Abstract
Evidence for the etiology of autism spectrum disorders (ASD) has consistently pointed to a strong genetic component complicated by substantial locus heterogeneity1,2. We sequenced the exomes of 20 sporadic cases of ASD and their parents, reasoning that these families would be enriched for de novo mutations of major effect. We identified 21 de novo mutations, of which 11 were protein-altering. Protein-altering mutations were significantly enriched for changes at highly conserved residues. We identified potentially causative de novo events in 4/20 probands, particularly among more severely affected individuals, in FOXP1, GRIN2B, SCN1A, and LAMC3. In the FOXP1 mutation carrier, we also observed a rare inherited CNTNAP2 mutation and provide functional support for a multihit model for disease risk3. Our results demonstrate that trio-based exome sequencing is a powerful approach for identifying novel candidate genes for ASD and suggest that de novo mutations may contribute substantially to the genetic risk for ASD.
ASD are characterized by pervasive impairment in language and communication, social reciprocity, and having restricted interests or stereotyped behaviors1. Several new candidate loci for ASD have recently been identified using genome-wide approaches that discover individually rare events of major effect2. A number of genetic syndromes with features of the ASD phenotype, collectively referred to as syndromic autism, have also been described4. Despite this progress, the genetic basis for the vast majority of ASD cases remains unknown. Several observations support the hypothesis that the genetic basis for ASD in sporadic cases may differ from that of families with multiple affected individuals, with the former more likely to result from de novo mutation events rather than inherited variants1,5–7. In this study, we sequenced the protein-coding regions of the genome (the exome)8 to test the hypothesis that de novo protein-altering mutations substantially contribute to the genetic basis of sporadic ASD. In contrast with array-based analysis of large de novo copy number variants (CNVs), this approach has greater potential to implicate single genes in ASD.
We selected 20 trios with idiopathic ASD, each consistent with sporadic ASD based on clinical evaluations (Supplementary Table 1), pedigree structure, familial phenotypic evaluation, family history, and/or elevated parental age. Each family was initially screened by array comparative genomic hybridization (CGH) using a customized microarray9. We identified no large (>250 kbp) de novo CNVs but did identify a maternally inherited deletion (~350 kbp) at 15q11.2 in one family (Supplementary Fig. 1). This deletion has been associated with increased risk for epilepsy10 and schizophrenia11,12 but has not been considered as causal for autism.
Similar to Vissers and colleagues13, who reported exome sequencing on 10 parent-child trios with sporadic cases of moderate to severe intellectual disability (ID), we performed exome sequencing on each of the 60 individuals separately, by subjecting whole-blood derived genomic DNA to in-solution hybrid capture and Illumina sequencing (Methods). We obtained sufficient coverage to call variants for ~90% of the primary target (26.4 Mb) (Table 1). Genotype concordance with SNP microarray data was high (99.7%) (Supplementary Table 2) and on average 96% of proband variant sites were also called in both parents (Supplementary Table 3). Given the expected rarity of true de novo events in the targeted exome (<1/trio) (Supplementary Table 4)14, we reasoned that most apparently de novo variants would result from undercalling in parents or systematic false positive calls in the proband. We therefore filtered variants previously observed in dbSNP, 1000 Genomes Pilot Project data15, and 1490 other exomes sequenced at the University of Washington (Supplementary Fig. 2). We performed Sanger sequencing on the remaining de novo candidates (<5/trio), validating 18 events within coding sequence and three additional events mapping to 3′ untranslated regions (Table 2). A list of predicted variant sites within these genes from the 1000 Genomes Pilot Project data15 is provided for comparison (Supplementary Table 5).
Table 1.
Summary of the exome sequencing results from of 20 sporadic ASD probands
| Family SSC/SAGE ID# | Proband Sex | Fa Age months | Mo Age months | Trio Bases‡ | % target +/− 2 bp | Coding SNV (− dbSNP/1kG) | Rare Disruptive SNV□ | Coding Indels (− dbSNP/1kG) | Rare Indels ≠‚3n | Protein Coding De Novo Events |
|---|---|---|---|---|---|---|---|---|---|---|
| 11048 | M | 358 | 358 | 23,901,726 | 88.19 | 14,095 (752) | 131 | 74 (44) | 27 | 0 |
| 11307 | M | 421 | 407 | 23,549,536 | 86.89 | 13,509 (583) | 75 | 64 (40) | 19 | 0 |
| 11580 | M | 443 | 305 | 23,823,712 | 87.90 | 13,912 (642) | 89 | 62 (36) | 24 | 1 |
| 11666 | M | 398 | 370 | 24,179,474 | 89.21 | 14,306 (622) | 77 | 59 (40) | 25 | 1 |
| 12325 | M | 363 | 313 | 24,088,772 | 88.88 | 13,866 (629) | 79 | 65 (43) | 24 | 1 |
| 12499 | M | 425 | 372 | 25,217,651 | 93.04 | 14,479 (634) | 86 | 80 (47) | 21 | 3 |
| 12575 | M | 351 | 317 | 24,259,870 | 89.51 | 14,568 (679) | 78 | 80 (55) | 26 | 0 |
| 12647 | M | 541 | 413 | 24,669,129 | 91.02 | 14,144 (830) | 78 | 68 (42) | 22 | 1 |
| 12680 | M | 502 | 471 | 24,437,989 | 90.16 | 14,124 (642) | 69 | 70 (42) | 24 | 2 |
| 12681 | F | 399 | 375 | 24,723,806 | 91.22 | 14,750 (691) | 93 | 68 (39) | 20 | 2 |
| 12817 | M | 485 | 430 | 24,520,475 | 90.47 | 14,364 (656) | 83 | 72 (38) | 24 | 2 |
| 12974 | M | 366 | 365 | 24,235,164 | 89.42 | 13,990 (555) | 52 | 54 (37) | 23 | 0 |
| 13095 | M | 337 | 322 | 24,460,239 | 90.25 | 14,605 (645) | 66 | 89 (54) | 29 | 0 |
| 13253 | M | 436 | 427 | 24,070,345 | 88.81 | 13,775 (610) | 96 | 41 (25) | 16 | 2 |
| 13284* | M | 300 | 302 | 24,911,060 | 91.91 | 17,806 (639) | 111 | 151 (79) | 53 | 1 |
| 13466 | M | 353 | 385 | 24,676,574 | 91.05 | 14,023 (591) | 72 | 58 (39) | 23 | 0 |
| 13683 | M | 470 | 402 | 24,139,439 | 89.06 | 14,419 (725) | 73 | 72 (49) | 22 | 0 |
| 13708 | M | 397 | 382 | 23,933,169 | 88.30 | 13,997 (686) | 77 | 78 (41) | 26 | 2 |
| 13970 | M | 313 | 234 | 24,465,009 | 90.26 | 14,293 (626) | 84 | 89 (58) | 31 | 0 |
| SAGE4022 | F | 271 | 283 | 24,130,743 | 89.03 | 14,538 (713) | 141 | 86 (56) | 29 | 0 |
| AVG | 18M:2F | 397 | 362 | 24,319,694 | 89.73 | 14,378 (658) | 86 | 74 (45) | 25 | 0.9 |
Paternal and maternal ages at time of conception were estimated based on month-year birth information assuming a 9-month pregnancy
Number of bases covered at 8x and Q30 in all three individuals
13284 Included Additional RefSeq Targets
Not observed in 1490 other exomes sequenced at the University of Washington
Simons Simplex Collection (SSC) or Study of Autism Genetics Exploration (SAGE) family number
Table 2.
Summary of confirmed de novo mutation events
| SNV Proband | Type | Chromosome: Position | Gene Symbol | Variant | AA Change | GERP Score | Grantham Score | PolyPhen-2 | CpG | Ts/TV | Mut Origin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 11580.p1 | missense | chr20:2239665 | TGM3 | R | V144I | 5.15 | 29 | probably damaging | Y | Ts | Mo |
| 11666.p1 | missense | chr9:132904111 | LAMC3* | R | D339G | 4.92 | 94 | probably damaging | N | Ts | Fa |
| 12325.p1 | 3′UTR | chr12:55708658 | MYO1A | R | 2.23 | N | Ts | ||||
| 12325.p1 | missense | chr16:19951169 | GPR139 | Y | S151G | 1.71 | 56 | benign | N | Ts | |
| 12499.p1 | missense | chr2:166556317 | SCN1A* | R | P1894L | 5.55 | 98 | probably damaging | N | Ts | Fa |
| 12499.p1 | synonymous | chr3:38033207 | PLCD1 | K | −8.24 | Y | Tv | ||||
| 12499.p1 | missense | chr6:152865504 | SYNE1 | Y | Y282C | 4.48 | 194 | probably damaging | N | Ts | |
| 12575.p1 | 3′UTR | chr9:32619906 | TAF1L | R | −1.02 | Y | Ts | ||||
| 12647.p1 | 3′UTR | chr16:23585994 | DCTN5 | Y | −0.989 | N | Ts | ||||
| 12647.p1 | missense | chr5:68453390 | SLC30A5 | S | S561R | 4.6 | 110 | possibly damaging | N | Tv | |
| 12680.p1 | synonymous | chr2:101992478 | IL1R2 | Y | −1.53 | N | Ts | ||||
| 12680.p1 | synonymous | chr5:132251451 | AFF4 | Y | −11.2 | Y | Ts | Fa | |||
| 12681.p1 | 3′ splice | chr12:13614220 | GRIN2B* | Y | 4.17 | 215# | N | Ts | Fa | ||
| 12681.p1 | synonymous | chr7:142274902 | EPHB6 | Y | −3.14 | Y | Ts | Fa | |||
| 12817.p1 | synonymous | chr2:143724639 | ARHGAP15 | R | 3.51 | N | Ts | ||||
| 13253.p1 | missense | chr3:39204494 | XIRP1 | Y | V483M | 2.04 | 21 | probably damaging | N | Ts | |
| 13253.p1 | synonymous | chr16:74121475 | CHST5 | Y | −3.22 | Y | Ts | ||||
| 13284.p1 | synonymous | chr2:179145956 | TTN | Y | 0.328 | Y | Ts | ||||
| 13708.p1 | missense | chr17:58033198 | TLK2 | Y | S595L | 5.43 | 145 | probably damaging | Y | Ts | |
| 13708.p1 | missense | chr3:30004687 | RBMS3 | Y | T383M | 5.44 | 81 | probably damaging | Y | Ts | |
| Indel | |||||||||||
| 12817.p1 | frameshift | chr3:71132860 | FOXP1* | +T | A339SfsX4 | 5.38‡ | 215# | NA | NA | Fa |
Disruptive de novo mutations that are potentially causative
Maximum Grantham score given for splice and frameshifting variants
Average GERP score for two sites flanking the insertion
We observed subtle differences with respect to mutation rate and characteristics when compared to Vissers and colleagues13 (Supplementary Note). The overall protein-coding de novo rate (0.9 events/trio) was slightly higher than expected14 (0.59 events/trio), suggesting that we are identifying the majority of de novo events in these trios (Supplementary Table 4). The transition to transversion ratio was highly skewed (18:2), with eight transitions mapping to hypermutable CpG dinucleotides14. The proportion of synonymous events was higher than expected based on a neutral model and may reflect selection against embryonic lethal nonsynonymous variants. We successfully determined the parent of origin for seven events, six of which occurred on the paternal haplotype (Table 2). Notably, the eight probands with two or more validated de novo events corresponded to families with higher parental age (Mann–Whitney U, Combined Age, One-Sided P<0.004).
Eleven of the 18 coding de novo events are predicted to alter protein function. Each of these mutations occurred at a different gene, precluding a statistical assessment for any specific locus despite their deleterious nature (e.g. PolyPhen-216). We assessed whether proband de novo mutations were enriched in the aggregate for disruptive events by considering two independent quantitative measures: the nature of the amino-acid replacement (Grantham matrix score17) and the degree of nucleotide-level evolutionary conservation (Genomic Evolutionary Rate Profiling (GERP)18,19) (Fig. 1a,b). For comparison, we sequenced 20 exomes from unrelated ethnically matched controls (HapMap) and applied the same filters to identify coding-sequence mutations that were common or private to each of the samples. These control DNA were isolated from immortalized lymphoblasts; however, the counts of private variants in the cases and controls were highly similar suggesting that suggesting that the contribution of novel somatic events is likely minimal (Supplementary Fig. 3).
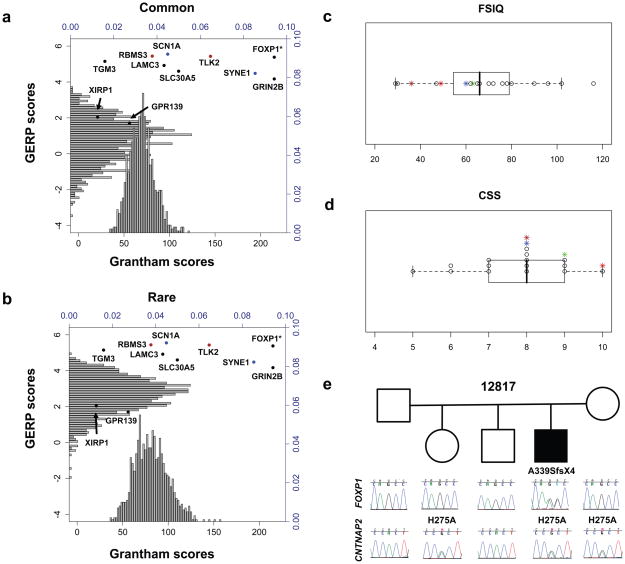
Figure 1.
Evaluation of de novo mutations by simulation, proband severity, and family 12817. a,b We compared the mean Grantham (black x-axis) and GERP scores (black y-axis) of the 10 proband de novo protein-changing substitutions to 20 HapMap control samples by building a distribution of the mean values of 10 randomly selected common or private variants over 1000 trials. Splice-site and nonsense events were given a maximum Grantham score (215) and indels were not included in the simulation. Histograms show the relative frequency (blue axes) of each distribution. Points show the proband variants, with variants from the same individual highlighted (blue=13708.p1, red=12499.p1). Proband mean values, GERP: 4.349 and Grantham: 104.3. *FOXP1 not included in proband mean values. a, Control common variants (GERP: p<0.001, Grantham: p=0.015). b, Control rare variants (GERP: p=0.026, Grantham: p=0.098). c,d We evaluated the disease severity of the mutation carriers 12817.p1-FOXP1 (brown), 12681.p1-GRIN2B (green), 12499-SCN1A (blue) and 11666.p1-LAMC3 (red). c, Box and whisker plot of Full Scale Intelligence Quotient (FSIQ) values. d, Box and whisker plot of Calibrated Severity Scores (CSS) based on the Autism Diagnostic Observation Schedule (ADOS). Data were available for 19/20 probands; CSS were estimated for two probands based on ADOS module 4 data. e, Pedigree for 12817 showing chromatogram traces surrounding FOXP1 (top) and CNTNAP2 (bottom) mutation events. Proband carries a de novo single-base (+A relative to mRNA) frameshifting mutation p.A339SfsX4 in FOXP1 and an inherited missense variant p.H275A in CNTNAP2.
We determined by simulation the expected mean GERP and Grantham distributions for 10 randomly selected common or private control single nucleotide variants (SNVs) (Methods). When we compared the observed means of the 10 de novo protein-altering ASD proband variants to the distribution of common control SNVs (Fig. 1a), they corresponded to more highly conserved (GERP: p<0.001) and disruptive amino acid mutations (Grantham: p=0.015). If we limited the analysis to the private control SNVs, which serve as a proxy for evolutionarily young mutation events (Fig. 1b), we again found the de novo events were at the right tail of these distributions. Only the mean GERP score, however, remained significant (GERP: p=0.02, Grantham: p=0.115). In total, these results suggest that these de novo mutation sites are subjected to stronger selection and likely to have functional impact.
We identified a subset of trios (4/20) with disruptive de novo mutations that are potentially causative, including genes previously associated with autism, ID, and epilepsy (Table 2 and Supplementary Note). We examined the available clinical data for each of these four families and found they were among the most severely affected individuals in our study based on intelligence quotient (IQ) measures and on calibrated severity score20 (CSS), which is largely independent from IQ and focuses specifically on autistic features with a score of 10 being most severe (Fig. 1c,d). For example, in proband 12681 we identified a single-base substitution (IVS9-2A>G, CCDS8662.1) at the canonical 3′ splice site of exon 10 in Glutamate receptor, ionotropic, N-methyl D-aspartate 2B (GRIN2B) (Supplementary Fig. 4a,b). She is severely affected (CSS 9), with evidence of early onset, possible regression, and comorbid for mild ID. Expression and association studies have suggested that glutamatergic neurotransmission may play a role in ASD4. Recently, Endele and colleagues21 described GRIN2A and GRIN2B as sites of recurrent de novo mutations in individuals with mild to moderate ID and/or epilepsy suggesting variable expressivity. Our data suggest that de novo mutations in GRIN2B may also lead to an ASD presentation.
Proband 12499 has a missense variant (p.P1894L, CCDS33316.1) predicted to be functionally deleterious and at a highly conserved position in Sodium channel, voltage-gated, type I, alpha subunit (SCN1A) (Supplementary Fig. 4c). He is severely affected (CSS 8) with evidence of early onset, possible regression, language delay, a diagnosis of epilepsy and mild ID. SCN1A was previously associated with epilepsy and suggested as an ASD candidate22,23, although limited screening has been conducted in idiopathic ASD. Hundreds of disease-associated mutations have been described in epilepsy and typically patients with de novo events show more severe phenotypes24. The proband also carries the maternally inherited 15q11.2 deletion increasing the risk for epilepsy10.
Proband 11666 has a missense variant (p.D399G, CCDS6938.1) predicted to be functionally deleterious and at a highly conserved position within the second laminin-type epidermal growth factor-like domain of Laminin, gamma 3 (LAMC3) (Supplementary Fig. 4d). He is severely affected (CSS 10) with evidence of early onset and moderate ID. LAMC3 is not known to be involved in neuronal development; however, human microarray data have shown expression in many areas of the cortex and limbic system25. Additional study is warranted since laminins have structural similarities to the neurexin and contactin-associated families of proteins, both of which have been associated with ASD2.
The fourth example of a potentially causative mutation is a single-base insertion in Forkhead box P1 (FOXP1), introducing a frameshift and premature stop codon (p.A339SfsX4, CCDS2914.1) in proband 12817 (Fig. 1e). He is severely affected (CSS 8) with evidence for regression, language delay, and comorbidity for moderate ID and nonfebrile seizures. Recently, rare occurrences of large de novo deletions and a nonsense variant disrupting FOXP1 were reported in individuals with mild to moderate ID and language defects, with or without ASD features26,27. FOXP1 encodes a member of the forkhead-box family of transcription factors and is closely related to FOXP2, a gene implicated in rare monogenic forms of speech and language disorder28–31. Functional evidence of heterodimer formation and overlapping neural expression patterns suggests that FOXP1 and FOXP2 can co-regulate gene expression in the brain32,33. We assessed relative levels of the mutant transcript in proband derived lymphoblasts finding strong evidence for nonsense-mediated decay (NMD) (Supplementary Fig. 5a). HEK293T cell-based functional assays further demonstrated that, if translated, the protein would be truncated and mislocalized from the nucleus to the cytoplasm—similar to results obtained with FOXP2 mutations31 (Supplementary Fig. 5b,c).
Remarkably, in addition to the FOXP1 mutation, proband 12817 also carried an inherited missense variant (p.H275A, CCDS5889.1) in Contactin associated protein-like 2 (CNTNAP2) predicted to be functionally deleterious and at a highly conserved position. This variant is likely to be extremely rare or private as it was not observed in 942 previously sequenced controls34 or in 1490 other exomes. CNTNAP2 is directly downregulated by FOXP235 and has been independently associated with ASD and specific language impairment34–37. In HEK293T cells, we found that wild-type FOXP1 significantly reduced expression of CNTNAP2 (p=0.0005), while the truncated protein was associated with a three-fold expression increase (p=0.0056) (Supplementary Note, Fig. 5d). Overall, we hypothesize that FOXP1 haploinsufficiency (due to NMD), combined with dysfunction of FOXP1 mutant proteins that escape this process, may yield overexpression of CNTNAP2 proteins, amplifying any deleterious effects of p.H275A in the proband.
Among the ~110 (85 SNVs, 25 indels) novel inherited protein-altering variants in each proband, we identified several rare inherited variants in genes overlapping the SFARI Gene38, a curated database of potential ASD candidate loci, but no excessive burden in cases relative to controls (Supplementary Table 6). While the numbers from our pilot study are few, we do observe two cases with a significant de novo event and a potential inherited risk variant (12817p1:FOXP1/CNTNAP2 and 12499.p1: SCN1A/15q11.2 deletion) highlighting that in some sporadic families a multihit model may be playing a role3 (Supplementary Table 7). In the future, this hypothesis could be further explored by comparing burden in a much larger number of affected/unaffected sibling pairs.
The probands with the four potentially causative de novo events met strict criteria for a diagnosis of autistic disorder (Supplementary Note). Our finding of de novo events in genes that have also been disrupted in children with ID without ASD, ID with ASD features, and epilepsy provides further evidence that these genetic pathways may lead to a spectrum of neurodevelopmental outcomes depending on the genetic and environmental context2,4. Recent data suggest that CNVs may also blur these lines with diverse conditions all showing association to the same loci2,4. Distinguishing primary from secondary effects will require a better understanding of the underlying biology and identification of interacting genetic and environmental factors within the phenotypic context of the family. The identification of de novo events along with disruptive inherited mutations underlying “sporadic” ASD has the potential to fundamentally transform our understanding of the genetic basis of ASD.
Supplementary Material
Acknowledgments
We would like to thank and recognize the following ongoing studies that produced and provided exome variant calls for comparison: NHLBI Lung Cohort Sequencing Project (HL 1029230), NHLBI WHI Sequencing Project (HL 102924), NIEHS SNPs (HHSN273200800010C), NHLBI/NHGRI SeattleSeq (HL 094976), and the Northwest Genomics Center (HL 102926). We also thank M-C. King and S. Stray for processing and managing DNA samples, B.H. King and E. Bliss for their work in patient recruitment and phenotype collection, E. Turner, C. Igartua, I. Stanaway, M. Dennis, and B. Coe for thoughtful discussions, M. State for providing SNP genotyping data, and especially the families that volunteered their time to participate in this research. This work was supported by NIH grant HD065285 (E.E.E. and J.S.), Wellcome Trust core award 075491/Z/04 (S.E.F. and P.D.), the Max Planck Society (S.E.F.), and the Simons Foundation Autism Research Initiative (E.E.E., R.B., S.E.F., and P.D.). E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions E.E.E., J.S., and B.J.O. designed the study and drafted the manuscript. E.E.E. and J.S. supervised the study. R.B. analyzed the clinical information and contributed to the manuscript. S.E.F and P.D. designed cell-based functional experiments, analyzed data, interpreted results, and contributed to the manuscript. S.G., C.B., and L.V. generated and analyzed array CGH data. C.L. performed Illumina GAIIx sequencing. B.J.O. and E.K. developed analysis pipeline and analyzed sequence data. A.P.M. and S.B.N. designed and optimized capture protocol. B.J.O., L.V., A.P.M., and S.B.N. constructed exome libraries. B.J.O., L.V., A.P.M., and J.J.S. performed mutation validation and haplotype characterization. B.J.O. and J.J.S. performed the evaluation of 12817 lymphoblast cell lines. P.D. performed functional experiments. M.J.R and D.A.N. performed sequencing of control samples.
Author Information E.E.E is on the scientific advisory board for Pacific Biosciences. J.S. is a member of the scientific advisory boards of Tandem Technologies, Stratos Genomics, Good Start Genetics, Halo Genomics, and Adaptive TCR. B.J.O. is an inventor on patent PCT/US2009/30620: Mutations in Contactin Associated Protein 2 are Associated with Increased Risk for Idiopathic Autism.
References
- 1.Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 2.O’Roak BJ, State MW. Autism genetics: strategies, challenges, and opportunities. Autism Research. 2008;1:4–17. doi: 10.1002/aur.3. [DOI] [PubMed] [Google Scholar]
- 3.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–55. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durkin MS, et al. Advanced parental age and the risk of autism spectrum disorder. Am J Epidemiol. 2008;168:1268–76. doi: 10.1093/aje/kwn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–7. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 10.de Kovel CG, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2009;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirov G, et al. Support for the involvement of large cnvs in the pathogenesis of schizophrenia. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010 doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–8. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–4. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 18.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper GM, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat Methods. 2010;7:250–1. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 2009;39:693–705. doi: 10.1007/s10803-008-0674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endele S, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010 doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 22.Claes L, et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–32. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss LA, et al. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol Psychiatry. 2003;8:186–94. doi: 10.1038/sj.mp.4001241. [DOI] [PubMed] [Google Scholar]
- 24.Mulley JC, et al. SCN1A mutations and epilepsy. Hum Mutat. 2005;25:535–42. doi: 10.1002/humu.20178. [DOI] [PubMed] [Google Scholar]
- 25.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 26.Hamdan FF, et al. De Novo Mutations in FOXP1 in Cases with Intellectual Disability, Autism, and Language Impairment. Am J Hum Genet. 2010;87:671–8. doi: 10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn D, et al. Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum Mutat. 2010;31:E1851–60. doi: 10.1002/humu.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 29.Feuk L, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–72. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDermot KD, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vernes SC, et al. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–67. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–22. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–63. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernes SC, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–45. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arking DE, et al. A Common Genetic Variant in the Neurexin Superfamily Member CNTNAP2 Increases Familial Risk of Autism. Am J Hum Genet. 2008;82:160–4. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alarcon M, et al. Linkage, Association, and Gene-Expression Analyses Identify CNTNAP2 as an Autism-Susceptibility Gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee-Basu S, Packer A. SFARI Gene: an evolving database for the autism research community. Dis Model Mech. 2010;3:133–5. doi: 10.1242/dmm.005439. [DOI] [PubMed] [Google Scholar]
- 39.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Hurley RS, Losh M, Parlier M, Reznick JS, Piven J. The broad autism phenotype questionnaire. J Autism Dev Disord. 2007;37:1679–90. doi: 10.1007/s10803-006-0299-3. [DOI] [PubMed] [Google Scholar]
- 41.Constantino JN, Todd RD. Intergenerational transmission of subthreshold autistic traits in the general population. Biol Psychiatry. 2005;57:655–60. doi: 10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–19. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 43.Itsara A, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–61. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Igartua C, et al. Targeted enrichment of specific regions in the human genome by array hybridization. Curr Protoc Hum Genet. 2010;Chapter 18(Unit 18):3. doi: 10.1002/0471142905.hg1803s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. The Sequence Alignment/Map format and SAM tools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–6. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat Methods. 2010;7:576–7. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andres AM, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.