Abstract
The predicted protein product of open reading frame slr0328 from Synechocystis sp. PCC 6803, SynPTP, possesses significant amino acid sequence similarity with known low molecular weight protein tyrosine phosphatases (PTPs). To determine the functional properties of this hypothetical protein, open reading frame slr0328 was expressed in Escherichia coli. The purified recombinant protein, SynPTP, displayed its catalytic phosphatase activity towards several tyrosine, but not serine, phosphorylated exogenous protein substrates. The protein phosphatase activity of SynPTP was inhibited by sodium orthovanadate, a known inhibitor of tyrosine phosphatases, but not by okadaic acid, an inhibitor for many serine/threonine phosphatases. Kinetic analysis indicated that the Km and Vmax values for SynPTP towards p-nitrophenyl phosphate are similar to those of other known bacterial low molecular weight PTPs. Mutagenic alteration of the predicted catalytic cysteine of PTP, Cys7, to serine abolished enzyme activity. Using a combination of immunodetection, mass spectrometric analysis and mutagenically altered Cys7SerAsp125Ala-SynPTP, we identified PsaD (photosystem I subunit II), CpcD (phycocyanin rod linker protein) and phycocyanin-α and -β subunits as possible endogenous substrates of SynPTP in this cyanobacterium. These results indicate that SynPTP might be involved in the regulation of photosynthesis in Synechocystis sp. PCC 6803.
Keywords: cyanobacteria, phycocyanin, substrate trapping, tyrosine phosphorylation
Reversible protein phosphorylation is one of the most fundamental regulatory mechanisms in signal transduction and information processing in cells for modulating the functional properties of proteins. This mechanism is evolutionarily conserved in every living cell among all three domains of life. This reversible phosphorylation/dephosphorylation process is catalysed by opposing reactions of protein kinases and protein phosphatases (1, 2). It has been well studied that in eukaryotes protein phosphorylation on tyrosine residues plays a key role in the regulatory mechanism of various cellular processes including growth, differentiation, cell cycle regulation and cytoskeletal function (3, 4), and as well as controls many diseases (5, 6) including infectious diseases (7, 8). In comparison to eukaryotes, the occurrence of protein tyrosine kinases (PTKs)/protein tyrosine phosphatases (PTPs) in bacteria including photosynthetic cyanobacteria, was suggested much later (9–15).
The PTPs are characterized by the presence of a common active site signature motif (CX5RS/T), namely phosphate binding loop, and an essential aspartate that serves as a catalytic acid/base (16, 17). The PTPs are classified into three superfamilies: (i) conventional PTPs, where the active site sequence preceding to histidine (HCX5RS/T) is located in the central portion of the catalytic domain and conserved aspartate is present in 25 to 45 residues to the N-terminal side of the catalytic cysteine (18); (ii) low molecular weight (LMW) PTPs, where the active site sequence precedes by either a leucine or valine residue and resides near the extreme N-terminus of the catalytic domain and the conserved aspartate is located 85 to 105 residues to the C-terminal side of catalytic cysteine followed by a proline (19); and (iii) in dual specific phosphatases (DSPs), which can catalyse phosphoesters from both tyrosine and serine/threonine residues, the active site sequence closely resembles to that of conventional PTPs, D-X45-CX5RS/T (16–19).
In photosynthetic cyanobacterium Synechocystis sp. strain PCC 6803, genome analysis (20–22) revealed that this organism contains three ORFs, which encode LMW protein tyrosine PTPs (23). One is slr0946, whose predicted protein product was characterized as arsenate reductase (24). The functional properties of the other two ORFs, slr0328 and slr1617, have not been reported yet. In Synechocystis sp. PCC 6803, a number of serine/threonine kinases (25, 26), protein serine/threonine phosphatases (PP2A) (27, 28), and serine/threonine phosphoproteins (29–32) are found. However, no specific PTKs, not any member of conventional or LMW PTPs, or any tyrosine-phosphorylated proteins have been detected yet.
In this study, we have characterized one open reading frame, slr0328, which encodes a hypothetical LMW PTP in Synechocystis sp. PCC 6803. We have described here that Slr0328, named as SynPTP functions as a PTP, not as a DSP. To understand the biological function of this phosphatase, we have identified several tyrosyl-phosphorylated proteins, which include PsaD, photosystem I subunit II, and CpcD, phycocyanin rod linker protein from whole cell extracts and phycocyanin alpha and beta subunits from soluble fractions of this organism as potential substrates of SynPTP. Results suggest that this tyrosine phosphatase in Synechocystis sp. may be involved in the regulation of photosynthesis.
Materials and Methods
Materials
All general laboratory reagents were purchased from Sigma (St Louis, MO) or Fisher Scientific (Pittsburgh, PA, USA). Chelating Sepharose Fast Flow was from GE Healthcare Life Sciences (Piscataway, NJ, USA), the pET 101/D-TOPO cloning kit was from Invitrogen (Carlsbad, CA, USA). Oligonucleotides were from Life Technologies, Inc. (Frederick, MD, USA). [γ-32P] ATP was from Perkin Elmer (Boston, MA, USA).
Growth of Synechocystis sp. PCC 6803 and isolation of genomic DNA
The cyanobacterium organism was cultured in 50 ml BG11 media at 25°C with continuous shaking at 100 rpm under white light at an intensity of ∼10 µmol photons m−2 s−1. Cells were grown until OD730nm reached 1.0 (exponential), and then harvested by centrifugation at 2700g for 20 min at 4°C. Pellets were washed 3 times with 15 ml sterile water and stored at −80°C until needed. Cells were lysed and genomic DNA was isolated as described by Li et al. (11). A mutant strain of Synechocystis sp. PCC 6803, which lacks the genes encoding phycocyanin α subunit (CpcA, Sll1577), phycocyanin β subunit (CpcB, Sll1578), 33 kDa rod linker protein (Cpc1, Sll1580) and 30 kDa rod linker protein (Cpc2, Sll1579), was obtained from Cyanobase. This mutant strain is called the CK mutant (Ghada Ajlani, unpublished work). This mutant strain was grown under similar conditions to wild type strain with 10µg/ml kanamycin added as a selection marker.
Cloning of ORF slr0328, expression of its recombinant protein product and purification of Slr0328
ORF slr0328 was cloned using pET101/D-TOPO cloning kit (Invitrogen) following the manufacturer’s protocol. The sequences of the forward and reverse primers used were 5′-caccatgaaattgttatttgtttgtttaggtaac-3′ and 5′-attaaccaattccttgcccaagc-3′, respectively. The correct orientation and sequences of the cloned gene were verified by DNA sequencing of the isolated plasmid. Site-directed mutagenesis of ORF slr0328 for Cys7Ser and Asp125Ala were performed using Quick change (Stratagene, CA, USA) according to manufacturer’s protocol. Using these plasmids, the predicted protein product of slr0328, named as SynPTP, and mutagenically altered SynPTP, Cys7Ser-SynPTP, were expressed using the competent cells of E. coli BL21 star (DE3) (Invitrogen), following the manufacturer’s protocol in 200 ml LB media at 25°C for 5 h and cells were harvested. The cell pellet from a 200 ml culture was thawed and resuspended in 5 ml of 50 mM Tris HCl (pH 7.5) containing 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 0.1 mg/ml of lysozyme and 5% (v/v) glycerol. The resulting solution was incubated on ice for 30 min. The cells were lysed by sonic disruption with four bursts of 20 S each at 50 watts of a Heat Systems-Ultrasonics Inc, Model W185 Sonifier Cell Disruptor equipped with a microprobe. The resulting lysate was centrifuged for 20 min at 10,000g and 4°C. The supernatant was filtered through a 0.45 µm filter to remove any particulate materials and applied to a metal ion affinity column for purification of the recombinant protein following established protocols (26). In brief, a 1 ml bed volume of column was prepared using Chelating Sepharose Fast Flow (Pharmacia Biotech, Uppsala, Sweden). The column was washed with 10 bed volumes of sterile water. Next, the column was charged with Ni2+ by applying 3 bed volumes of 200 mM NiSO4, followed by 10 bed volumes of sterile water. Equilibration of the column was performed with 10 ml buffer A, which contains 50 mM Tris HCl, pH 7.5, 50 mM NaCl and 5% (v/v) glycerol. The supernatant, prepared from the cell lysate as described above, was passed through the column twice. The column then was washed with 2 × 10 bed volumes of buffer containing 10 mM imidazole. Bound proteins were eluted in a step gradient with the same buffer in which imidazole concentration was increased from 10 mM to 250 mM. Eluted fractions were collected in 1 ml volume and analysed for the presence of recombinant SynPTPs by SDS–PAGE in 12% (w/v) acrylamide gels and Western blot analysis was performed using anti-V5 antibody (Invitrogen).
Preparation of 32P-labelled phosphoprotein substrates
The protein substrates, which are casein, myelin basic protein (MBP) and reduced, carboxymethylated and maleylated lysozyme [RCML, prepared as described by Tonks et al. (33)]. These proteins were phosphorylated on either serine or tyrosine residues as described previously (14).
Assay of protein phosphatase activity
Assay of protein phosphatase activity was performed as described previously (14). In brief, SynPTP, 0.1 ∼2.0 µg, was incubated at 30°C for 20 min in a volume of 40 µl containing 100 mM MES, pH 6.5, 2 mM DTT, 1 mM EDTA, 0.5 mg/ml bovine serum albumin (BSA) and 4 µM protein bound [32P] phosphate. At the end of the incubation period, the reaction was stopped by adding 200 µl of chilled 20% (w/v) trichloroacetic acid. Following vigorous agitation on a vortex mixer, the assay solution was centrifuged for 10 min at 14,000g. A 40 µl sample of the supernatant liquid was removed and dispersed into 1 ml of scintillation fluid, and the liberated 32Pi was measured in a scintillation counter.
Phosphohydrolase activity of SynPTP towards pNPP
Protein phosphatase assay towards pNPP was performed by the following method. SynPTP, 0.1–1.0 µg, was incubated at 30°C for 30 min in a volume of 200 µl. The reaction mixture contained 100 mM MES, pH 6.5, 2 mM DTT, 1 mM EDTA and 10 mM pNPP. The reaction was terminated by the addition of 700 µL of 0.5 M NaOH, and the absorbance of the resulting solution was measured at 410 nm. The amount of p-nitrophenolate released was quantified by using a molar extinction coefficient of 18,300 M−1 cm−1.
Preparation of soluble fractions of Synechocystis sp. PCC 6803
The cell pellet of Synechocystis sp. PCC 6803, grown from a 250 ml culture, was thawed and resuspended in 15 ml cell lysis buffer (50 mM MES, pH 6.5, 1 mM DTT, 1 mM EDTA, 0.1 mM sodium fluoride, 10 mM NaCl and protease inhibitor cocktail). Suspended cells were lysed by three passes through a chilled French pressure cell at 100 MPa. Cell debris were removed by centrifugation at 5000g for 10 min. The whole-cell extracts were then centrifuged at 100,000g, using a Beckman 50Ti rotor, for 2 h at 4°C. Supernatant liquid was saved as the soluble fraction. The pellet was collected as the membrane fraction and stored at −20°C for further analysis.
Preparation of substrate trapping column
Hi-trap N-hydroxysuccinimide (NHS)-activated Sepharose (GE Healthcare Life Sciences) beads and 2 ml bed volume in a Falcon tube were used to immobilize 5 mg purified active SynPTP or mutagenically altered Cys7SerinAsp125Ala SynPTP according to the manufacturer’s protocol. Following that, the soluble fractions of Synechocystis sp., prepared as mentioned in above, containing 15 mg of total protein in 10 ml supernatant liquid was applied to the Sepharose beads (active SynPTP or mutagenically altered SynPTP bound Sepharose beads) and incubated at 30°C overnight. Following day, beads were centrifuged at 500g for 5 min and washed with three times with 5 ml buffer containing 50 mM MES (pH 6.5), 10 mM NaCl, 1 mM DTT, 1 mM EDTA and protease inhibitor cocktail for 10 min by gentle agitation in a tube rotator. Bound proteins were eluted in a step gradient with 50–250 mM NaCl in the same buffer. Each eluted fraction was collected in a 2 ml volume. Sepharose beads were further washed with 250 mM NaCl or 1 M NaCl in the same buffer containing 50 mM phosphotyrosine and collected in 2 ml volume each. Eluted fractions were analysed on SDS–PAGE and immunoblotted with a rabbit monoclonal anti-phosphotyrosine antibody, RC20, conjugated with horseradish peroxidase (B.D. Transduction Laboratory, Rockville, MD, USA). An enhanced chemiluminiscence system (Pierce, Rockford III) was used for the detection of immunoreactive proteins.
2D gel electrophoresis and immunodetection of phosphotyrosine proteins from whole-cell extracts of Synechocystis
A 7 cm long IPG strip (GE Healthcare Life Sciences), pH 3–10, linear, was used to perform 2D gel electrophoresis according to manufacturer’s protocol with whole-cell extracts of Synechocystis. After completion of the second dimension, proteins were transferred to a PVDF membrane. The membrane was incubated either with or without 100 µg of active SynPTP in 10 ml buffer containing 20 mM MES, pH 6.5, 2 mM DTT, 1 mM EDTA and protease inhibitor cocktail (Sigma, St Louis, MA, USA) overnight at 30°C. Following that, membranes were blocked for 1 h at room temperature in TBST buffer containing 10 mM Tris HCl (pH 7.5), 150 mM of NaCl, 0.05% (v/v) Tween-20 and 1% (w/v) bovine serum albumin. Immunodetection was performed using anti-phosphotyrosine antibody, RC20, at a dilution of 1 : 50,000 in 25 ml TBST buffer with 1% BSA for 1 h at room temperature. Following incubation, membrane was washed for several times and an enhanced chemiluminiscence system was used to detect the immunoreactive proteins.
Mass spectral analysis
The procedure for in-solution trypsin digestion and extraction of tryptic peptides was followed as described in Lower et al. (34). In brief, protein samples were concentrated by precipitation. Chilled methanol was added to the sample in the ratio of 4 : 1(v/v). The mixture was incubated on ice for at least 1 h and then centrifuged at 14,000g for 30 min at 4°C. The pellet was resuspended in 200 µl of freshly prepared 100 mM ammonium bicarbonate. To enhance the solubilization of protein pellets, 100% acetonitrile was added to a final concentration of 5% (v/v) to the mixture. Following that, sequencing grade trypsin (Promega, Madison, WI, USA) was added to the mixture in 1 : 25 (w/w) ratio to total protein (1 µg trypsin; 25 µg protein). The mixture was incubated overnight at 37°C. Next, 5% (v/v) trifluoroacetic acid (TFA) stock solution was added to the mixture to bring the final concentration of TFA to 0.2% (v/v) and the pH of the solution was adjusted to ≤3 by adding 98% formic acid. Next, the tryptic peptides were eluted by using OMIX C18 tips (Varian Inc, CA, USA) and analysed by mass spectral analysis using a nano-electrospray source from Proxeon (Odense, Denmark) attached to a Finnigan TSQ Quantum Ultra AM mass spectrometer (Thermo Electron Corp., West Palm Beach, FL, USA).
Results and Discussion
Determination of the phosphatase activity of recombinant SynPTP and mutagenically altered Cys7Ser SynPTP
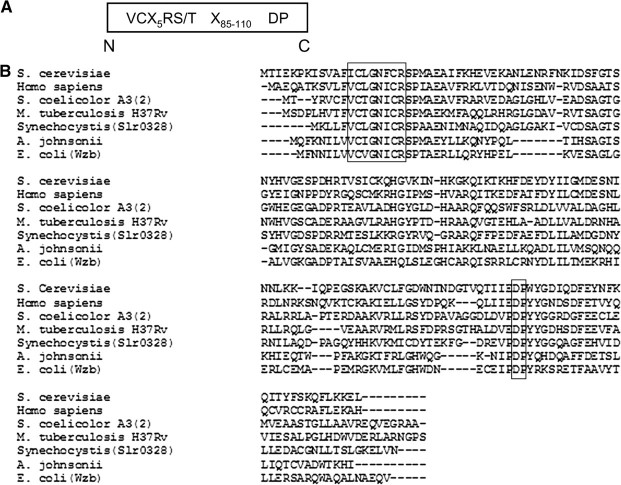
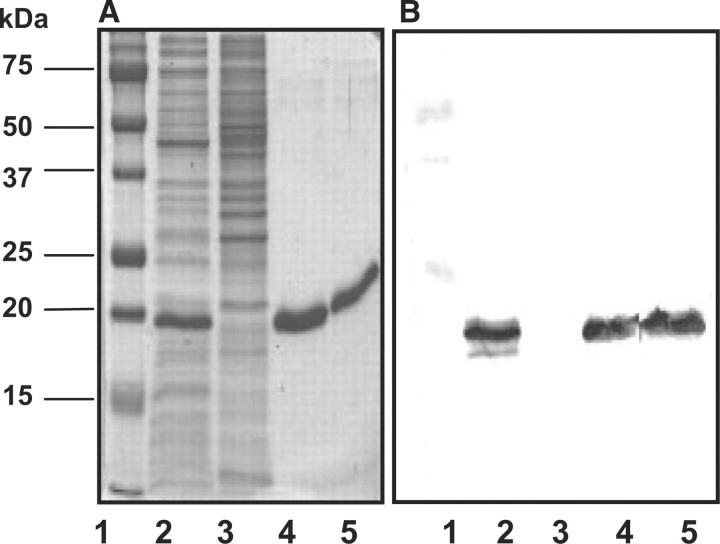
The amino acid sequence of ORF slr0328 contains common signature motif (Fig. 1) for LMW protein tyrosine phosphatases as previously described (23). Full-length protein product of slr0328, named as SynPTP or mutagenically altered Cys7Ser-SynPTP was expressed in E. coli as a recombinant protein with a C-terminal histidine tag, purified by metal ion column chromatography, and the purified fractions were analysed by SDS–PAGE (Fig. 2A). The presence of fusion proteins was confirmed by Western blot analysis using anti-V5 antibody (Fig. 2B). Next, we examined whether this recombinant protein possesses any phosphatase activity. Para-nitrophenyl phosphate (pNPP), a common substrate for many protein phosphatases and casein (32P-Tyr) or reduced carboxymethylated and maleylated lysozyme (RCML, 32P-Tyr) as protein substrates were used to check the phosphatase activity of SynPTP and Cys7Ser-SynPTP. It was observed that SynPTP, but not mutagenically altered Cys7Ser-SynPTP, exhibits phosphohydrolase activity towards pNPP and protein tyrosine phosphatase activity towards phospho-protein substrates in the detectable amount (Table 1). Next, a comparison of kinetic parameters, Km and Vmax, for SynPTP, was made with the values of other known bacterial LMW PTPs towards pNPP as a substrate (Table 2). Prior to the kinetic analysis of SynPTP, the assay conditions for phosphohydrolase activity of SynPTP for pNPP were optimized with respect to time (20 min), pH (6.5), temperature (30°C) and enzyme concentration (200 ng) (Supplementary Fig. S1). The result indicated that Km and Vmax values of SynPTP are comparable in similar order of magnitude with those values for other bacterial LMW PTPs (11, 12, 35–39). However, it was observed that Vmax value of SynPTP is almost same with that of yeast PTP Ltp1 and 4-fold lower than that of another yeast PTP Stp1 and 35-fold lower than the mammalian PTP, BHPTP (39). Therefore, our results indicated that SynPTP exhibited similar pattern of its catalytic function towards pNPP like other known bacterial LMW PTPs, not as higher eukaryotes like PTPs.
Fig. 1.
The amino acid sequence alignment between the predicted protein product of ORF slr0328 and other known LMW PTPs. (A) The conserved sequence for LMW PTP. (B) The alignment of the deduced amino acid sequence of Slr0328 (SynPTP) with other LMW PTPs, was constructed by using the CLUSTALW program of Biology Workbench. The active site sequence VC-X5-RS/T (bold and in box) is present near the extreme N-terminus of the catalytic domain, and conserved aspartate is located at 110 residues to the C-terminal side of conserved serine followed by a proline. The DNA-derived amino acid sequences of LMW PTPs are: S. cerevisiae, GenBank accession no. P40347; Homo sapiens, GenBank accession no. P24666; S. coelicolor A3 (2), GenBank accession no. P53433; M. tuberculosis H37Rv, GenBank accession no. CAA94656; Synechocystis sp. PCC 6803 (Slr0328), GenBank accession no. Q55535; A. johnsonii, GenBank accession no. CAA75430; E. coli (Wzb), GenBank accession no. P77153.
Fig. 2.
Analysis of the purified recombinant SynPTP by SDS–PAGE. (A) Recombinant SynPTP expressed in E. coli with a C-terminal His tag was analysed by SDS–PAGE. Each lane contained equal amount of protein (3 mg). Lane 1: protein markers; lane 2: a soluble fraction of cell lysate of SynPTP expressed with a C-terminal His tag after induction with 1 mM IPTG; lane 3: a soluble fraction of cell lysate without induction as a negative control; lane 4: a purified fraction of the recombinant SynPTP with a C-terminal His tag after elution from metal ion column; lane 5: a purified fraction of mutagenically altered Cys7Ser SynPTP with a C-terminal His tag. Gels were stained with Coomassie Blue. (B) The corresponding result of Western blot analysis of a duplicate gel from SDS–PAGE. Anti-V5 antibody was used to detect the epitope of fusion proteins.
Table I.
Activity of rSynPTP towards exogenous substrates.
| Specific activity |
|||
|---|---|---|---|
| Recombinant protein | RCML (32P-Tyr) (pmol/min/mg) | Casein (32P-Tyr) (pmol/min/mg) | pNPP (µmol/min/mg) |
| SynPTP | 800 ± 20 | 1380 ± 70 | 3.1 ± 0.16 |
| Cys7Ser-SynPTP | ND | ND | ND |
The catalytic activity of rSynPTP or mutagenically altered SynPTP was determined using 500 ng of each recombinant protein and 4 µM Casein (32P-Tyr)/RCML (32P-Tyr) or 10 mM pNPP as exogenous substrates as described in the ‘Materials and Methods’ section. ND, not detected, ≤2%.
Table II.
Comparison of kinetic constants of SynPTP with those of known LMW PTPs.
| Organism | Enzyme | Km (mM) | Vmax (µmol/min/mg) | Reference |
|---|---|---|---|---|
| Streptomyces coelicolor A3(2) | PtpA | 0.75 | 4.8 | Li and Strohl (11) |
| Staphylococcus aureus | PtpB | 1.5 | 1.4 | Soulat et al. (35) |
| Staphylococcus aureus | PtpA | 1.2 | 33.6 | Soulat et al. (35) |
| Acinetobacter johnsonii | PTP | 5.0 | 9.7 | Grangeasse et al. (12) |
| E. coli | Wzb | 1.0 | 4.6 | Vincent et al. (36) |
| Synechocystis sp. PCC 6803 | SynPTP | 0.6 | 3.2 | This report |
| Schizosaccharomyces pombe | Stp1 | 0.21 | 12.8 | Zhang et al. (37) |
| Schizosaccharomyces cerevisiae | Ltp1 | 0.02 | 3.2 | Ostanin et al. (38) |
| Bovine heart phosphatase | BHPTP | 3.5 | 103 | Wo et al. (39) |
SynPTP functions as a protein tyrosine phosphatase not as a dual-specific phosphatase
Since SynPTP has phosphatase activity, next it was examined whether SynPTP is specific to protein tyrosine phosphatase activity or it possesses dual specificity. We took three strategies to address this question. First, we examined the phosphatase hydrolase activity of SynPTP using several LMW organophosphoesters including isomers of naphthyl phosphate. Relative enzymatic activity towards these substrates is summarized in Table 3. The naphthyl phosphate isomers (Supplementary Fig. S2) were of particular interest, since they represent potential ‘diagnostic’ substrates for differentiating PTPs from DSPs in vitro (14, 40–42). From the data, it was observed that SynPTP was catalytically active towards β-naphthyl phosphate but not towards α-naphthyl phosphate, which indicated that the geometry of the substrate-binding pocket of SynPTP is similar to that of a protein tyrosine phosphatase.
Table III.
Hydrolysis of low molecular weight organophosphate substrates by SynPTP.
| Substrates | Specific activity (µmol/min/mg) | Relative activity (% pNPP) |
|---|---|---|
| p-Nitrophenyl-P | 3.2 ± 0.26 | 100 |
| α-Naphthyl-P | ND | ND |
| β-Naphthyl-P | 3.6 ± 0.42 | 117 |
| Glucose-6-P | ND | ND |
| 1, 4, 5 Inositol phosphate | ND | ND |
| Phosphotyrosine | 3.3 ± 0.41 | 104 |
| Phosphoserine | ND | ND |
Phosphohydrolase activity of SynPTP towards the listed organophosphoesters was measured as described in the ‘Materials and Methods’ section using Malachite Green assay. The amount of enzyme used in the reaction mixture was 500 ng. All organophosphates were present at the final concentration of 10 mM in the reaction mixture. The enzyme activity is reported relative to that observed with pNPP, 3.2 µmol/min/mg, which was set equal to 100%. ND indicates not detectable, <2% or 0.07 µmol/min/mg. All values are averages of triplicate determinations plus or minus standard error.
Second, we examined the substrate specificity of SynPTP using several exogenous protein substrates, which were phosphorylated on either serine or tyrosine residues as described in the ‘Materials and Methods’ section. The data are summarized in Table 4. From the results, it was evident that SynPTP displayed catalytic activity towards all tyrosyl-phosphorylated proteins, but failed to dephosphorylate the same seryl-phosphorylated substrates. The results indicated that SynPTP exhibited its catalytic function similar to a PTP not as a DSP.
Table IV.
Catalytic activity of SynPTP against exogenous protein substrates.
| Substrates | Specific activity (pmol/min/mg) | relative activity (% RCML 32P-Tyr) |
|---|---|---|
| RCML (32P-Tyr) | 800 ± 16 | 100 |
| Casein (32P-Tyr) | 1217 ± 60 | 162 |
| MBP (32P-Tyr) | 283 ± 6 | 38 |
| RCML (32P-Ser) | ND | ND |
| Casein (32P-Ser) | ND | ND |
| MBP (32P-Ser) | ND | ND |
The phosphatase activity of SynPTP, 500 ng, was measured towards the 32P-labelled phosphoproteins listed above as described in the ‘Materials and Methods’ section. All substrates were present at a final concentration of 4 µM protein bound [32P] phosphate. Enzyme activity is reported in terms of both specific activity (pmol/min/mg) and relative to that observed with RCML (32P-Tyr), which was set equal to 100%. ND represents not detectable, <2%, or 16 pmol/min/mg. All values are the results of triplicate determination plus or minus standard error.
Third, we examined the catalytic activity of SynPTP by analysing effect of various phosphatase inhibitors to determine whether SynPTP displayed similar pattern of sensitivity towards these inhibitors as other known LMW PTPs (11, 12, 33, 35, 42). Ammonium molybdate, sodium orthovanadate and Zn2+ are known to be potent inhibitors of PTPs (12, 33, 35), and our results suggested that these inhibitors strongly inhibited the activity of SynPTP (Table 5). Since sulphate (SO2−4) might mimic phosphate (PO3−4), we examined effect of sodium sulphate on the catalytic activity of SynPTP. It was observed that sodium sulphate had no inhibitory effect on phosphatase activity of SynPTP, which confirmed that Zn2+ behaves as a potent inhibitor for SynPTP. On the other hand, it was observed that sodium fluoride, a non-specific inhibitor (33, 42) for phosphatases and okadaic acid, a potent inhibitor (25) of many PP2A, failed to inhibit the catalytic activity of SynPTP. These results suggested that the sensitivity of the catalytic function of SynPTP for these inhibitors followed the pattern that was similar to other known LMW PTPs. Taken together, these results indicate that SynPTP is a LMW PTP not a DSP.
Table V.
Influence of various phosphatase inhibitors on the catalytic activity of SynPTP.
| Inhibitors | Concentration (mM) | Specific activity (pmole/min/mg) |
|---|---|---|
| Control | 0 | 560 ± 22 |
| Ammonium molybdate | 5 | ND |
| Sodium orthovanadate | 5 | 44 ± 2.5 |
| Zinc sulphate | 5 | ND |
| Sodium sulphate | 5 | 640 ± 35 |
| Sodium fluoride | 5 | 680 ± 14 |
| Okadaic acid | 0.005 | 720 ± 22 |
All assays were carried out as described in the ‘Materials and Methods’ section. RCML (32P-Tyr) was used as substrate at a final concentration of 2 µM of protein bound phosphate. Potential inhibitors were present at the indicated final concentration. The enzyme activity is reported relative to that observed with RCML(32P-Tyr) in the absence of any inhibitor, which was 560 pmol/min/mg, and set equal to 100%. ND represents not detectable, <2%, or 12 pmol/min/mg.
Identification of endogenous substrates for SynPTP
To identify endogenous physiological substrates, we performed ‘substrate trapping’ experiments. Members of the protein tyrosine phosphatase (PTPs) family share a common mechanism for phosphomonoester hydrolysis (20, 43). Detailed studies have shown that mutagenic alteration of either the catalytic cysteine, conserved aspartate, or both renders a PTPs incapable of dephosphorylating substrates, but these mutagenically altered PTPs still retain the ability for binding substrates (44–46). Consequently, these mutationally altered forms of PTPs have been named ‘Substrate Trapping Mutant(s)’ (47) and are commonly used as affinity reagents to isolate and identify physiological substrates for various PTPs.
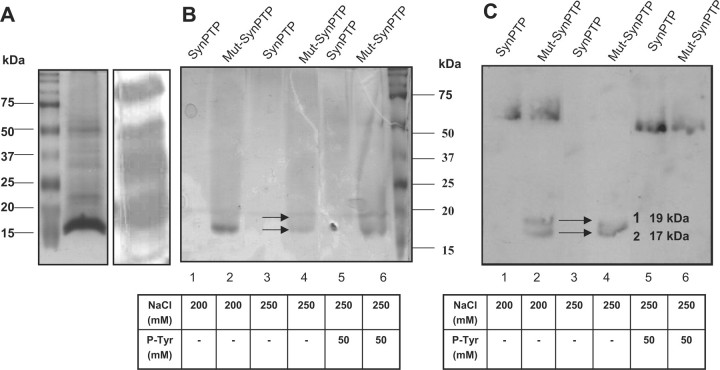
In this study, we used mutagenically altered Cys7SerAsp125Ala-SynPTP as a ‘Substrate Trapping Mutant’ to isolate potential substrates for SynPTP from soluble extracts of Synechocystis sp. PCC 6803. Active SynPTP was used as a negative control. The soluble fractions (15 mg) of Synechocystis sp. PCC 6803 cell extracts were applied to columns, on which either active SynPTP or mutagenically altered inactive SynPTP had been immobilized and substrate trapping experiment was performed as described in the ‘Materials and Methods’ section. Bound proteins from the eluted fractions from each step of the two columns were analysed by SDS–PAGE and western blot analysis with anti-phosphotyrosine antibody. Our data suggested that there is no immunoreaction in 50 mM to 150 mM eluates (data was not shown) but the presence of two putative phosphotyrosine-containing proteins in the molecular mass range of 15–20 kDa was observed in the eluates of 200 mM NaCl and 250 mM NaCl from mutagenically altered SynPTP column (Fig. 3B and C, lane: 2 and 4). These two proteins were not observed in the eluates of either 200 mM NaCl or 250 mM NaCl (Fig. 3B and C, lane: 1, 3) from active SynPTP column. Therefore, these two proteins were considered as potential substrates for SynPTP. The eluted fractions of both active SynPTP and mutagenically altered inactive SynPTP bound Sepharose column with 250 mM NaCl were used for in-solution Trypsin digestion and these tryptic peptides were analysed by mass spectrometry to identify the proteins as described in the ‘Materials and Methods’ section. Mass peptide profiles were matched to the potential protein sources using NCBInr (http://www.ncbi.nlm.nih.gov/BLAST) database through Mascot search developed by www.matrixscience.com. The results of mass spectral analysis matched the following proteins from Synechocystis sp. PCC 6803: (i) Sll1577 (CpcB), β-subunit of phycocyanin, molecular weight 18.8 kDa, accession no. 1652309; and (ii) Sll1578 (CpcA), α-subunit of phycocyanin, molecular weight 17.8 kDa, accession no. 1652308. The results of mass spectral analysis are summarized in Tables 6 and 7.
Fig. 3.
Identification of potential protein substrates for SynPTP from soluble fractions of Synechocystis sp. PCC 6803 by substrate trapping experiment. Soluble fractions of Synechocystis cell extracts were applied to active SynPTP bound Sepharose or mutagenically altered inactive SynPTP bound Sepharose. Bound proteins were eluted with 200 mM NaCl, 250 mM NaCl, and 250 mM NaCl containing 50 mM phosphotyrosine. Eluted proteins were subjected to SDS-PAGE and immunoblotted with anti-phosphotyrosine antibody. (A) Presence of phosphotyrosine proteins in the soluble fractions of Synechocystis cell extracts was analyzed by SDS-PAGE (Coomassie blue stained gel, left panel) and Western blot analysis (right panel). (B) Results of substrate trapping experiment, analyzed by SDS-PAGE, stained with Coomassie blue. Lane 1, 3, 5 contained proteins eluted from active SynPTP column with 200 mM NaCl, 250 mM NaCl, and 250 mM NaCl with 50 mM phosphotyrosine respectively. Lane 2, 4, 6 contained proteins eluted from mutagenically altered inactive SynPTP column with 200 mM NaCl, 250 mM NaCl, and 250 mM NaCl with 50 mM phosphotyrosine respectively. (C) Western blot analysis of a duplicate gel from SDS-PAGE. Arrowheads indicate the positions of potential substrates for SynPTP.
Table VI.
A proteomic analysis of potential substrates for SynPTP, observed from substrate trapping experiment, in soluble fractions of Synechocycistis sp. PCC 6803 cell extracts.
| Protein ID | No. of peptides matched/total | Predicted Mr | Observed Mr |
|---|---|---|---|
| Phycocyanin β | 5/13 | 18.8 | 19 |
| Phycocyanin α | 3/13 | 17.8 | 17 |
Predicted molecular mass (Mr) was deduced from amino acid sequence, calculated by using the PROTEIN CALCULATOR v3.2 program (http://www.scripps.edu/∼cdputnam/protcalc.html). Observed molecular mass was determined from Coomassie blue stained SDS gel.
Table VII.
Mass Spectral analysis of partial sequence for ∼19 kDa and 17 kDa proteins, potential substrates of SynPTP.
| Protein | Peptide no | Observed mass (Da) | Calculated mass (Da) | Sequence | Individual ions scorea |
|---|---|---|---|---|---|
| Phycocyanin βa (19 kDa) | 1 | 914.59 | 914.53 | MFDVFTR.V | 34 |
| 13 | 2109.17 | 2109.02 | R.GEYLSGSQLDALSATVAEGNK.R | 97 | |
| 11 | 1906.90 | 1906.89 | R.YVTYATFTGDASVLEDR.C | 44 | |
| 9 | 1816.09 | 1815.97 | R.ETYVALGVPGASVAAGVQK.M | 36 | |
| 7 | 1598.55 | 1596.81 | K.EAALDIVNDPNGITR.G | 38 | |
| Phycocyanin αa (17 kDa) | 5 | 1467.25 | 1467.77 | R. FLSSTELQIAFGR.L | 84 |
| 6 | 1532.37 | 1531.75 | K. TPLTEAVSTADSQGR.F | 42 | |
| 8 | 1654.77 | 1654.82 | R.TFDLSPSWYVEALK.Y | 54 |
aProteins were isolated from substrate trapping experiment after immobilization with SynPTP. Shown are the partial sequences of amino acids deduced from peptides mass fingerprint obtained after fragmentations of tryptic peptides by MS/MS analysis. Observed mass and calculated mass of each peptide was determined from Mascot search results of www.matrixscience.com. Individual ions score >30 indicated homology. Individual ions score >52 indicated identity or extensive homology.
However, no tyrosine-phosphorylated peptide was detected by mass spectral analysis, therefore, we performed additional experiments to determine whether phycocyanin contains phosphotyrosine. Phycocyanin was isolated from soluble fractions of Synechocystis sp. PCC 6803 cell extracts by anion exchange column chromatography followed by size exclusion column chromatography (30). The blue coloured fractions, whose ratio of A620/A280 was in the range of 3.8 to 4.2 [phycocyanin-rich fractions (29, 30)], were collected from size exclusion column chromatography and analysed on SDS–polyacrylamide gel and immunodetection using anti-phosphotyrosine antibody (Supplementary Fig. S3A–C). For each fraction, two protein bands were observed on the SDS gel in the molecular size range of 15–20 kDa, the expected molecular size of phycocyanin β- and α-subunits. It was also observed that these fractions were immunoreactive with the anti-phosphotyrosine antibody. Results indicated that these two proteins are tyrosine phosphorylated.
Now, the question of whether phycocyanin is phosphorylated either on serine or tyrosine residue has been controversial. It has been reported (29) that β-phycocyanin in Synechocystis sp. PCC 6803 was phosphorylated in vitro using [γ-32P]ATP. Site-directed mutagenesis revealed that Ser-50 of β-phycocyanin was the site of phosphorylation (29). Similar work has been done both in vivo and in vitro in the cyanobacterium Synechococcus sp. PCC 6301 using 32P orthophosphate or [γ-32P] ATP (30, 31, 48). It was repeatedly observed that major phosphorylated bands appeared at 18.5 kDa and 17 kDa on SDS gels. It was suggested that these protein bands corresponded to polypeptides of the phycobilisome complex (49). The 18.5 kDa protein was proposed as β-phycocyanin (29–31). Nevertheless, no tyrosine phosphorylation of phycocyanin was reported from previous studies. However, only Piven and co-workers (28) could not detect any phosphorylation in β-phycocyanin using phosphoprotein staining gel dye Pro-Q Diamond or phospho-threonine antisera. Since phycocyanin auto-fluoresces due to the covalent attachment of bilin groups, which are open-chain tetrapyrroles (50, 51), Piven and co-workers suggested that the signals obtained for phycocyanin from Pro-Q staining arose due to its self-fluorescence. Therefore, we further examined our data whether the signals of phycocyanin from the Western blot analysis obtained due to its phosphorylation, in this case tyrosine phosphorylation, or its self-fluorescence.
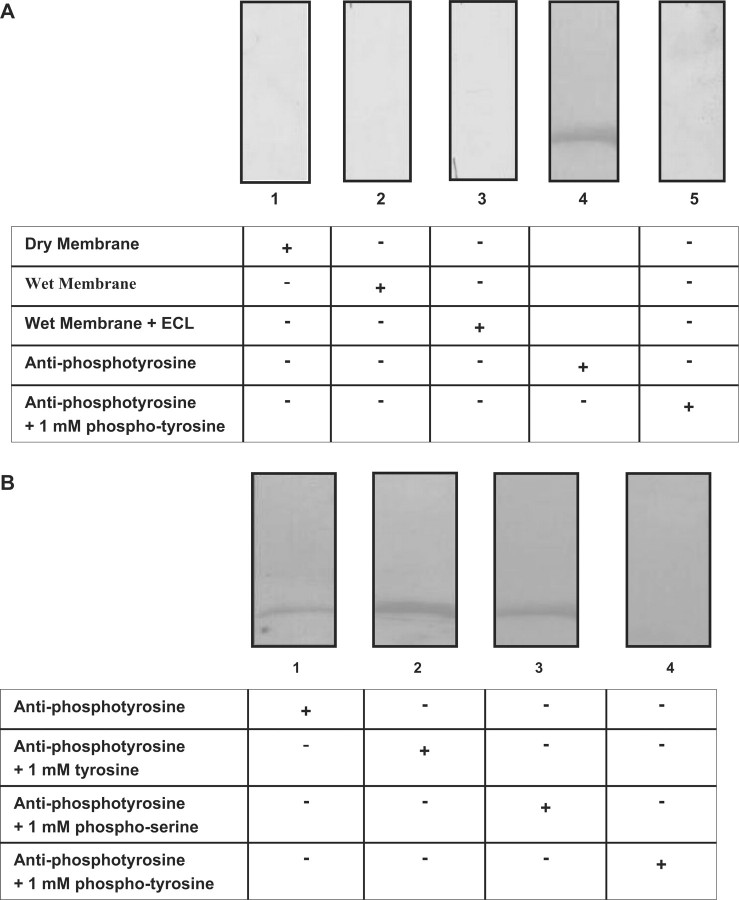
The purified fractions of phycocyanin eluted from the size exclusion chromatography were resolved on a 12% (w/v) SDS gel and the proteins were transferred to a PVDF membrane. Following that, an X-ray film was exposed to the membrane (with transferred proteins) under different conditions and the film was developed. First, the film was developed after exposure to a dry membrane. Second, the film was exposed to the wet membrane in TBS buffer (Tris-base saline, pH 7.5) and the corresponding film was developed. Third, a duplicate wet membrane from second condition was incubated with ECL reagent (Pierce, Rockford III) for 5 min and then exposed to the film. Fourth, the membrane was incubated with an anti-phosphotyrosine antibody conjugated with horseradish peroxidase and imnmunodetection was performed. Finally, the anti-phopshotyrosine antibody was pre-incubated with 5 mM free phosphotyrosine for 1 h at room temperature, and then immunodetection was carried out. No luminescent band for phycocyanin was visualized after exposing the film under the first three conditions (Fig. 4A, lanes: 1–3). Protein band for phycocyanin was only visualized when the membrane was incubated with anti-phosphotyrosine antibody, which indicated that only the chemiluminescence of immunoreactive species was detected (Fig. 4A, lane: 4). The immunoreactivity of the antibody was blocked when it was pre-incubated with 5 mM free phosphotyrosine (Fig. 4A, lane: 5). These results confirmed that phycocyanin is tyrosine phosphorylated.
Fig. 4.
Comparison of the intensity of the intrinsic autofluorescence of phycocyanin with Chemiluminescence produced by western blot analysis. (A) Purified phycocyanin eluted from size exclusion column was analysed on SDS gel and proteins were transferred to a PVDF membrane. Lane1: an X-ray film exposed on the dry membrane; lane 2: the wet membrane in TBS buffer, which was exposed to the X-ray film; lane 3: a duplicate of the wet membrane that was incubated with ECL reagent and then exposed to the X-ray film; lane 4: immunoblot probed with anti-phosphotyrosine antibody; lane 5: immunoblot that was prepared with pre-incubated anti-phosphotyrosine antibody in presence of 5 mM free phosphotyrosine. (B) Western blot analysis of phycocyanin for competition of antibody with free phosphoamino acids. Purified phycocyanin, obtained from size exclusion column chromatography, 6 mg, was resolved by SDS–PAGE and transferred to a PVDF membrane. Antibody was pre-incubated at room temperature for 1 h with 1 mM specified compound as mentioned below. Lane 1: antibody alone, no phosphoamino acid was added; lane 2: 1 mM tyrosine; lane 3: 1 mM phospho-serine; lane 4: 1 mM phospho-tyrosine.
Next, we examined the specificity of the anti-phosphotyrosine antibody, RC20, used in all immunodetection experiments to detect phosphotyrosine proteins. We asked whether immunoreactivity could be competed with free phosphotyrosine, free tyrosine or free phosphoserine.
Fractions containing purified phycocyanin from size exclusion column chromatography were resolved on a 12% (w/v) SDS gel and transferred to a PVDF membrane. The membrane was probed with anti-phosphotyrosine antibody or antibody that had been pre-incubated with either phospho-serine, phospho-tyrosine or tyrosine (1 mM each). It was observed that the intensity of signal for immunodetection of phycocyanin was abolished only when membrane was probed with antibody that had been pre-incubated with 1 mM free phosphotyrosine (Fig. 4B, lane: 4). Immunoreactivity of the antibody was not blocked when it was pre-incubated with free tyrosine or phosphoserine (Fig. 4B, lanes: 2 and 3), and appeared to be similar in intensity to the immunoblot that had been incubated with antibody alone (Fig. 4B, lane: 1). Therefore, the data indicated that antibody was specific to recognize the phosphotyrosine epitope.
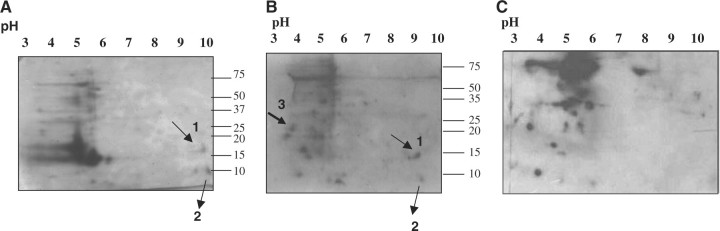
Next, we compared the intensity of the immunoreactivity of antibody for phycocyanin between wild type strain and a phycocyanin less mutant strain (CK mutant) of Synechocystis sp. PCC 6803. If phycocyanin is not present in the mutant strain, then signal for immunoreaction of antibody with phycocyanin will be weak or less than that of wild type strain. The wild type and CK mutant strain of Synechocystis sp. PCC6803 were grown in BG-11 media as described in the ‘Materials and Methods’ section. It was observed that the growth of CK mutant strain was poor in comparison to wild type strain (Supplementary Fig. S4A). The whole cell extracts for both wild type and CK mutant strains were prepared as described in the ‘Materials and Methods’ section with the exception that cell lysis buffer included 0.1mM sodium orthovanadate, 0.1% Tween-20 (v/v), and 0.1% Triton-X-100 (v/v). The whole cell extracts from both wild type and CK mutant, 30 µg protein each, were resolved on 12% (w/v) SDS–PAGE and immunoblotted with anti-phosphotyrosine antibody (Supplementary Fig. S4B and C). It was noticed that the intensity of immunoreactivity of antibody for phycocyanin (position of phycocyanin was indicated by arrow) was much weaker in mutant strain than that in the wild-type strain as expected. Taken together, our results indicated that phycocyanin is tyrosyl-phosphorylated. However, we could not detect phycocyanin alpha or beta subunit from the whole cell extracts of Syenchocystis due to the presence of many proteins in the same pI range of phycocyanin, 4.5–5.5 (48), in the Commassie blue stained 2D gel (Fig. 5A), which prevented to isolate the right protein.
Fig. 5.
Identification of potential substrates for SynPTP from whole-cell extracts of Synechocystis sp. PCC 6803. (A) Coomassie-stained SDS gel of 2D gel electrophoresis of whole cell extracts of Synechocystis sp. PCC 6803. (B) Immunoblot was incubated in the absence of active SynPTP. (C) Immunoblot was incubated in the presence of active SynPTP. Arrowheads in (B) indicate the positions of the potentials substrates, which were immunoreactive with antibody in the absence of enzyme, but not following incubation in the presence of SynPTP (C).
It has been reported that phycocyanin is a major bilin-containing protein in the phycobilisome complex of Synechocystis sp. PCC 6803 and functions as a light harvesting antenna for the transfer of the excitation energy to photosynthetic reaction centres (50, 52). It is known that in green plants, phosphorylation of proteins in the light harvesting complex II (LHCII) regulate the distribution of absorbed excitation energy between photosystem I (PSI) and photosystem II (PSII) such that light-limited photosystem receives more energy, while the light-saturated photosystem receives less (53). The transition between two states is described as light-state transition (54). Phosphorylation of LHCII alters its three-dimensional structure (54). This conformational change induces the dissociation of the phospho-LHCII trimer into monomeric phospho-LHCII. The monomers are free to migrate from PSII to PSI (55). After dephosphorylation, unphosphorylated LHCII monomers may trimerize at the periphery of PSII. Phosphorylation of LHCII leads the adjustment of the stoichiometry of two photosystems (51). We propose that the possible role of SynPTP in dephosphorylation of phycocyanin in cyanobacterium Synechocystis sp. PCC 6803 might be involved in the regulation of excitation energy distribution similar to that of LHCII in higher plants. At the state of adjustment to PSII light, phosphorylation of phycocyanin might induce dissociation of phosphorylated phycobilisome trimers or hexamers to monomeric phosphorylated phycobilisomes. The dissociation of phosphorylated phycobilisomes may induce lateral movement of monomeric phosphorylated phycobilisomes to bind preferentially to PSI at the state of adjustment to PSI light. After dephosphorylation by SynPTP, monomeric unphosphorylated phycobilisomes may assemble in trimer or hexamer form and reassociate with PSI at the state of adjustment to PSI light.
Identification of the potential substrates from whole-cell extracts of Synechocystis sp. PCC 6803
Whole cell extracts of Synechocystis sp. PCC 6803 were prepared as described in the ‘Materials and Methods’ section with the exception that cell lysis buffer included 0.1 mM sodium orthovanadate, 0.1% Tween-20 (v/v), and 0.1% Triton-X-100 (v/v). 2D gel electrophoresis was performed as described in the ‘Materials and Methods’ section to detect the phosphotyrosine proteins from whole cell extracts of Synechocystis. Several phosphotyrosine-containing proteins appeared on the immunoblot incubated without SynPTP. The presence of three phosphotyrosine-containing proteins (indicated by arrow in Fig. 5B) was not observed in the immunoblot incubated with SynPTP (Fig. 5C). Results indicated that these three proteins were inactive for immunoreaction with antibody due to dephosphorylation by SynPTP and therefore could be the potential substrates for SynPTP.
Two of these proteins (protein #1 and protein #2) were identified by mass spectral analysis. Peptide mass profiles were matched to the potential protein sources using NCBInr database through Mascot search developed by www.matrixscience.com. The results matched with the following proteins from Synechocystis sp. PCC 6803, which are: (i) Slr0737 (PsaD), Photosystem I subunit II, molecular weight 15.6 kDa, pI 8.9 and the GenBank accession no. 16329280. Predicted molecular mass and pI of this protein were matched with those of observed values. (ii) Ssl3093 (CpcD), phycocyanin-associated linker protein, molecular weight 9.3 kDa, pI 9.4 and GenBank accession no. 16329820.
However, the molecular mass and pI of this protein determined by 2D gel electrophoresis were matched with those of predicted values. Third protein (Protein # 3) of the probable substrates was not identified since it was difficult to isolate the correct protein from the Coomassie blue-stained acrylamide gel from the surrounding other proteins. Results of mass spectrometric analysis are summarized in Table 8. Observed pI values for PsaD and CpcD were compared and found consistent with those values previously reported by Wang and Chitnis, 2000 (54).
Table VIII.
Proteomic analysis of PsaD and Ssl3093.
| Protein ID | No. of peptides matched/total | Predicted Mr | Observed Mr |
|---|---|---|---|
| PsaD | 22/281 | 15.6 | 16 |
| Ssl3093 | 1/6 | 9.3 | 10 |
Predicted molecular mass was deduced from amino acid sequence, calculated by using PROTEIN CALCULATOR v3.2 program (http://www.scripps.edu/∼cdputnam/protcalc.html). Observed molecular mass was determined from Coomassie blue stained acrylamide gel of 2D gel electrophoresis.
However, detailed studies of PsaD in Synechocystis sp. PCC 6803 suggested that this photosystem I subunit plays multiple functions, which include formation and stabilization of PSI complex by binding to other subunits (A. B, C and L) and playing the major docking site for ferredoxin (fd) (56–58). Further study is required to determine how phosphorylation of PsaD affects these functions. Previously, phosphorylation of linker proteins (33 kDa, 35 kDa, 99 kDa and 27 kDa) on threonine in Synechocystis sp. PCC 6803 and the possible function of these linker proteins by phosphorylation/dephosphorylation as a signal for protein degradation during assembly/disassembly of the phycobilisome complex were reported by Piven et al. (28). Future studies need to be performed to firmly establish the effect of tyrosine phosphorylation of this 9.5 kDa phycocyanin-associated rod linker protein, CpcD (Ssl3093), as a substrate for SynPTP and to identify the tyrosine phosphorylation sites on phycocyanin, PsaD and CpcD. It is also important to study the stoichiometry of phosphorylated/non-phosphorylated states of these potential substrates for their physiological significance of SynPTP in Synechocystis. However, we could not measure the stoichiometry of tyrosyl-phosphorylated and non-phosphorylated form of substrates in our studies due to the low abundance of the tyrosine-phosphorylated substrates. It is primarily because the phosphorylation states of the proteins depend on the intensity of light at which Synechocystis sp. was grown (13, 15). In our studies, Synechocystis was grown at low light intensity, 10 µmol photons m−2 s−1, a condition that is not conducive for elevated levels of tyrosine phosphorylation.
Taken together, our results indicated that the possible substrates of SynPTP include phycocyanin, PsaD and CpcD and these proteins are the subunits of photosystem complex. Therefore, we speculate that SynPTP might be involved in the regulation of photosynthesis.
Supplementary Data
Supplementary Data are available at JB Online.
Funding
Grant GM 55067 from the National Institute of Health.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
The authors thank Dr William Keith Ray for his help with mass spectral analysis.
Glossary
Abbreviations
- LMW
low molecular weight
- PTPs
protein tyrosine phosphatases
- pNPP
para-nitrophenyl phosphate
- ORF
open reading frame
References
- 1.Cozzone AJ. Protein phosphorylation in prokaryotes. Ann. Rev. Microbiol. 1988;42:97–125. doi: 10.1146/annurev.mi.42.100188.000525. [DOI] [PubMed] [Google Scholar]
- 2.Gary LJ, Razvan L. Mitogen activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinase. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 3.Ye W, Zhang L. Heme controls the expression of cell cycle regulators and cell growth in HeLa cells. Biochem. Biophys. Res. Commun. 2004;315:546–554. doi: 10.1016/j.bbrc.2004.01.092. [DOI] [PubMed] [Google Scholar]
- 4.Chiarugi P, Cirri P, Taddei L, Giannoni E, Camici G, Manno G, Raugei G, Ramponi G. The low Mr protein tyrosine phosphatases is involved in Rho mediated cytoskeleton rearrangement after integrin and platelet derived growth factor. J. Biol. Chem. 2000;275:4640–4646. doi: 10.1074/jbc.275.7.4640. [DOI] [PubMed] [Google Scholar]
- 5.Rao GS, Ramachandran MV, Bajaj JS. In silico structure-based design of a potent and selective small peptide inhibitor of protein tyrosine phosphatase 1B, a novel therapeutic target for obesity and type 2 diabetes mellitus: a computer modeling approach. J. Biomol. Struct. Dyn. 2006;23:377–384. doi: 10.1080/07391102.2006.10531233. [DOI] [PubMed] [Google Scholar]
- 6.Lee G, Thangavel R, Sharma VM, Litersky JM, Bhaskar K, Fang SM, Do LH, Andreadis A, Van Hoesen G, Ksiezak-Reding H. Phosphorylation of tau by fyn: implications for Alzheimer’s disease. Neurosci. J. 2004;24:2304–2312. doi: 10.1523/JNEUROSCI.4162-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozzone AJ, Grangeasse C, Doublet P, Bertrand D. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 2004;181:171–181. doi: 10.1007/s00203-003-0640-6. [DOI] [PubMed] [Google Scholar]
- 8.Bliska JB, Galan JE, Falkow S. Signal transduction in the mammalian cell during bacterial attachment and entry. Cell. 1993;73:903–920. doi: 10.1016/0092-8674(93)90270-z. [DOI] [PubMed] [Google Scholar]
- 9.Manai M, Cozzone AZ. Analysis of the protein kinase activity of Escherichia coli cells. Biochem. Biophys. Res. Commun. 1979;91:819–826. doi: 10.1016/0006-291x(79)91953-3. [DOI] [PubMed] [Google Scholar]
- 10.Guan KL, Dixon JE. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Strohl WR. Cloning, purification and properties of a phosphotyrosine protein tyrosine phosphatase from Streptomyces coelicolor A3(2) J. Bacteriol. 1996;178:136–142. doi: 10.1128/jb.178.1.136-142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grangeasse C, Doublet P, Vincent C, Vaganay E, Riberty M, Duclos B, Cozzone AZ. Functional characterization of the low-molecular-mass phosphotyrosine-protein phosphatase of Acinetobacter johnsonii. J. Mol. Biol. 1998;278:339–347. doi: 10.1006/jmbi.1998.1650. [DOI] [PubMed] [Google Scholar]
- 13.Warner KM, Bullerjahn GS. Light-dependent tyrosine phosphorylation in the cyanobacterium Prochlorothrix hollandica. Plant Physiol. 1994;105:629–633. doi: 10.1104/pp.105.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell D, Griffiths C, Slade LW, Potts M, Kennelly PJ. Substrate specificity of IphP, a cyanobacterial dual specificty protein phosphatase with MAP kinase phosphatase activity. Biochemistry. 1996;35:7566–7572. doi: 10.1021/bi9600409. [DOI] [PubMed] [Google Scholar]
- 15.McCartney B, Howell D, Kennelly PJ, Potts M. Protein tyrosine phosphorylation in the cyanobacterium Anabaena sp. strain PCC7120. J. Bacteriol. 1997;179:2314–2318. doi: 10.1128/jb.179.7.2314-2318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ZY. Protein tyrosine phosphatases: biological function, structural characteristic, and mechanism of catalysis. Crit. Rev. Biochem. Mol. Biol. 1998;33:1–52. doi: 10.1080/10409239891204161. [DOI] [PubMed] [Google Scholar]
- 17.Kennelly PJ. Protein phosphatases—a phylogenetic perspective. Chem. Rev. 2001;101:2291–2312. doi: 10.1021/cr0002543. [DOI] [PubMed] [Google Scholar]
- 18.Fauman EB, Saper MA. Structure and the function of the protein tyrosine phosphatases. Trends Biochem. Sci. 1996;21:413. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- 19.Ramponi G, Stefani M. Structural, catalytic and functional properties of low Mr phosphotyrosine protein phosphatases. Evidence for a long evolutionary history. Int. J. Biol. Chem. Cell Biol. 1997;29:279–292. doi: 10.1016/s1357-2725(96)00109-4. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;30:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CC. Bacterial signalling involving eukaryotic-type protein kinases. Mol. Microbiol. 1996;20:9–15. doi: 10.1111/j.1365-2958.1996.tb02483.x. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Kennelly PJ, Potts M. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 1998;22:229–253. doi: 10.1111/j.1574-6976.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Haile J, Kennelly PJ. An arsenate reductase from Synechocystis sp. strain PCC 6803 exhibits a novel combination of catalytic characteristics. J. Biol. Chem. 2003;185:780–789. doi: 10.1128/JB.185.23.6780-6789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamei A, Yusa T, Geng X, Ikeuchi M. Biochemical examination of the potential eukaryotic-type protein kinase genes in the complete genome of the unicellular cyanobacterium Synechocystis sp. PCC 6803. DNA Res. 2002;9:71–78. doi: 10.1093/dnares/9.3.71. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Bischoff KM, Kennelly PJ. The icfG gene cluster of Synechocystis sp. strain PCC 6803 encodes an Rsb/Spo-like protein kinase, protein phosphatase, and two phosphoproteins. J. Bacteriol. 1999;181:4761–4767. doi: 10.1128/jb.181.16.4761-4767.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irmler A, Forchhammer K. A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803. PNAS. 2001;98:12978–12983. doi: 10.1073/pnas.231254998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li R, Potters MB, Shi L, Kennelly PJ. The protein phosphatases of Synechocystis sp. strain PCC 6803: open reading frames sll1033 and sll1387 encode enzymes that exhibit both protein-serine and protein-tyrosine phosphatase activity in vitro. J. Bacteriol. 2005;187:5877–5884. doi: 10.1128/JB.187.17.5877-5884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piven I, Ajlani G, Sokolenko A. Phycobilisome linker proteins are phosphorylated in Synechocystis sp. PCC 6803. J. Biol. Chem. 2005;280:21667–21672. doi: 10.1074/jbc.M412967200. [DOI] [PubMed] [Google Scholar]
- 29.Mann HN, Newman J. The Phototrophic Prokaryotes. New York: Kluwer Academic/Plenum Publisher; 1999. Phosphorylation of β-phycocyanin in Synechocystis sp. PCC 6803; pp. 71–75. [Google Scholar]
- 30.Sanders CE, Allen JF. The 18.5 kDa phosphoproteins of the Cyanobacterium Synechococcus 6301: A component of phycobilisome. Progress Photosynth. Res. 1987;2:761–764. [Google Scholar]
- 31.Sanders CE, Melis A, Allen JF. In vivo phosphorylation of proteins in the cyanobacterium Synechococcus 6301 after chromatic acclimation to Photosystem I or Photosystem II light. Biochim. Biophys. Acta. 1989;976:168–172. [Google Scholar]
- 32.Li R, Debella HJ, Carmichael WW. Isolates identifiable as Athrospira maxima and Athrospira fusiformis (Oscillatoriales, Cyanobacteria) appear identical on the basis of a morphological study in culture and 16S rRNA sequences. Phycologia. 2001;40:367–371. [Google Scholar]
- 33.Tonks NK, Diltz CD, Fischer EH. Characterization of the major protein tyrosine phosphatases of human placenta. J. Biol. Chem. 1988;263:6731–6737. [PubMed] [Google Scholar]
- 34.Lower BH, Kennelly PJ. Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein serine kinase activity. J. Bacteriol. 2003;185:3436–3445. doi: 10.1128/JB.185.11.3436-3445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulat D, Vaganay E, Duclos B, Genestier AL, Cozzone AJ. Staphylococcus aureus contains two low molecular mass phosphotyrosine phosphatases. J. Bacteriol. 2002;184:5194–5199. doi: 10.1128/JB.184.18.5194-5199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone AJ, Duclos B. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang ZY, Zhou G, Denu JM, Wu L, Tang X, Mondesert O, Russel P, Butch E, Guan KL. Purification and characterization of the low molecular weight protein tyrosine phosphatse Stp1, from the fission yeast Schizosaccharomyces pombe. Biochemistry. 1995;34:10560–10568. doi: 10.1021/bi00033a031. [DOI] [PubMed] [Google Scholar]
- 38.Ostanin K, Polasky C, Wang S, Etten RLV. Cloning and characterization of a Saccharomyces cerevisiae gene encoding the low molecular weight protein tyrosine phosphatase. J. Biol. Chem. 1995;270:18491–18499. doi: 10.1074/jbc.270.31.18491. [DOI] [PubMed] [Google Scholar]
- 39.Wo YP, Zhou MM, Panayiotis S, June PD, Zhang ZY, Van Etten RL. Cloning, expression, and catalytic mechanism of the low molecular weight phosphotyrosyl protein phosphatase from Bovine Heart. Biochemistry. 1992;31:1712–1721. doi: 10.1021/bi00121a019. [DOI] [PubMed] [Google Scholar]
- 40.Zhang ZY, Malachowski WP, Van Etten RL, Dixon JE. Nature of the rate-determining steps of the reaction catalyzed by the Yersinia protein-tyrosine phosphatase. J. Biol. Chem. 1994;269:8140–8145. [PubMed] [Google Scholar]
- 41.Savle PS, Shelton TE, Meadows CA, Potts M, Gandour RD, Kennelly PJ. N-(cyclohexanecarboxyl)-O-phospho-L-serine, a minimal substrate for the dual specificity protein phosphate IphP. Arch. Biochem. Biophys. 2000;376:439–448. doi: 10.1006/abbi.2000.1750. [DOI] [PubMed] [Google Scholar]
- 42.Koul A, Choidas A, Treder M, Tyagi AK, Singh Y, Ulrich A. Cloning and characterization of secretory tyrosine phosphatases from Mycobacterium tuberculosis. J. Bacteriol. 2000;182:5425–5432. doi: 10.1128/jb.182.19.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Denu JM, Stuckey JA, saper MA, Dixon JE. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 44.Agazie YM, Hayman MJ. Development of an efficient substrate trapping mutant of Src homology phosphotyrosine phosphatase-2 and identification of the epidermal growth factor receptor, gab1 and three other proteins as target substrates. J. Biol. Chem. 2003;278:13592–13598. doi: 10.1074/jbc.M210670200. [DOI] [PubMed] [Google Scholar]
- 45.Jeon SJ, Fujiwara S, Takagi M, Tanaka T, Imanaka T. Tk-PTP, protein tyrosine/serine phosphatase from hyperthermophilic archeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Comm. 2002;295:508–514. doi: 10.1016/s0006-291x(02)00705-2. [DOI] [PubMed] [Google Scholar]
- 46.Xie L, Zhang YL, Zhang ZY. Design and characterization of an improved protein tyrosine substrate-trapping mutant. Biochemistry. 2002;41:4032–4039. doi: 10.1021/bi015904r. [DOI] [PubMed] [Google Scholar]
- 47.Gardon AJ, Flint AJ, Tonks NA. Identification of p130cas as a substrate for the cytosolic protein tyrosine phosphatase PTP-PSET. Mol. Cell. Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrison MA, Tsinoremas NF, Allen JF. Cyanobacterial thylakoid membrane proteins are reversibly phosphorylated under plastoquinone-reducing conditions in vitro. FEBS. 1991;282:295–299. doi: 10.1016/0014-5793(91)80499-s. [DOI] [PubMed] [Google Scholar]
- 49.Allen JF, Sanders CE, Holmes NG. Correlation of membrane protein phosphorylation with excitation energy distribution in the cyanobacterium Synechococcus 6301. FEBS Lett. 1985;193:271–275. [Google Scholar]
- 50.MacColl R. Cyanobacterial phycobilisomes. J. Struct. Biol. 1998;124:311–333. doi: 10.1006/jsbi.1998.4062. [DOI] [PubMed] [Google Scholar]
- 51.Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- 52.Forsberg J, Allen JF. Protein tyrosine phosphorylation in the transition to light state 2 of chloroplast thylakoids. Photosynth. Res. 2001;68:71–79. doi: 10.1023/A:1011891017067. [DOI] [PubMed] [Google Scholar]
- 53.Allen JF, Nilsson A. Redox signaling and the structural basis of regulation of photosynthesis by protein phosphorylation. Physiol. Plant. 1997;100:863–868. [Google Scholar]
- 54.Wang Y, Chitnis P. Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC 6803. Electrophoresis. 2000;21:1746–1754. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1746::AID-ELPS1746>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 55.Bennett J. Regulation of photosynthesis by reversible phosphorylation of the light-harvesting chlorophyll a/b protein. Biochem. J. 1983;212:1–13. doi: 10.1042/bj2120001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruip J, Chitnis P, Lagoutte B, Rogner M, Boekema E. Structural organization of the major subunits in cyanobacterial Photosystem I. J. Biol. Chem. 1997;272:17061–17069. doi: 10.1074/jbc.272.27.17061. [DOI] [PubMed] [Google Scholar]
- 57.Chitnis VP, Chitnis PR. PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS. 1993;336:330–334. doi: 10.1016/0014-5793(93)80831-e. [DOI] [PubMed] [Google Scholar]
- 58.Lagoutte B, Hanley J, Bottin H. Multiple functions for the C-terminus of the PsaD subunit in the cyanobacterial Photosystem I complex. Plant Physiol. 2001;126:307–316. doi: 10.1104/pp.126.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.