Blumberg H. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci U S A. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sincheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- Tazi L, Pérez-Losada M, Gu W, Yang Y, Xue L, Crandall K, Viscidi R. Population dynamics of Neisseria gonorrhoeae in Shanghai, China: a comparative study. BMC Infect Dis. 2010;10:13. doi: 10.1186/1471-2334-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations. Trends in total migrant stock 1960–2000. New York: United Nations Population Division; 2004. [Google Scholar]
- United Nations Population Division. World population prospects, 2008 revision. New York: United Nations; 2008. [Google Scholar]
- Waldron D, Lindsay J. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188:5578–5585. doi: 10.1128/JB.00418-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger P. Staphylococcus aureus positive skin infections and international travel. Wien Klin Wochenschr. 2010;122(Suppl 1):31–33. doi: 10.1007/s00508-010-1333-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.