Abstract
Although nonsteroidal antiinflammatory drugs (NSAIDs) show great promise as therapies for colon cancer, a dispute remains regarding their mechanism of action. NSAIDs are known to inhibit cyclooxygenase (COX) enzymes, which convert arachidonic acid (AA) to prostaglandins (PGs). Therefore, NSAIDs may suppress tumorigenesis by inhibiting PG synthesis. However, various experimental studies have suggested the possibility of PG-independent mechanisms. Notably, disruption of the mouse group IIA secretory phospholipase A2 locus (Pla2g2a), a potential source of AA for COX-2, increases tumor number despite the fact that the mutation has been predicted to decrease PG production. Some authors have attempted to reconcile the results by suggesting that the level of the precursor (AA), not the products (PGs), is the critical factor. To clarify the role of AA in tumorigenesis, we have examined the effect of deleting the group IV cytosolic phospholipase A2 (cPLA2) locus (Pla2g4). We report that ApcMin/+, cPLA2−/− mice show an 83% reduction in tumor number in the small intestine compared with littermates with genotypes ApcMin/+, cPLA2+/− and ApcMin/+, cPLA2+/+. This tumor phenotype parallels that of COX-2 knockout mice, suggesting that cPLA2 is the predominant source of AA for COX-2 in the intestine. The protective effect of cPLA2 deletion is thus most likely attributed to a decrease in the AA supply to COX-2 and a resultant decrease in PG synthesis. The tumorigenic effect of sPLA2 mutations is likely to be through a completely different pathway.
The therapeutic benefits of nonsteroidal antiinflammatory drugs (NSAIDs) in colon cancer treatment are impressive. However, the mechanisms by which NSAIDs act to reduce tumorigenesis remain unclear. NSAIDs are known to bind and inhibit the cyclooxygenase (COX) enzymes, COX-1 and COX-2, which produce prostaglandins (PGs). Therefore, it has been inferred that NSAIDs protect against tumorigenesis by down-regulating PG synthesis. A role for PGs in promoting tumorigenesis is also supported by the observations that, relative to normal tissue, human and mouse tumors contain high levels of both PGs and COX-2 (1–3). Further evidence of the importance of the COX-2 pathway, in particular, is provided by recent selective COX-2 inhibitors that retain or exceed the potent antitumor activity of NSAIDs (4). Nevertheless, some authors claim that NSAIDs act in a manner independent of PG synthesis (5–7).
Some important insights into the pathogenesis of colon cancer have been gained from the study of the ApcMin strain, one of the most widely used mouse models for colon cancer. Mice carrying the ApcMin mutation on a C57BL/6J background spontaneously develop ≈100 early stage tumors (adenomas or polyps) caused by a dominant, germ-line mutation in the adenomatous polyposis coli (Apc) gene, the mouse homologue of the human APC gene (8). APC is a key tumor-suppressor gene mutated in familial colon cancer syndromes as well as the vast majority of sporadic cases, making ApcMin a clinically relevant model and an excellent system to unravel the underlying genetic interactions. One gene that has emerged from the study of ApcMin is Modifier of Min-1 (Mom1) locus. Moser et al. (9) observed in 1992 that strain background can modulate ApcMin tumor phenotype. The major modifying locus was genetically mapped to distal mouse chromosome 4 and dubbed Mom1 (10). Strains carrying a sensitive Mom1 allele (such as C57BL/6J) developed high tumor numbers, whereas those carrying a resistant allele (such as AKR, CAST, and BALB/c) developed low tumor numbers.
The group IIA secretory phospholipase A2 gene (pla2g2a) was proposed as a candidate gene for Mom1 because it mapped to the initial Mom1 interval and carried a strain-specific loss-of-function mutation (11). Specifically, Mom1-sensitive strains carried a frame-shift mutation in the pla2g2a gene that abolished pla2g2a (also known as group IIA sPLA2) expression. Mom1-resistant strains, in contrast, did not carry the pla2g2a mutation and expressed high levels of sPLA2 in the intestinal tract. sPLA2 was considered an attractive candidate for Mom1 because it was suggested that it functioned directly upstream from COX-2. Specifically, sPLA2 is a member of the phospholipase A2 family, a group of enzymes that catalyze the hydrolysis of membrane glycerophospholipids to generate free fatty acids. Certain of the phospholipase A2 family, including sPLA2, are thought to be capable of generating arachidonic acid (AA), the substrate used by COX-2 to synthesize PGs. Therefore, it was suggested that the group IIA sPLA2 functioned in a pathway widely considered to be important to tumorigenesis, and thus was a good candidate to be Mom1. In fact, we tested this suggestion by constructing recombinant and transgenic strains, and demonstrated that the mutational status of the sPLA2 locus does indeed account for a significant portion of the Mom1 effect (12, 13).
Although sPLA2 was suggested (and subsequently confirmed) as a candidate for Mom1 because of its supposed connection with COX-2, this connection is not well supported and the hypothesis is quite problematic. Specifically, the COX-2 and sPLA2 loss-of-function phenotypes are fundamentally opposed in nature. Targeted deletion of COX-2 on an Apc-mutant background has been shown to be strongly protective, reducing tumor number by 86% in COX-2-knockout homozygotes (14). By contrast, loss of function of sPLA2 (such as with the sPLA2 mutation in the C57BL/6 strain) increases ApcMin-induced tumor number (15); restoring sPLA2 expression through transgenic constructs decreases tumor number (12). In short, COX-2 activity enhances tumorigenesis on an Apc-mutant background, whereas sPLA2 activity suppresses it. At the least, the results are incompatible with mutations in sPLA2 and COX-2 acting by decreasing PG levels.
Chan et al. (7) attempted to resolve this paradox by proposing that the key to tumorigenesis is, instead, the level of AA. Loss-of-function mutation of sPLA2 would be predicted to decrease AA levels, whereas loss-of-function mutation of COX-2 would be predicted to increase AA. They noted that addition of either the NSAID sulindac or exogenous AA induces apoptosis in certain human colon cancer cell lines. Furthermore, they proposed a model in which AA induces apoptosis by stimulating the conversion of sphingomyelin to ceramide. Deletion of sPLA2 would promote tumorigenesis because of a decrease in AA and a concomitant failure to correctly initiate cell death. Inhibition of COX-2 would be protective because of a net increase in AA and enhanced apoptosis.
Alternatively, the paradox could be explained if the role of sPLA2 in tumorigenesis is unrelated to supplying AA to COX-2 for PG synthesis and, instead, involves a different, unrelated pathway.
To distinguish between these two possibilities, we have examined the effect of deleting a different phospholipase A2, the group IV cytosolic phospholipase A2 (cPLA2, encoded by Pla2g4). Unlike the group IIA sPLA2 (whose physiological contribution to AA production is unclear), group IV cPLA2 is well characterized as a major AA-producing enzyme; deletion of cPLA2 abolishes PG synthesis in a number of cPLA2−/− cell types. Deletion of cPLA2 tests the AA vs. PG hypothesis of NSAID action. If NSAIDs work by increasing AA levels, one would predict that loss-of-function mutations in cPLA2 would have the same effect as those in sPLA2—namely, to increase tumor number. If, on the other hand, NSAIDs work by decreasing PG synthesis, one would expect a decrease in tumor number in the cPLA2−/− background, similar to the effect of the deletion of COX-2. In fact, we find that deletion of cPLA2 suppresses ApcMin-induced polyp number, supporting the notion that PGs promote tumorigenesis, and arguing against a protective role for AA.
Materials and Methods
Construction and Genotyping of ApcMin and cPLA2 Mutant Mice.
C57BL/6J ApcMin/+ mice were obtained from The Jackson Laboratory. Construction of the cPLA2 knockout line has been previously described (16). The cPLA2 knockout line was backcrossed six generations onto C57BL/6J before matings. As both the ApcMin/+ and cPLA2−/− mutant phenotypes greatly compromise breeding efficacy in female mice, these alleles were propagated through the male germ line. Apc+/+ cPLA2+/− females were mated to ApcMin/+, cPLA2+/+ males to obtain ApcMin/+, cPLA2+/− males, which were then crossed to Apc+/+, cPLA2+/− females to produce both ApcMin/+, cPLA2+/− and ApcMin/+, cPLA2−/− males. These males were then mated to Apc+/+, cPLA2+/− females, and their ApcMin/+-bearing progeny were scored for tumor number. Mice were maintained on a breeder diet containing 10% fat (Harlan Teklad). All offspring were genotyped by PCR of tail DNA. Tail DNA was isolated as previously described, with the exception that a phenol/chloroform extraction using 2 ml of Heavy Phase Lock gels (Eppendorf) was included before isopropyl alcohol precipitation (17). ApcMin genotyping has been described (10). The cPLA2 PCR assay involved the use of three primers—two forward primers: cPLA3F (5′-TGTGTACAATCTTTGTGTTGTTTCA-3′) and pgkNeo (5′-GGGAACTTCCTGACTAGGGG-3′), and one reverse primer: cPLA604 (5′-CGACTCATACAGTGCCTTCATCAC-3′). cPLA3F and cPLA604 amplify an ≈100-bp fragment from the endogenous, wild-type cPLA2 locus. pgkNeo and cPLA604 amplify an ≈300-bp product from the targeted knockout allele. All three primers were added simultaneously at a concentration of 500 nM each in a 20-μl PCR mixture containing 1.5 mM MgCl2, 200 μM each dNTP, 50 ng of genomic tail DNA, and 0.75 units of AmpliTaq Gold (Perkin–Elmer) in standard PCR buffer. PCR was performed under the following conditions: 94°C for 9 min, followed by 25 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. PCR products were resolved on a 4% Metaphor agarose gel (Bio Whittaker) run in the cold room at 20 V/cm.
Tumor Phenotyping.
Mice were killed at 90 ± 3 days of age by CO2 asphyxiation. The entire intestinal tract was removed, placed in PBS, and flushed with 70% ethanol from a 20-ml syringe fitted with a 23 gauge needle. Intestines were then cut open longitudinally and vigorously washed to dislodge any remaining debris. Intestines were then rocked overnight in 4% formaldehyde in PBS for further fixation. Polyps were scored by visual inspection of fixed tissue with an Olympus SZX12 dissecting microscope at 16–20× magnification. Polyp size was estimated by measuring the maximum polyp diameter with a calibrated eyepiece reticule. All polyps were counted by a single observer who was blind to the genotype of the samples.
Statistical Analysis.
One-sided P values for intestinal polyp numbers and sizes were determined by using the Mann–Whitney U test performed with statview software (SAS Institute, Cary, NC). All reported values are mean ± SEM.
Western Blot Analysis.
For COX-1 and COX-2, membrane fractions were prepared as previously described (2). Total protein concentrations were determined with the Bio-Rad DC Protein Assay. Membrane fractions were mixed with 2× Laemmli sample buffer (Bio-Rad), boiled for 5 min, and analyzed by SDS/PAGE according to the method of Laemmli. Proteins were then electrophoretically transferred to nitrocellulose according to standard methods. All hybridizations were performed at room temperature in Tris-buffered saline/Tween 20 with 5% milk. Primary antibodies against COX-1 and COX-2 (Santa Cruz Biotechnology) were both diluted to 1:500 and hybridized for 6 h at room temperature. The actin antibody (Santa Cruz Biotechnology) was used at a final dilution of 1:2,000. The horseradish peroxidase-conjugated secondary anti-goat IgG antibody (Santa Cruz Biotechnology) was used at a dilution of 1:1,000. Immunodetection was performed by using enhanced chemiluminescence according to the manufacturer's instructions (Amersham Pharmacia).
For cPLA2, tissues were lysed in buffer G (20 mM β-glycerophosphate/20 mM NaF/2 mM EDTA/0.2 mM Na3VO4/10 mM benzamidine) with protease inhibitors, incubated on ice for 30 min, and vortexed vigorously. Each sample was then sonicated with 10 pulses and homogenized with a hand-held homogenizer. Samples were then spun 10 min at 16,000 × g, and the total protein contents of supernatants were quantitated with the Bio-Rad assay. Western blot analyses were performed as described above with the following exceptions. Primary anti-cPLA2 antibody (rabbit polyclonal, a gift from A. Cybulsky) and horseradish peroxidase-conjugated secondary anti-goat IgG antibody (Dako) were used at a dilution of 1:5,000.
Construction of Phylogenetic Trees.
Protein sequences were obtained from GenBank. Phylogenetic trees (branch-and-bound, maximum parsimony, strict consensus) were calculated in the GCG suite of programs by using default parameters in paupsearch and paupdisplay (wisconsin package version 10.1). GenBank accession numbers for sPLA2 family members are IID Mus 5359708, IID Human 6912596, IIA Human 129483, IIA Mus 984837, IIE Mus 6164697, IIE Human 7108922, V Human 4505853, V Mus 6755096, X Human 4505845, X Mus 6525307, IIC Mus 6679367, IIF Mus 6755094, IB Mus 6755090, and IB Human 4505847. GenBank accession numbers for cPLA2 family members are cPLA2α Mus 110807, cPLA2α Human 107294, cPLA2γ Human 3452314, and cPLA2β Human 4886977.
Results
Homozygous Deletion of cPLA2 Reduces Polyp Number and Size in the Small Intestine.
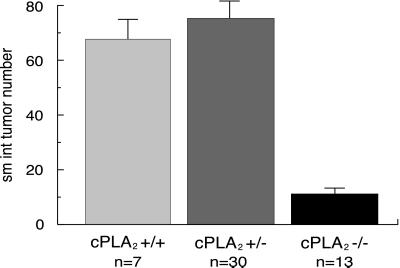
Mice carrying the ApcMin mutation on a C57BL/6 background typically develop on the order of ≈100 small adenomas or polyps, the vast majority of which occur in the small intestine. The number of polyps in the small intestine of ApcMin/+, cPLA2+/− double heterozygotes was statistically indistinguishable from that of ApcMin/+, cPLA2+/+ animals (75.3 ± 6.4 vs. 67.7 ± 7.0, respectively, P = 0.70; Fig. 1). In contrast, ApcMin/+, cPLA2−/− homozygotes showed an impressive 83% reduction in small intestine polyp number (11.2 ± 1.9 vs. 67.7 ± 7.0, P < 0.0001; Fig. 1). This reduction occurred in all sections of the small intestine. In the large intestine polyp number, a slight trend in the opposite direction was observed; however, this difference did not reach statistical significance (P = 0.10; Fig. 2).
Figure 1.
Effect of deletion of cPLA2 on ApcMin-induced polyp number in the small intestine. Mice were analyzed at 90 days of age and tumors were counted from the entire small intestine. Small intestine polyp number in ApcMin/+, cPLA2+/− double heterozygotes was statistically indistinguishable from that of ApcMin/+, cPLA2+/+ animals (P = 0.70). In contract, ApcMin/+, cPLA2−/− homozygotes showed an 83% reduction in small intestine polyp number (P < 0.0001). n = number of mice; values plotted are mean ± SEM.
Figure 2.
Effect of deletion of cPLA2 on ApcMin-induced polyp number in the large intestine. Overall, a trend in the opposite direction from that seen in the small intestine was observed. However, the differences observed did not reach statistical significance (P = 0.10). n = number of mice; values plotted are mean ± SEM.
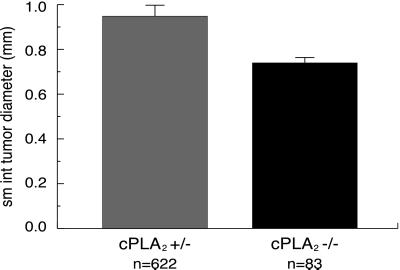
Furthermore, polyps in the small intestine of ApcMin/+, cPLA2−/− animals were approximately 22% smaller than those of ApcMin/+, cPLA2+/− littermates (average polyp diameter of 0.74 ± 0.09 mm versus 0.95 ± 0.02 mm, respectively; P = 0.0006; Fig. 3). The morphology of these smaller polyps was otherwise quite similar to that described in the comments about ApcMin mice. Histological sections of ApcMin/+, cPLA2−/− tissue were also examined for the presence of microadenomas which may have eluded visual inspection; none were observed (K.H.H., data not shown). No difference in size was observed in the polyps of the large intestine among any cPLA2 genotypes.
Figure 3.
Effect of deletion of cPLA2 on ApcMin-induced polyp size in the small intestine. Tumor diameter was estimated by measuring the maximum diameter of each tumor with a calibrated eyepiece reticle. The polyps from the small intestines of ApcMin/+, cPLA2−/− animals were approximately 22% smaller than those of ApcMin/+, cPLA2+/− littermates (P = 0.0006). n = number of mice; values plotted are mean ± SEM.
cPLA2 Levels Elevated in ApcMin cPLA2+/+ Polyps.
We observed an increase in cPLA2 immunoreactivity in polyps relative to normal adjacent tissue from the small intestines of ApcMin/+, cPLA2+/+ mice (Fig. 4). However, this up-regulation was not seen in polyps from ApcMin/+, cPLA2+/− double heterozygotes, which developed as many tumors as ApcMin/+, cPLA2+/+ mice. In the large intestine, where deletion of cPLA2 was not protective, no elevation was observed in the tumors relative to normal tissue (Fig. 4). Therefore, whereas up-regulation of cPLA2 is seen in polyps of wild-type ApcMin/+ mice, this event does not seem to be necessary for tumorigenesis. A minimal threshold of cPLA2-protein levels may suffice.
Figure 4.
Expression of the cPLA2 protein and normal tissues of the small and large intestines by Western blot analyses. All tissues were collected from ApcMin-bearing mice; cPLA2 genotypes are indicated above each section. T, tumor; N, normal. Blots were first probed with a cPLA2 antibody and then stripped and probed with an actin antibody to provide a loading control. No cPLA2 protein was detected in ApcMin/+, cPLA2−/− tissue from either the small or large intestine (data not shown).
COX-2 Remains Up-Regulated in ApcMin/+, cPLA2−/− Mice.
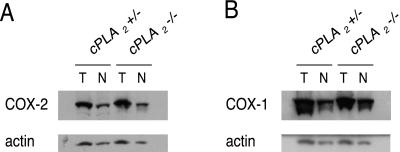
COX-2 is an inducible enzyme which is normally expressed at low levels in the mucosa of the small intestine. However, the protein levels have been observed to be highly elevated in both human and mouse tumors (1–3). Failure of COX-2 induction during tumorigenesis, for example in the case of COX-2 knockout mice, dramatically decreases tumor number. Importantly, COX-2 induction has been observed to be considerably lower in cells derived from cPLA2−/− mice (18). Therefore, failure to induce COX-2 in the ApcMin/+, cPLA2−/− background could account for the protective phenotype we observed. To check this possibility, we examined COX-2 and COX-1 levels in our mice by Western blot analysis. COX-2 levels remained elevated in polyps relative to normal mucosa in ApcMin/+, cPLA2−/− animals (Fig 5A). COX-1 was highly expressed in both polyp and normal tissue, consistent with previous reports (Fig 5B). Thus, we did not observe alterations in either COX-2 or COX-1 expression between cPLA2+/− and cPLA2−/− mice carrying ApcMin.
Figure 5.
Expression of COX-2 (A) and COX-1 (B) in tumor and normal tissue of the small intestines of ApcMin/+, cPLA2+/− mice and ApcMin/+, cPLA2−/− mice. T, tumor; N, normal. Western blots were first probed with either COX-2 or COX-1 antibodies. The blots were then stripped and rehybridized with an actin antibody to provide a loading control.
Discussion
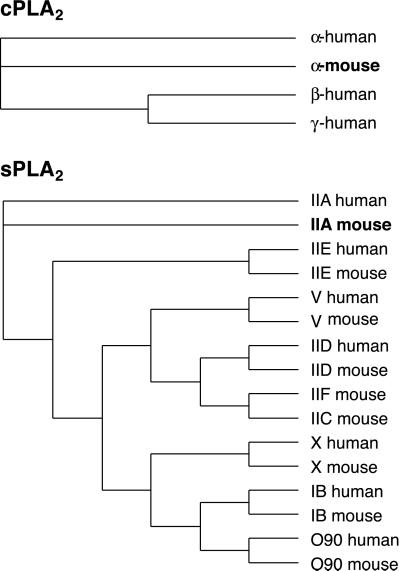
Group IV cPLA2 and group IIA sPLA2 are prominent members of a burgeoning superfamily of mammalian phospholipases A2. Previously unknown family members related to each archetype have been identified by recent sequencing efforts (Fig. 6). Moreover, in the case of the sPLA2 family, the precise biochemical roles of all of the isoforms are not clearly understood and may overlap. The presence of a panoply of related low-molecular-weight secretory phospholipase may account for the lack of obvious phenotypes in animals carrying mutations in the group IIA sPLA2.
Figure 6.
Phylogenetic trees of the cPLA2 and sPLA2 families of enzymes. Enzymes discussed in this article (group IV cPLA2 and group IIA sPLA2) are shown in bold. (Upper) Multiple sequence alignment (clustalx) and phylogenetic tree analysis (GCG/paup) indicate that the α, β, and γ members of the cPLA2 protein family are approximately equally distinct from one another. The input alignment was trimmed to a common core before the tree calculation to exclude the N-terminal 484 residues of cPLA2-β and the N-terminal 143 residues of cPLA2-α. Excluding these two cPLA2 sequences (which are 93% identical), the sequences are all approximately 28% identical in this core region. (Lower) sPLA2 family members cluster into four evolutionary subgroups, as calculated by using paupdisplay in GCG. The sPLA2 multiple sequence alignment was trimmed to a core, to exclude N- and C-terminal overhangs on some sequences. Other heuristic, neighbor-joining and identity-based trees calculated in GCG/paup, clustalx, and belvu support the inclusion of IIA and IIE into one subgroup (data not shown), probably owing to their 51% sequence identity in the core of the overlay.
The group IIA sPLA2 was examined as a candidate for Mom1 due in large part to the fact that it had been proposed as a prominent source of AA and placed upstream of Cox-2. However, the discovery of additional isoforms and antibody crossreactivity with these other enzymes has prompted a reevaluation of the role of the group IIA sPLA2 in AA generation. For example, work with bone marrow mast cells from C57BL/6J mice, which are homozygous for a disruption in the Pla2g2a gene, shows that the group V sPLA2—not the group IIA sPLA2—is required for both immediate- and delayed-phase AA production (19, 20). In contrast, cPLA2 is undisputed as a central player in AA generation. Both immediate- and delayed-phase PGD2 production is abolished in a number of cell types derived from cPLA2−/− mice (18, 21). In addition, both cPLA2−/− and COX-2−/− female mice have reproductive defects because of the failure to produce PGF2α, again suggesting a pathway with cPLA2 upstream of COX-2 (16, 22).
The striking observation of this article is that both phospholipases can modify the ApcMin tumor phenotype, but in opposite directions (Table 1). Mice carrying the ApcMin mutation on a C57BL/6J background are group IIA sPLA2−/− and develop higher tumor numbers than equivalent strains with functional sPLA2 expression. Restoring sPLA2 expression in transgenic or recombinant lines confers an ≈28% decrease in polyp number in the small intestine, as well as a decrease in polyp size (13). Thus, expression of sPLA2 protects against tumorigenesis, although the effect is relatively modest in the small intestine.
Table 1.
Reduction in mouse tumor number and genotypes
In contrast, cPLA2 expression in the small intestine promotes tumorigenesis. We have demonstrated here that homozygous deletion of cPLA2 produces an 83% decrease in small intestinal polyp number with a concomitant decrease in polyp size. This phenotype is strikingly similar to that of the COX-2 knockout in the ApcΔ716/+ background (14). Notably, after this work was completed, a separate study involving a different cPLA2 knockout allele and ApcΔ716 was reported by Takaku et al. (23). Whereas both studies show a protective effect of cPLA2 deletion, the effect seen by Takaku et al. was limited to a reduction in polyp size. The difference with our work may be because of the different Apc alleles used, as the cPLA2 targeted deletion alleles are quite similar. In general, ApcΔ716 mice develop many more tumors (over threefold more) than ApcMin mice (24). Further work will be required to understand the differing interactions between cPLA2 and these Apc alleles.
Our work also underscores the difference between ApcMin-induced tumorigenesis in the small and large intestines. Whereas homozygous deletion of cPLA2 produces a significant reduction in the tumor number in the small intestine, no effect or at most a slight effect in the opposite direction was observed in the large intestine. Regional differences have also been observed with respect to the efficacy of NSAID treatment. Whereas treatment of ApcMin/+ with piroxicam results in a 95% reduction in tumor number, the distribution of residual tumors suggests that tumors of the duodenum and the colon are more resistant to chemosuppression (25). In addition, ApcMin/+ tumors derived from the colon are resistant to the effects of sulindac treatment, which rapidly induces the regression of 70–80% of ApcMin/+ tumors in the small intestine (26). Transgenic expression of group IIA sPLA2, in contrast, has the same directional effect in both the small and the large intestine; however, the magnitude of the effect is much larger in the colon (13). Unlike the 24–34% reduction seen in the small intestine, transgenic lines overexpressing sPLA2 confer a 65–90% reduction in tumor number in the large intestine (13). These results cumulatively suggest that the pathogenesis of tumors of the small intestine and tumors of the colon differ significantly—at least in ApcMin/+ mice and possibly in humans as well.
Because mutations in the group IV cPLA2 and the group IIA sPLA2 have opposite effects on tumorigenesis, it is difficult to reconcile both observations in terms of a simple model involving AA metabolism. Indeed, we favor the notion that sPLA2 may work in a separate pathway, unrelated to cPLA2 and COX-2. For instance, in the intestine, sPLA2 may play a role in mucosal barrier function. sPLA2 is specifically expressed in Paneth cells in the small intestine and goblet cells in the large intestine; both cell types are thought to protect the mucosa from injury. cPLA2 and COX-2, on the other hand, mostly likely act through the same mechanism, that of decreasing PG production.
The original hypothesis of NSAIDs working through decreased PG production is likely to be correct, and the sPLA2 effect may simply work through another mechanism. In part, the proposed role of AA levels on tumorigenesis is called into question by our results. One approach to clarifying the situation further would be to measure AA and PGE2 levels in the small intestines of cPLA2−/− mice. In preliminary experiments with normal intestinal tissue, we did not detect alterations of basal levels of PGE2 or AA in ApcMin/+, cPLA2−/− animals relative to their heterozygous littermates. However, these results are difficult to interpret, because the gross nature of the assays, the complexity of the tissue, and the multiple phospholipases A2 present in the intestine may obscure potential differences.
Interestingly, work from other laboratories supports a role for AA levels in tumorigenesis but runs counter to the AA hypothesis of Chan et al. (7). Direct dietary manipulations of AA content in ApcMin animals suggest that AA promotes tumorigenesis and is not protective (27). Eicosapentaenoic acid (EPA) competes with AA for incorporation into phospholipids. Dietary supplementation with EPA decreases intestinal AA content and PG production, and results in a 68% reduction in ApcMin polyp number. Supplementing with AA in combination with EPA reverses this antitumorigenic effect. Supplementation with AA alone, although increasing intestinal AA content, has no effect on polyp number.
Furthermore, recent work with the PGE2 receptor EP1 underscores the importance of PGs in general, and PGE2 in particular, in tumorigenesis. PGE2 signals via binding to specific membrane receptors, subtypes EP1 to EP4. Targeted deletion of the EP1 receptor reduced colonic lesions by ≈60% in an azoxymethane-induced colon cancer model. More relevant to this study, ApcMin-induced polyp formation was also reduced 57% by treatment with a specific EP1 antagonist (28).
Therefore, a number of animal studies involving Apc-mutant mice are converging on a central pathway involving cPLA2–COX-2–PGE2 in intestinal tumorigenesis. NSAIDs mostly likely act via inhibition of this signaling pathway. However, we are left with the mystery of how the group IIA sPLA2 (the Mom1 suppressor) actually acts to reduce tumorigenesis.
Acknowledgments
We thank Steve Johnson for constructing the phylogenetic trees, Robert Tyszkowski for technical assistance with immunofluorescence, François Gaudet and Adam Sapirstein for helpful discussions, and Nancy Hong for editorial assistance with the manuscript. Finally, we thank Timothy Wang and Jonathan Arm for critical review of this manuscript and many helpful comments. This work was supported by National Institutes of Health Grants DK39773, DK38452, and NS10828 (to J.V.B.).
Abbreviations
- NSAIDs
nonsteroidal antiinflammatory drugs
- COX
cyclooxygenase
- PG
prostaglandin
- sPLA2
secretory phospholipase A2
- cPLA2
cytosolic phospholipase A2
- AA
arachidonic acid
References
- 1.Williams C S, Luongo C, Radhika A, Zhang T, Lamps L W, Nanney L B, Beauchamp R D, DuBois R N. Gastroenterology. 1996;111:1134–1140. doi: 10.1016/s0016-5085(96)70083-5. [DOI] [PubMed] [Google Scholar]
- 2.Kargman S L, O'Neill G P, Vickers P J, Evans J F, Mancini J A, Jothy S. Cancer Res. 1995;55:2556–2559. [PubMed] [Google Scholar]
- 3.Bamba H, Ota S, Kato A, Adachi A, Itoyama S, Matsuzaki F. Int J Cancer. 1999;83:470–475. doi: 10.1002/(sici)1097-0215(19991112)83:4<470::aid-ijc6>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 4.Jacoby R F, Seibert K, Cole C E, Kelloff G, Lubet R A. Cancer Res. 2000;60:5040–5044. [PubMed] [Google Scholar]
- 5.Whelan J, Chiu C H, McEntee M F. Adv Exp Med Biol. 1999;469:607–615. doi: 10.1007/978-1-4615-4793-8_88. [DOI] [PubMed] [Google Scholar]
- 6.Chiu C H, McEntee M F, Whelan J. Cancer Res. 1997;57:4267–4273. [PubMed] [Google Scholar]
- 7.Chan T A, Morin P J, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1998;95:681–686. doi: 10.1073/pnas.95.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moser A R, Pitot H C, Dove W F. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 9.Moser A R, Dove W F, Roth K A, Gordon J I. J Cell Biol. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich W F, Lander E S, Smith J S, Moser A R, Gould K A, Luongo C, Borenstein N, Dove W. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- 11.MacPhee M, Chepenik K P, Liddell R A, Nelson K K, Siracusa L D, Buchberg A M. Cell. 1995;81:957–966. doi: 10.1016/0092-8674(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 12.Cormier R T, Hong K H, Halberg R B, Hawkins T L, Richardson P, Mulherkar R, Dove W F, Lander E S. Nat Genet. 1997;17:88–91. doi: 10.1038/ng0997-88. [DOI] [PubMed] [Google Scholar]
- 13.Cormier R T, Bilger A, Lillich A J, Halberg R B, Hong K H, Gould K A, Borenstein N, Lander E S, Dove W F. Oncogene. 2000;19:3182–3192. doi: 10.1038/sj.onc.1203646. [DOI] [PubMed] [Google Scholar]
- 14.Oshima M, Dinchuk J E, Kargman S L, Oshima H, Hancock B, Kwong E, Trzaskos J M, Evans J F, Taketo M M. Cell. 1996;87:803–809. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy B P, Payette P, Mudgett J, Vadas P, Pruzanski W, Kwan M, Tang C, Rancourt D E, Cromlish W A. J Biol Chem. 1995;270:22378–22385. doi: 10.1074/jbc.270.38.22378. [DOI] [PubMed] [Google Scholar]
- 16.Bonventre J V, Huang Z, Taheri M R, O'Leary E, Li E, Moskowitz M A, Sapirstein A. Nature (London) 1997;390:622–6225. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- 17.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujishima H, Sanchez Mejia R O, Bingham C O, 3rd, Lam B K, Sapirstein A, Bonventre J V, Austen K F, Arm J P. Proc Natl Acad Sci USA. 1999;96:4803–4807. doi: 10.1073/pnas.96.9.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bingham C O, 3rd, Murakami M, Fujishima H, Hunt J E, Austen K F, Arm J P. J Biol Chem. 1996;271:25936–25944. doi: 10.1074/jbc.271.42.25936. [DOI] [PubMed] [Google Scholar]
- 20.Reddy S T, Winstead M V, Tischfield J A, Herschman H R. J Biol Chem. 1997;272:13591–13596. doi: 10.1074/jbc.272.21.13591. [DOI] [PubMed] [Google Scholar]
- 21.Gijon M A, Spencer D M, Siddiqi A R, Bonventre J V, Leslie C C. J Biol Chem. 2000;275:20146–20156. doi: 10.1074/jbc.M908941199. [DOI] [PubMed] [Google Scholar]
- 22.Uozumi N, Kume K, Nagase T, Nakatani N, Ishii S, Tashiro F, Komagata Y, Maki K, Ikuta K, Ouchi Y, et al. Nature (London) 1997;390:618–622. doi: 10.1038/37622. [DOI] [PubMed] [Google Scholar]
- 23.Takaku K, Sonoshita M, Sasaki N, Uozumi N, Doi Y, Shimizu T, Taketo M M. J Biol Chem. 2000;275:34013–34016. doi: 10.1074/jbc.C000585200. [DOI] [PubMed] [Google Scholar]
- 24.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Proc Natl Acad Sci USA. 1995;92:4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritland S R, Gendler S J. Carcinogenesis. 1999;20:51–58. doi: 10.1093/carcin/20.1.51. [DOI] [PubMed] [Google Scholar]
- 26.McEntee M F, Chiu C H, Whelan J. Carcinogenesis. 1999;20:635–640. doi: 10.1093/carcin/20.4.635. [DOI] [PubMed] [Google Scholar]
- 27.Petrik M B, McEntee M F, Chiu C H, Whelan J. J Nutr. 2000;130:1153–1158. doi: 10.1093/jn/130.5.1153. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe K, Kawamori T, Nakatsugi S, Ohta T, Ohuchida S, Yamamoto H, Maruyama T, Kondo K, Ushikubi F, Narumiya S, et al. Cancer Res. 1999;59:5093–5096. [PubMed] [Google Scholar]