Abstract
Because selected xenobiotic-metabolizing enzymes process pro-carcinogens that could initiate ovarian carcinogenesis, we hypothesized that single-nucleotide polymorphisms (SNPs) in the genes encoding xenobiotic-metabolizing enzymes are associated with risk of ovarian cancer. Cases with invasive epithelial ovarian cancer (N = 1,571 including 956 of serous sub-type) and controls (N = 2,046) from three studies were genotyped at 11 SNPs in EPHX1, ADH4, ADH1A, NQO2, NAT2, GSTP1, CYP1A1, and NQO1, following an initial SNP screen in a subset of participants. Logistic regression analysis of genotypes obtained via Illumina GoldenGate and Sequenom iPlex technologies revealed the following age- and study-adjusted associations: EPHX1 rs1051740 with increased serous ovarian cancer risk (per-allele odds ratio (OR) 1.17, 95% confidence interval (95% CI) 1.04–1.32, p = 0.01), ADH4 r1042364 with decreased ovarian cancer risk (OR 0.90, 95% CI 0.81–1.00, p = 0.05), and NQO1 rs291766 with increased ovarian cancer risk (OR 1.11, 95% CI 1.00–1.23, p = 0.04). These findings are consistent with prior studies implicating these genes in carcinogenesis and suggest that this collection of variants is worthy of follow-up in additional studies.
Keywords: Gynecologic neoplasia, carcinogenesis, epidemiology
INTRODUCTION
Ovarian cancer is the leading cause of gynecologic cancer death among women in developed countries [1]. Known risk factors for ovarian cancer overall or for particular subtypes include age, family history, smoking, fertility drug use, and postmenopausal hormone therapy [2]. Polymorphisms have been associated with ovarian cancer risk in the 9p22.2 chromosomal region [3] and in genes that regulate DNA repair [4].
Environmental carcinogens, such as chemicals in cigarettes, are processed by numerous xenobiotic-metabolizing enzymes including alcohol dehydrogenases (ADHs), cytochrome P450s (CYPs), epoxide hydrolases (EPHXs), glutathione S-transferases (GSTs), N-acetyltransferases (NATs), and NAD(P)H dehydrogenases (quinone) (NQOs) [5]. The CYP enzyme family is key to detoxification of the polycyclic aromatic hydrocarbons, N-nitrosamine, and aromatic amines found in cigarette smoke [6,7]. ADHs convert ethanol to acetaldehyde, a known carcinogen that interferes with DNA synthesis and repair during the first step of alcohol metabolism [8,9]. GSTs, NATs, NQOs, and EPHXs are critical to the metabolism of xenobiotics and potential pro-carcinogens that may be involved in cancer initiation [5].
Here, we hypothesize that, due to their important role in processing pro-carcinogens, inherited genetic variants in xenobiotic-metabolizing genes may be associated with risk of ovarian cancer. Following an initial screen of 163 single-nucleotide polymorphisms (SNPs) in 16 xenobiotic metabolizing genes, 11 SNPs in eight genes were evaluated in a combined analysis of three case-control study populations.
MATERIALS AND METHODS
An initial association screen was conducted within an ongoing two-site candidate gene study described in detail previously [10,11]. Briefly, 930 epithelial ovarian cancer cases and 1,037 frequency-matched (by age, race, and residence) controls from the Mayo Clinic Ovarian Cancer Study (MAY) and the North Carolina Ovarian Cancer Study (NCO) were genotyped at 163 SNPs in 16 xenobiotic-metabolizing genes using an Illumina GoldenGate assay. MAY and NCO participants were enrolled between 1999 and 2006, provided informed consent, and contributed a blood sample; risk factor information was collected through in-person interviews, and clinical data was obtained via medical record. Ascertainment of MAY participants was clinic-based and limited to a six-state catchment area that representing >85% of cases seen at the Mayo Clinic. We selected MAY controls from women seeking general medical evaluation. The NCO study was population based with a rapid case ascertainment network covering a 48-county region of North Carolina. List-assisted random digit dialing and Health Care Financing Administration roster methods were used to identify controls. Included SNPs tagged European variants (r2 ≥ 0.8, MAF ≥ 0.05) or were non-synonymous, within 1 kb upstream, within a 5′ UTR, or within a 3′ UTR (Supplemental Table 1) [12]. Inclusion of within-gene tagSNPs across the chromosome 4 ADH gene cluster enabled evaluation of inter-genic LD and suggested efficiency of the tagSNP selection strategy used (Supplemental Figure 1). Robust quality control measures were applied (Supplemental Table 2) [13]. Association-testing used logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs) assuming an ordinal, recessive, dominant, or co-dominant model for risk of invasive/borderline ovarian cancer and for risk of invasive serous sub-type adjusted for race, age, study site, body mass index, hormone therapy use, oral contraceptive use, parity, and age at first birth. These covariates were included based on association with MAY+NCO case-control status following step-wise regression.
Pooled analysis included MAY, NCO, and an additional collection of 904 invasive cases and 1,105 controls from the Australian Ovarian Cancer Study and the Australian Cancer Study, Ovarian (AUS) [14] genotyped using the Sequenom iPlex (Supplemental Table 3). AUS cases were diagnosed from 2002 to 2007; recruited through surgical treatment centers throughout Australia & cancer registries of Queensland, South Australia, West Australia, New South Wales, and Victoria. AUS controls were randomly selected from Commonwealth electoral rolls and frequency matched for age & geographical region. Eleven initially-screened SNPs which had p < 0.10 in at least one genetic model and which could be genotyped by Sequenom iPlex technology were assessed. A pooled approach was used because it has been shown to be more powerful than separate replication studies [15], and a simplified series of analyses was conducted to reduce multiple testing issues. Association testing used logistic regression to estimate ORs and 95% CIs assuming an ordinal model for risk of invasive ovarian cancer and for risk of invasive serous sub-type among white non-Hispanic women (97% of the pooled study population), adjusted for study site and age. Interactions between genotype and alcohol intake (never, monthly, daily/weekly) and between genotype and two characterizations of tobacco use (never smokers, former smokers, current smokers; 0 pack-years, ≤ 20 pack-years, > 20 pack-years) were explored for SNPs in relevant genes and evaluated using likelihood-ratio testing. Differences in risk by histological sub-type (serous, endometrioid, clear cell, mucinous) were examined using polytomous logistic regression. All analyses were conducted in SAS (SAS Institute, Cary, NC, Version 8, 1999), and, because of the a priori nature of the candidate gene hypotheses being tested, no corrections were made for multiple-testing.
RESULTS
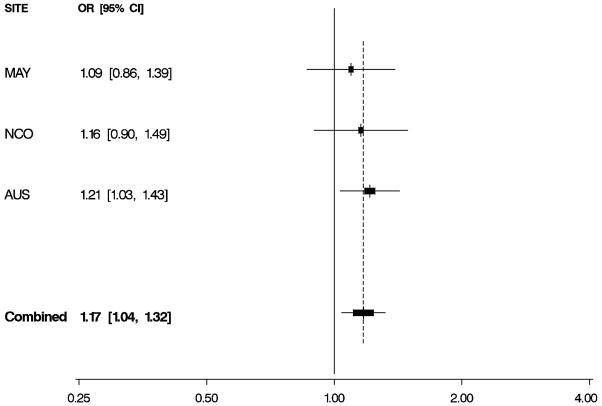
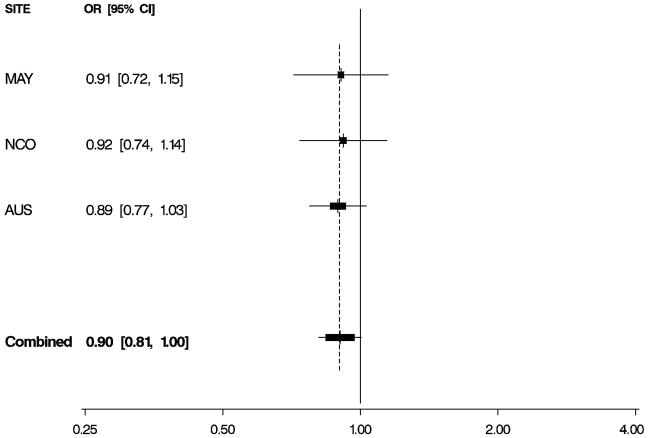
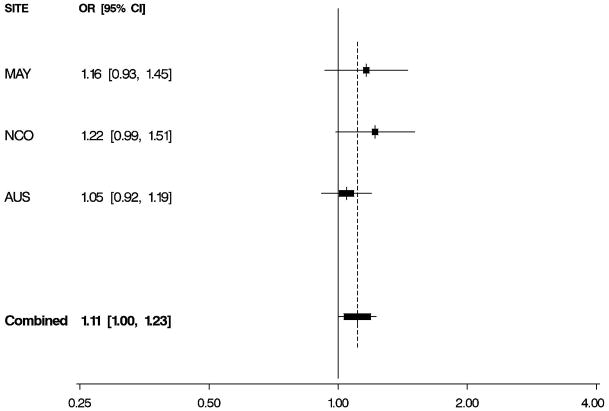
Characteristics of 1,571 white non-Hispanic invasive epithelial ovarian cancer cases and 2,046 controls are shown by study site in Table 1. Approximately 60% of the cases had serous histology, and trends in known risk factors were as expected; in addition, the prevalence of alcohol and tobacco use was sufficiently high to permit assessment of interactions with the candidate genes of interest. Eleven SNPs in EPHX1, ADH4, ADH1A, NQO2, NAT2, GSTP1, CYPA1A, and NQO1 were evaluated for association with risk of invasive ovarian cancer and for serous sub-type (Table 2), following initial screening (Supplemental Table 2). We found evidence that a non-synonymous SNP rs1051740 in EPHX1 was associated with increased invasive ovarian cancer risk (p = 0.03), particularly for serous sub-type (p = 0.01), that a 3′ UTR SNP rs1042364 in ADH4 was associated with decreased invasive ovarian cancer risk (p = 0.05), and that an upstream, possibly promoter-related, SNP rs2917666 in NQO1 was associated with increased invasive ovarian cancer risk (p = 0.04) (Table 2). These results were consistent across studies (Figure 1, Supplemental Table 4).
Table 1.
Characteristics of Study Participants
| Mayo Clinic Ovarian Cancer Study (MAY) | North Carolina Ovarian Cancer Study (NCO) | Australian Ovarian Cancer Study and Australian Cancer Study, Ovarian (AUS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (N=328) | Cases (N=339) | Cases (N=904) | ||||||||
| Histology | Serous | 201 (61.3%) | 201 (59.3%) | 554 (62.0%) | ||||||
| Endometrioid | 62 (18.9%) | 54 (15.9%) | 109 (12.2%) | |||||||
| Clear Cell | 22 (6.7%) | 28 (8.3%) | 64 (7.2%) | |||||||
| Mucinous | 10 (3.0%) | 20 (5.9%) | 29 (3.2%) | |||||||
| Mixed/Other | 33 (10.1%) | 36 (10.6%) | 138 (15.4%) | |||||||
| Unknown | 0 | 0 | 10 | |||||||
| Stage | I | 56 (17.2%) | 81 (24.3%) | 151 (18.4%) | ||||||
| II | 22 (6.7%) | 31 (9.3%) | 72 (8.8%) | |||||||
| III | 192 (58.9%) | 207 (62.2%) | 523 (63.7%) | |||||||
| IV | 56 (17.2%) | 14 (4.2%) | 75 (9.1%) | |||||||
| Unknown | 2 | 6 | 83 | |||||||
| Cases (N=328) | Controls (N=462) | p-value | Cases (N=339) | Controls (N=479) | p-value | Cases (N=904) | Controls (N=1,105) | p-value | ||
| Age | Mean (S.D.) | 61 (12.61) | 60 (13.02) | 0.31 | 56.8 (10.55) | 54.8 (11.97) | 0.01 | 59.2 (10.69) | 57.3 (11.64) | <0.001 |
| Age at Menarche | <12 | 45 (18.3%) | 67 (15.8%) | 0.67 | 84 (24.9%) | 85 (17.7%) | 0.07 | 136 (15.0%) | 193 (17.5%) | 0.29 |
| 12 | 62 (25.2%) | 97 (22.9%) | 99 (29.3%) | 143 (29.9%) | 207 (22.9%) | 229 (20.7%) | ||||
| 13 | 65 (26.4%) | 124 (29.2%) | 81 (24.0%) | 139 (29.0%) | 227 (25.1%) | 294 (26.6%) | ||||
| ≥14 | 74 (30.1%) | 136 (32.1%) | 74 (21.9%) | 112 (23.4%) | 334 (36.9%) | 389 (35.2%) | ||||
| Unknown | 82 | 38 | 1 | 0 | 0 | 0 | ||||
| Oral contraceptive use | Never | 154 (49.7%) | 164 (38.6%) | <0.001 | 115 (34.6%) | 147 (30.9%) | 0.15 | 288 (33.1%) | 219 (19.9%) | <0.001 |
| 1–48 months | 78 (25.2%) | 90 (21.2%) | 104 (31.3%) | 134 (28.2%) | 239 (27.4%) | 254 (23.1%) | ||||
| >48 months | 78 (25.2%) | 171 (40.2%) | 113 (34.0%) | 194 (40.8%) | 344 (39.5%) | 627 (57.0%) | ||||
| Unknown | 18 | 37 | 7 | 4 | 33 | 5 | ||||
| Number of live births | Nulliparous | 55 (17.2%) | 64 (14.7%) | 0.61 | 69 (20.4%) | 62 (12.9%) | 0.02 | 156 (17.7%) | 118 (10.7%) | <0.001 |
| 1–2 | 112 (35.1%) | 153 (35.2%) | 170 (50.1%) | 268 (55.9%) | 351 (39.9%) | 459 (41.5%) | ||||
| ≥3 | 152 (47.6%) | 218 (50.1%) | 100 (29.5%) | 149 (31.1%) | 372 (42.3%) | 528 (47.8%) | ||||
| Unknown | 9 | 27 | 0 | 0 | 25 | 0 | ||||
| Alcohol Use | Never | 51 (27.9%) | 55 (12.6%) | <0.001 | 137 (40.4%) | 171 (35.7%) | 0.35 | 188 (25.3%) | 195 (17.8%) | <0.001 |
| Monthly | 87 (47.5%) | 172 (39.5%) | 102 (30.1%) | 149 (31.1%) | 49 (6.6%) | 66 (6.0%) | ||||
| Weekly/Daily | 45 (24.6%) | 208 (47.8%) | 100 (29.5%) | 159 (33.2%) | 507 (68.1%) | 837 (76.2%) | ||||
| Unknown | 145 | 27 | 0 | 0 | 160 | 7 | ||||
| Smoking status | Never | 200 (64.7%) | 279 (64.3%) | 0.23 | 185 (54.6%) | 239 (49.9%) | <0.001 | 474 (58.9%) | 659 (59.7%) | 0.02 |
| Former | 83 (26.9%) | 131 (30.2%) | 125 (36.9%) | 153 (31.9%) | 214 (26.6%) | 330 (29.9%) | ||||
| Current | 26 (8.4%) | 24 (5.5%) | 29 (8.6%) | 87 (18.2%) | 117 (14.5%) | 115 (10.4%) | ||||
| Unknown | 19 | 28 | 0 | 0 | 99 | 1 | ||||
| Smoking, pack- Years | None | 200 (65.8%) | 279 (68.0%) | 0.34 | 188 (57.8%) | 248 (54.0%) | 0.56 | 474 (59%) | 659 (59.7%) | 0.45 |
| ≤20 yrs | 57 (18.8%) | 83 (20.2%) | 79 (24.3%) | 120 (26.1%) | 214 (26.7%) | 308 (27.9%) | ||||
| >20 yrs | 47 (15.5%) | 48 (11.7%) | 58 (17.8%) | 91 (19.8%) | 115 (14.3%) | 137 (12.4%) | ||||
| Unknown | 24 | 52 | 14 | 20 | 101 | 1 | ||||
Includes data from white non-Hispanics and invasive ovarian cancer cases only; data are counts (percentage) unless otherwise indicated; p-values are from within-sites tests of case-control differences; continuous variables (t-test) and categorical variables (Chi square test); bold indicates site-specific case-control p-value < 0.05; covariates used in adjusted logistic regression models included age and study site for combined analysis of MAY+NCO+AUS and race, age, study site, body mass index, hormone therapy use, oral contraceptive use, parity, and age at first birth for initial MAY+NCO analyses..
Table 2.
SNPs and Risk of Epithelial Ovarian Cancer
| Gene | SNP | Location | Alleles | All Sub-types (1,571 cases, 2,046 controls) | Serous Sub-type (951 cases, 2,046 controls) | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | ||||
| EPHX1 | rs1051740 | ns Y113H | T>C | 1.12 (1.01,1.24) | 0.03 | 1.17 (1.04,1.32) | 0.01 |
| ADH4 | rs1042364 | 3′ UTR | G>A | 0.90 (0.81,1.00) | 0.05 | 0.91 (0.81,1.04) | 0.16 |
| ADH1A | rs2276332 | intron | T>G | 1.14 (0.96,1.36) | 0.12 | 1.11 (0.91,1.36) | 0.30 |
| rs13134764 | 5′ (31) | T>A | 0.96 (0.88,1.05) | 0.37 | 1.00 (0.90,1.11) | 0.98 | |
| NQO2 | rs927340 | 3′ (3,724) | G>A | 0.90 (0.80,1.02) | 0.10 | 0.92 (0.80,1.06) | 0.27 |
| NAT2 | rs2410556 | intron | T>C | 1.00 (0.89,1.14) | 0.95 | 0.98 (0.85,1.14) | 0.80 |
| GSTP1 | rs17593068 | 5′ (354) | C>A | 0.96 (0.86,1.07) | 0.45 | 1.03 (0.90,1.16) | 0.69 |
| CYP1A1 | rs4646421 | intron | G>A | 0.96 (0.82,1.13) | 0.64 | 0.96 (0.80,1.15) | 0.67 |
| rs2470893 | 5′ (1,572) | C>T | 1.06 (0.96,1.18) | 0.23 | 1.01 (0.89,1.13) | 0.91 | |
| NQO1 | rs1800566 | ns P187S | G>A | 1.08 (0.96,1.22) | 0.22 | 1.02 (0.88,1.18) | 0.80 |
| rs2917666 | 5′ (3,427) | C>G | 1.11 (1.00,1.23) | 0.04 | 1.06 (0.94,1.19) | 0.32 | |
White non-Hispanic participants from MAY, NCO, and AUS; position from genome build 36.3; Refseq release 29 (May 4, 2008); ns indicates non-synonymous SNP, other indications represent location and distance in base-pairs from gene; alleles represents major>minor; per-allele (ordinal) odds ratio and 95% confidence intervals adjusted for age and study site; GSTP1 rs17593068 failed for NCO samples; bold indicates p-value < 0.05.
Figure 1. Odds Ratios by Study and Combined.
Per-allele odds ratios (ORs) and 95% confidence intervals (CIs) by study and combined, adjusted for age and study site. Boxes indicate ORs and are proportionally sized relative to the number of particpants; horizontal bars represent 95% CIs. A vertical dashed line indicates the combined OR estimate. A. EPHX1 rs1051740 and risk of invasive serous ovarian cancer. B. ADH4 rs1042364 and risk of invasive ovarian cancer. C. NQO1 rs2917666 and risk of invasive ovarian cancer.
No compelling trends in risk were observed in analyses stratified by alcohol use (examined for ADH4 and AHD1A) or tobacco use (examined for EPHX1, NQO2, NAT2, GSTP1, CYPA1A, and NQO1; Supplemental Table 5). Finally, no significant (p < 0.05) heterogeneity of risks was observed across histological sub-types (Supplemental Table 6), including at EPHX1 rs1051740.
DISCUSSION
Xenobiotic-metabolizing enzymes are clearly important in processing of pro-carcinogens, and several studies have observed associations and interactions with non-genetic factors in the etiology of cancer. Here, we used a multi-site study to examine the hypothesis that inherited variation which may alter xenobiotic metabolism relates to ovarian cancer risk. We found that rs1051740 in EPHX1 was associated with increased invasive ovarian cancer risk (particularly serous sub-type); this SNPs results in an amino acid change at position 113 from the polar hydrophilic tyrosine to the electrically-charged (positive) histidine. This change is predicted to be damaging to the function of epoxide hydrolase [16] and has been shown to alter epoxide hydrolase’s processing of several carcinogens [17–19]. Genotypes at this SNP have been studied previously in relation to ovarian cancer risk in two smaller study populations [20,21], and association with risk was observed in one study (OR 2.6, 95% CI: 1.3 – 5.0) [20]. The modest EPHX1 rs1051740 risk estimate we observed may only be apparent with large sample size. Associations between genotypes at this SNP and increased risk of squamous cell esophageal cancer [22] and colorectal cancer [23] have also been reported; notably the “slow” phenotype was associated with increased risk of colorectal adenomatous and hyperplastic polyps in individuals exposed to cigarette smoke and high red meat consumption [24]. It is of particular interest that, in the current study, this SNP was selected for pooled analysis over other EPHX1 SNPs based on initial screening results regardless of its functional or tagging status.
We also report that an ADH4 SNP was associated with decreased ovarian cancer risk and that an NQO1 SNP was associated with increased risk. Alcohol dehydrogenase 4 (class II), pi polypeptide (ADH4) appears to be important to the initial metabolism of ethanol and also in the synthesis of retinoic acid [25]. Our findings in ADH4 are consistent with prior evidence indicating that the retinoic acid pathway plays a role in ovarian carcinogenesis [26,27]. NAD(P)H:quinone oxidoreductase 1 (NQO1) metabolizes quinones, aromatic compounds found in benzene and chemotherapeutics, and acts as an antioxidant that protects against the production of DNA- damaging and protein-damaging reactive oxygen species. Additionally, NQO1 expression is up-regulated in tumor tissues as a result of cancer-induced hypoxia [28] which could also be involved in ovarian carcinogenesis. Our observation of increased risk is consistent with the minor allele conferring a decrease in NQO1 function.
Strengths of the current approach include the use of three ovarian cancer study populations and examination of risk by alcohol and tobacco use and across histological subtypes. This study is limited by the inclusion of only 11 variants in pooled analysis, the inability to assess generalizability of associations across multiple ethnicities, multiple tests performed, and reduced power in subset and interaction analyses. Although only modest differences in relative risk are conferred by the risk alleles found here, they are consistent with risk estimates at other confirmed loci studied in over 8,000 cases and 8,000 controls [3,12,29]. As we note that a Bonferroni correction for multiple testing would suggest that no association is significant at α=0.05, follow-up in a larger sample will rule out both false positives and false negatives. In conclusion, further examination of EPHX1 rs1051740, ADH4 r1042364, and NQO1 rs291766 in additional study populations and an evaluation of interactions with relevant carcinogenic exposures in larger collections are needed to make more definitive conclusion on the role of inherited variation in xenobiotic metabolism in ovarian cancer etiology.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R01 CA 122443 and R01 CA 88868, as well as NIH grant CA 15083 which supports the Mayo Clinic College of Medicine Genotyping Shared Resource.
The AOCS Management Group (D. Bowtell, G. Chenevix-Trench, A. deFazio, D. Gertig, A. Green, P. Webb) gratefully acknowledges the contribution of all the clinical and scientific collaborators (see http://www.aocstudy.org/). The AOCS and ACS Management Group (A. Green, P. Parsons, N. Hayward, P. Webb, D. Whiteman) thank all of the project staff, collaborating institutions, and study participants. We are also grateful to the family and friends of Kathryn SladekSmith for their generous support of the Ovarian Cancer Association Consortium through their donations to the Ovarian Cancer Research Fund.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Morch LS, Lokkegaard E, Andreasen AH, Kruger-Kjaer S, Lidegaard O. Hormone Therapy and Ovarian Cancer. JAMA. 2009;302(3):298–305. doi: 10.1001/jama.2009.1052. [DOI] [PubMed] [Google Scholar]
- 3.Song H, Ramus SJ, Tyrer J, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41(9):996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schildkraut JM, Goode EL, Clyde MA, et al. Single nucleotide polymorphisms in the TP53 region and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2009;69(6):2349–2357. doi: 10.1158/0008-5472.CAN-08-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh MS, Micheal M. Role of Xenobiotic Metabolic Enzymes in Cancer Epidemiology. In: Verma M, editor. Cancer Epidemiology. Vol. 2. New York: Humana Press; 2009. pp. 243–264. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez FJ, Gelboin HV. Role of Human Cytochromes P450 in the Metabolic Activation of Chemical Carcinogens and Toxins. Drug Metabolism Reviews. 1994;26(1–2):165–183. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- 7.Rendic S, Di Carlo FJ. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab Rev. 1997;29(1–2):413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 8.Obe G, Ristow H. Mutagenic, cancerogenic and teratogenic effects of alcohol. Mutat Res. 1979;65(4):229–259. doi: 10.1016/0165-1110(79)90004-6. [DOI] [PubMed] [Google Scholar]
- 9.Dirk WL, Fotis K, Jürgen R. Carcinogenicity of acetaldehyde in alcoholic beverages: risk assessment outside ethanol metabolism. Addiction. 2009;104(4):533–550. doi: 10.1111/j.1360-0443.2009.02516.x. [DOI] [PubMed] [Google Scholar]
- 10.Sellers TA, Huang Y, Cunningham J, et al. Association of single nucleotide polymorphisms in glycosylation genes with risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(2):397–404. doi: 10.1158/1055-9965.EPI-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelemen LE, Sellers TA, Schildkraut JM, et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008;68(7):2498–2506. doi: 10.1158/0008-5472.CAN-07-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goode EL, Maurer MJ, Sellers TA, et al. Inherited determinants of ovarian cancer survival. Clin Cancer Res. 2010;16(3):995–1007. doi: 10.1158/1078-0432.CCR-09-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham JM, Sellers TA, Schildkraut JM, et al. Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1781–1789. doi: 10.1158/1055-9965.EPI-07-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spurdle AB, Purdie DM, Chen X, Chenevix-Trench G. The prohibitin 3′ untranslated region polymorphism is not associated with risk of ovarian cancer. Gynecol Oncol. 2003;90(1):145–149. doi: 10.1016/s0090-8258(03)00193-8. [DOI] [PubMed] [Google Scholar]
- 15.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 16.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassett C, Aicher L, Sidhu JS, Omiecinski CJ. Human microsomal epoxide hydrolase: genetic polymorphism and functional expression in vitro of amino acid variants. Hum Mol Genet. 1994;3(3):421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastorelli R, Guanci M, Cerri A, et al. Impact of inherited polymorphisms in glutathione S-transferase M1, microsomal epoxide hydrolase, cytochrome P450 enzymes on DNA, and blood protein adducts of benzo(a)pyrene-diolepoxide. Cancer Epidemiol Biomarkers Prev. 1998;7(8):703–709. [PubMed] [Google Scholar]
- 19.Kim S, Lan Q, Waidyanatha S, et al. Genetic polymorphisms and benzene metabolism in humans exposed to a wide range of air concentrations. Pharmacogenet Genomics. 2007;17(10):789–801. doi: 10.1097/FPC.0b013e3280128f77. [DOI] [PubMed] [Google Scholar]
- 20.Lancaster JM, Brownlee HA, Bell DA, et al. Microsomal epoxide hydrolase polymorphism as a risk factor for ovarian cancer. Mol Carcinog. 1996;17(3):160–162. doi: 10.1002/(SICI)1098-2744(199611)17:3<160::AID-MC8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 21.Spurdle AB, Purdie DM, Webb PM, Chen X, Green A, Chenevix-Trench G. The microsomal epoxide hydrolase Tyr113His polymorphism: association with risk of ovarian cancer. Mol Carcinog. 2001;30(1):71–78. doi: 10.1002/1098-2744(200101)30:1<71::aid-mc1015>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Jain M, Tilak AR, Upadhyay R, Kumar A, Mittal B. Microsomal epoxide hydrolase (EPHX1), slow (exon 3, 113His) and fast (exon 4, 139Arg) alleles confer susceptibility to squamous cell esophageal cancer. Toxicology and Applied Pharmacology. 2008;230(2):247–251. doi: 10.1016/j.taap.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Kiss I, Orsos Z, Gombos K, et al. Association between allelic polymorphisms of metabolizing enzymes (CYP 1A1, CYP 1A2, CYP 2E1, mEH) and occurrence of colorectal cancer in Hungary. Anticancer Res. 2007;27(4C):2931–2937. [PubMed] [Google Scholar]
- 24.Ulrich CM, Bigler J, Whitton JA, Bostick R, Fosdick L, Potter JD. Epoxide hydrolase Tyr113His polymorphism is associated with elevated risk of colorectal polyps in the presence of smoking and high meat intake. Cancer Epidemiol Biomarkers Prev. 2001;10(8):875–882. [PubMed] [Google Scholar]
- 25.Yin S-J, Chou C-F, Lai C-L, Lee S-L, Han C-L. Human class IV alcohol dehydrogenase: kinetic mechanism, functional roles and medical relevance. Chemico-Biological Interactions. 2003:143–144. 219–227. doi: 10.1016/s0009-2797(02)00167-9. [DOI] [PubMed] [Google Scholar]
- 26.Tung K-H, Wilkens LR, Wu AH, et al. Association of Dietary Vitamin A, Carotenoids, and Other Antioxidants with the Risk of Ovarian Cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(3):669–676. doi: 10.1158/1055-9965.EPI-04-0550. [DOI] [PubMed] [Google Scholar]
- 27.Williams SJ, Cvetkovic D, Hamilton TC. Vitamin A metabolism is impaired in human ovarian cancer. Gynecologic Oncology. 2009;112(3):637–645. doi: 10.1016/j.ygyno.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guha N, Chang JS, Chokkalingam AP, Wiemels JL, Smith MT, Buffler PA. NQO1 Polymorphisms and De Novo Childhood Leukemia: A HuGE Review and Meta-Analysis. Am J Epidemiol. 2008;168(11):1221–1232. doi: 10.1093/aje/kwn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolton KL, Tyrer J, Song H, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42(10):880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.