SUMMARY
Autophagy is of increasing interest as a target for cancer therapy. We find that leucine deprivation causes the caspase-dependent apoptotic death of melanoma cells because it fails to appropriately activate autophagy. Hyperactivation of the RAS-MEK pathway, which is common in melanoma, prevents leucine deprivation from inhibiting mTORC1, the main repressor of autophagy under nutrient-rich conditions. In an in vivo tumor xenograft model, the combination of a leucine-free diet and an autophagy inhibitor synergistically suppresses the growth of human melanoma tumors and triggers widespread apoptosis of the cancer cells. Together, our study represents proof of principle that anti-cancer effects can be obtained with a combination of autophagy inhibition and strategies to deprive tumors of leucine.
INTRODUCTION
While it is not completely understood how cancer cells survive and grow in nutrient limiting conditions, recent studies support a central role for autophagy (Klionsky, 2007; Kroemer and Levine, 2008; Levine and Kroemer, 2008; White et al., 2010). Autophagy is a lysosome-dependent cellular degradation pathway that is triggered by nutrient deprivation and requires the evolutionarily conserved ATG proteins. These proteins regulate the formation and expansion of a cup-shaped structure, termed the isolation membrane or phagophore, which eventually encloses a portion of cytoplasm in a double membrane vesicle called an autophagosome. In the late stages of autophagy the outer membrane of an autophagosome fuses with a lysosome to produce an autophagolysosome, which leads to the degradation of the enclosed cytoplasmic material by lysosomal enzymes and the recycling of metabolites that cannot be synthesized de novo, such as essential amino acids.
The development of the isolation membrane has two major steps: nucleation and elongation. The nucleation step requires the ATG1/ULK1 kinase and the type III PI3K/VPS34 kinase complex, and the elongation step the ATG8/LC3- and ATG12-conjugation systems (Levine and Kroemer, 2008; Nakatogawa et al., 2009). A key regulator of the nucleation of the isolation membrane is the mTOR complex 1 (mTORC1) signaling pathway. Under nutrient-rich conditions, mTORC1 suppresses autophagy by inhibiting, in a poorly understood fashion, the ATG1/ULK1 kinase complex (Hosokawa et al., 2009; Jung et al., 2009). The mTORC1 pathway is sensitive to amino acid levels (Hara et al., 1998) and amino acid deprivation activates autophagy (Mortimore and Schworer, 1977; Schworer et al., 1981). How amino acids activate mTORC1 is not well understood, but recent work has revealed an essential role for the amino acid-stimulated translocation of mTORC1 to the lysosomal surface (Sancak et al., 2010; Sancak et al., 2008).
Cells can differ in which amino acids they require for survival, and oncogenic transformation may make them liable to the deficiency of a particular amino acid. For example, human fibroblasts with activated c-MYC depend on glutamine (Yuneva et al., 2007), lymphoblastic leukemia cells require tryptophan, methionine, and valine (Gong et al., 2000; Kreis et al., 1980; Ohtawa et al., 1998; Woolley et al., 1974), and several types of solid tumor cells require arginine (Scott et al., 2000). In most cases however, the cellular underpinnings behind the particular amino acid requirements of a cancer cell type are largely unknown, making it difficult to exploit such information to implement anticancer therapeutics. Here, we investigated which essential amino acids are necessary for the survival of human melanoma cells and identified an oncogenic signaling pathway that determines their sensitivity to leucine deprivation.
RESULTS
Leucine deprivation triggers the apoptotic cell death of human melanoma cells
We examined the survival of four patient-derived melanoma cell lines (A-2058, SK-MEL-3, SK-MEL-5, SK-MEL-28) as well as the non-transformed but immortalized human Mel-ST melanocyte line (Figures 1A and 1B). We used the cleavage of caspase-3 as a readout for the caspase-dependent apoptosis (Galluzzi et al., 2009; Kroemer et al., 2009; Taylor et al., 2008). Caspase-3 cleaves an array of apoptosis-related proteins, including PARP (Figure S1A).
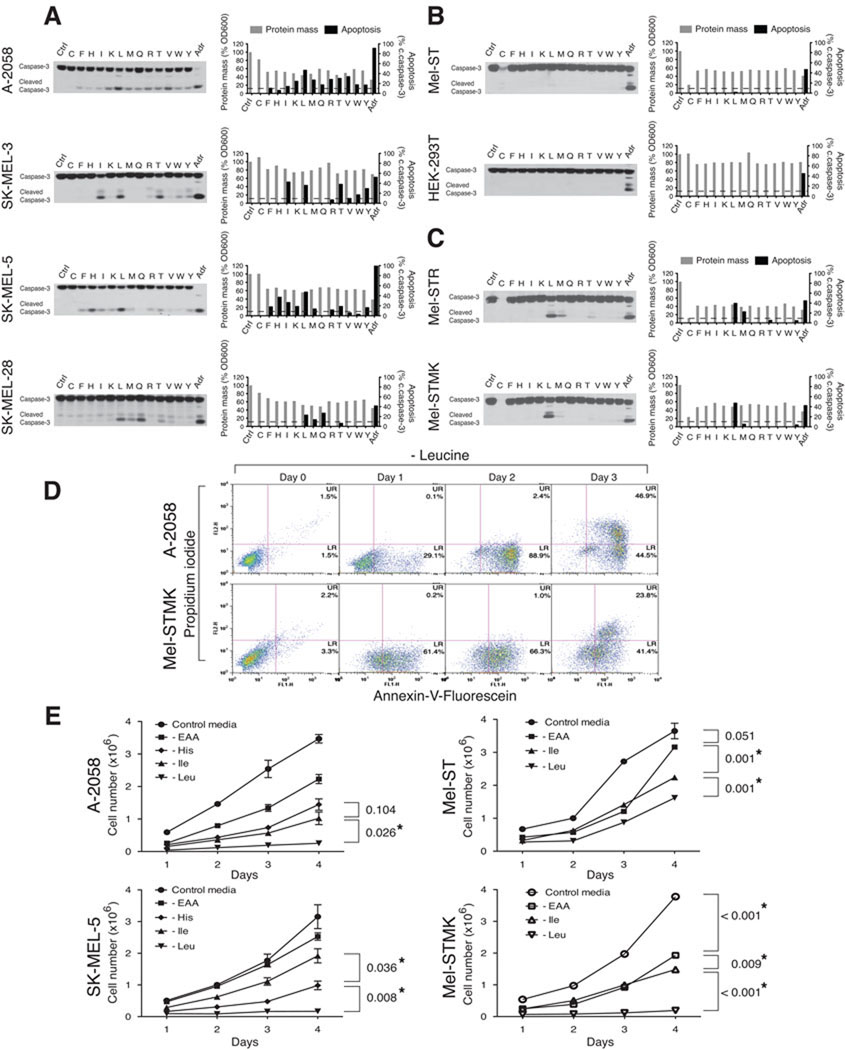
Figure 1. Leucine deprivation induces apoptosis in human melanoma cells.
Survey of patient derived melanoma cells (A), immortalized human melanocytes and the non-melanocyte-derived line (B), and transformed melanocytes (C). Immunoblot analyses for intact and cleaved caspase-3 of indicated cell lines following a 48 hr deprivation for individual essential amino acids. Bar graphs indicate relative changes in protein mass (a readout for cell growth and proliferation) and percent activation of caspase-3 (ratio of cleaved to full-length caspase-3). Dotted lines indicate 10% activation of caspase-3. Ctrl, control RPMI-1640 media; C-F-H-I-K-L-M-Q-R-T-V-W-Y, deprivation of the indicated single amino acid (single-letter code for amino acid); Adr, adriamycin at 2 µg/ml. (D) Annexin-V assay for apoptosis induction. FL1; Annexin-V-Fluorescein, FL2; Propidium Iodide, UR; upper right quadrant, LR; lower right quadrant. (E) Cell survival assay. Cells were deprived of all essential amino acids (-EAA), histidine (-His), isoleucine (-Ile), or leucine (-Leu) for 2 days and re-seeded into control RPMI-1640 media, and changes in cell number were measured at indicated time points. Data are represented as mean ± s.d. and * indicates values that are significantly different from controls. See also Figure S1.
Cells were deprived, one amino acid at a time, of the thirteen amino acids that are considered universally (F, I, K, L, M, T, V, W) or conditionally (C, H, Q, R, Y) essential in humans (Berg, 2007; Eagle, 1959). We deprived the melanoma cells of only essential amino acids because the cell lines could have differing capacities to synthesize non-essential amino acids, which would greatly confound the interpretation of the results. Unsurprisingly, upon the deprivation of any single essential amino acid, all cell lines halted proliferating and had a concomitant decrease in cyclin D1 levels and protein mass (Figures 1A and 1B, and Figure S1A). In contrast, the melanoma cell lines differed in which particular set of amino acids, when individually omitted from the media, trigger the cleavage of caspase-3 (Figure 1A). Interestingly, in all the melanoma lines, the only constant was that leucine deprivation triggered cleavage of caspase-3 and the corresponding caspase-dependent cleavage of PARP (Figure S1A). Leucine deprivation did not, however, induce caspase-3 cleavage in non-transformed Mel-ST melanocytes or non-melanocyte-derived HEK-293T cells (Figure 1B). The DNA-damaging agent adriamycin did induce caspase-3 cleavage in these latter two lines, like in the patient-derived melanoma cells (Figures 1A and 1B).
Consistent with the caspase-3 cleavage results, an Annexin-V assay (Galluzzi et al., 2009; Kroemer et al., 2009) revealed, upon leucine deprivation, a time-dependent increase in phosphatidylserine (PS) on the outer leaflet of the plasma membrane of A-2058 cells (Figure 1D). The increase in Annexin-V staining preceded the eventual loss of plasma membrane integrity, which was detected by an increase in propidium iodide staining at the later time points (Figure 1D).
Hyperactivation of the RAS-MEK pathway renders melanocytes dependent on leucine for survival
Because all the melanoma lines in our study have activating mutations in the RAS-MAPK pathway (COSMIC database, Wellcome Trust Sanger Institute) (Bamford et al., 2004), we asked if Ras pathway hyperactivation could confer on melanocytes the capacity to induce caspase-3 activation upon leucine deprivation. Indeed, Mel-STR cells, an engineered melanoma cell generated by transforming Mel-ST melanocyte with oncogenic RAS-G12V (Gupta et al., 2005), very strongly induced caspase-3 cleavage when deprived of leucine (Figure 1C). Mel-STMK cells, which are Mel-ST cells expressing an activated allele of MEK1 (MEK1-Q56P) (Bottorff et al., 1995; Marks et al., 2008), behaved very similarly to Mel-STR cells in the caspase-3 cleavage assay and to A-2058 cells in the Annexin-V assay (Figures 1C and 1D). These data support the notion that the RAS-MEK pathway is responsible for the sensitivity of melanocytes to leucine deprivation. Consistent with this interpretation, U-0126 (a small molecule inhibitor of MEK1/2) (Davies et al., 2000; Favata et al., 1998), but not KT5720 (an inhibitor of PKA), prevented the cleavage of caspase-3 in Mel-STR cells deprived of leucine (Figure S1B).
Of the several components of the RAS-MAPK pathway found mutated in human cancers, BRAF is a critical oncogene in melanoma. 50% – 70% of disease cases have activating mutations in it (Garnett and Marais, 2004; Gray-Schopfer et al., 2007) and all the patient-derived melanoma lines in our survey carry a mutant allele of BRAF (Bamford et al., 2004). Therefore, we asked if expression of oncogenic BRAF-V600E, the most common BRAF mutant allele found in melanoma (Davies et al., 2002), mimics the effects of RAS-G12V and MEK1-Q56P in sensitizing melanocytes to apoptosis upon leucine deprivation. Indeed, expression of BRAF-V600E as well as BRAF-Δ3-V600E, a variant that cannot interact with CRAF (Karreth et al., 2009), promoted the cleavage of caspase-3 upon leucine withdrawal (Figure S1C). Expression of wild-type BRAF or the BRAF-Δ3 variant without the V600E mutation did not have the same effects (Figure S1C). Together, these results support a key role for the RAS-BRAF-MEK1 signaling axis in determining the liability of the cells to leucine deprivation.
We also determined the capacity of cells to resume proliferation upon reseeding equal number of cells at ~80% cell confluency into complete media after being deprived of leucine, isoleucine, or all amino acids. Mel-STMK, A-2058, and SK-MEL-5, but not Mel-ST, cells deprived of leucine failed to show proliferation when re-seeded (Figure 1E). Just changing the media into complete media without re-seeding also showed concordant results with those in which cells were re-seeded, which excludes the possibility of a change in plating efficiency accounting for the results observed (Figure S1D). In contrast to the results observed with leucine deprivation, all cell lines deprived of isoleucine or all essential amino acids successfully resumed proliferation (Figure 1E). In SK-MEL-5 and A-2058 cells deprived of histidine, the extent of cell survival inversely correlated with that of caspase-3 cleavage (Figures 1A and 1E).
Caspase activity regulated by the mitochondrial apoptotic pathway is necessary for cell death caused by leucine deprivation
To investigate if the apoptotic caspases are required for leucine deprivation to trigger cell death, we used the pan-caspase inhibitors Q-VD-OPH (Caserta et al., 2003; Chauvier et al., 2007) and Z-VAD-fmk (Slee et al., 1996). Q-VD-OPH inhibits a spectrum of caspases with high specificity while Z-VAD-fmk may also inhibit other types of proteases, including calpains and lysosomal cathepsins (Caserta et al., 2003; Chauvier et al., 2007; Kroemer et al., 2009). Q-VD-OPH inhibited the morphological changes characteristic of apoptosing cells, as well the caspase-dependent cleavage of PARP in cells deprived of leucine (Figure 2A and 2B). Critically, Q-VD-OPH and Z-VAD-fmk greatly increased the survival of melanoma cells deprived of leucine (Figure 2C). As Q-VD-OPH did not completely rescue cells from cell death induced by leucine deprivation, it is possible that other mechanisms in addition to caspase-dependent apoptosis may also contribute to the death of the cells.
Figure 2. Activation of caspase cascade through the mitochondrial apoptotic pathway is necessary for leucine deprivation-induced death.
(A) Micrographs showing morphological changes following deprivations of all essential amino acids (-EAA), isoleucine, or leucine in the presence or absence of 20 µM Q-VD-OPH. Scale bar = 100 µm. (B) Immunoblot analyses showing the dose-dependent inhibitory effect of increasing concentrations of Q-VD-OPH (0 µM, 5 µM, 20 µM, and 100 µM) on caspase mediated processes. (C) Cell survival assay. Cells were deprived of leucine (-Leu) for 2 days in the presence or absence of pan-caspase inhibitors, 20 µM Q-VD-OPH or 100 µM Z-VAD-fmk. (D) Flow cytometric analyses showing changes in MOMP using JC-1 dye. FL1; J-monomer (JC-1 green fluorescence), FL2; J-aggregates (JC-1 red fluorescence). (E) Immunoblot analyses show the effect of Bcl-xL expression on caspase-3 activation upon leucine deprivation. (F) Flow cytometric analyses showing changes in MOMP. (G) Cell survival assay. Data are represented as mean ± s.d. and * indicates values that are significantly different from controls.
The caspase inhibitor experiments also hinted at how leucine deprivation activates the caspase cascade. Not only did Q-VD-OPH inhibit the cleavage of PARP, a known substrate of caspase-3, but also the self-cleavage at Asp-315 of caspase-9. As an initiator caspase of the mitochondrial pathway, caspase-9 regulates executioner caspases, including caspase-3 (Figure 2B). Consistent with this finding, leucine deprivation increased the mitochondrial outer membrane permeabilization (MOMP) of A-2058 and Mel-STMK, but not Mel-ST, cells (Figures 2D and 2F). To determine if the increase in MOMP is necessary for the cell death caused by leucine deprivation, we established A-2058 cells overexpressing Bcl-xL (Figure 2E). In contrast to the parental line, Bcl-xL-overexpressing cells did not increase MOMP or trigger cleavage of caspase-3 when deprived of leucine (Figures 2E and 2F). Under no leucine conditions the decrease in caspase-3 activation in Bcl-xL-overexpressing cells directly correlated with an increase in their survival (Figure 2G). These results support an important role for the mitochondrial apoptotic pathway in triggering cell death upon leucine deprivation.
Leucine deprivation does not activate autophagy in melanocyte-derived cells with constitutively active RAS-MEK signaling
To investigate why the deprivation of leucine, but not other essential amino acids, is a universal inducer of apoptosis in melanoma cells, we examined the effects of leucine deprivation and RAS signaling on autophagy activity. It is increasingly appreciated that autophagy is critical for cells to survive nutrient deprivation and that amino acids are major regulators of this process (Klionsky, 2007; Levine and Kroemer, 2008).
We used a fluorescent protein tagged LC3 reporter to quantitate autophagy activity. This dual color DsRed-LC3-GFP reporter is a modified form of the classical GFP-tagged LC3 reporter (Kabeya et al., 2000), and provides two readouts for autophagy activity: the number of DsRed-LC3 puncta and a flow cytometric measurement we call the autophagy index. Like previous results obtained using a GFP-LC3 reporter (Kabeya et al., 2000), our reporter showed an increase in the number of DsRed-LC3 puncta upon a PBS incubation or amino acid deprivation (Figure 3). The reporter has GFP separated from the C-terminus of LC3 by a recognition site for the autophagic protease, ATG4, and loss of GFP fluorescence can be monitored by flow cytometry (see Figure S2 for details). As expected, deletion of the ATG4 recognition sequence abrogated the sensitivity of the reporter to low nutrient conditions (Figures S2CS2E). In comparison to the control media condition, deprivation of all amino acids significantly decreased the levels of the full length DsRed-LC3-GFP reporter as detected by immunoblotting (Figure S2B). To represent results obtained by flow cytometry, we introduced an autophagy index, which normalizes the change in GFP fluorescence to that in DsRed-LC3 fluorescence (see Methods and Figure S2). With the autophagy index, potential changes in the synthesis of the reporter following amino acid deprivations can be normalized. Importantly, our autophagy index tightly correlated with the number of DsRed-LC3 puncta, an established measure of autophagy (Figures 3A–3D and Figure S2).
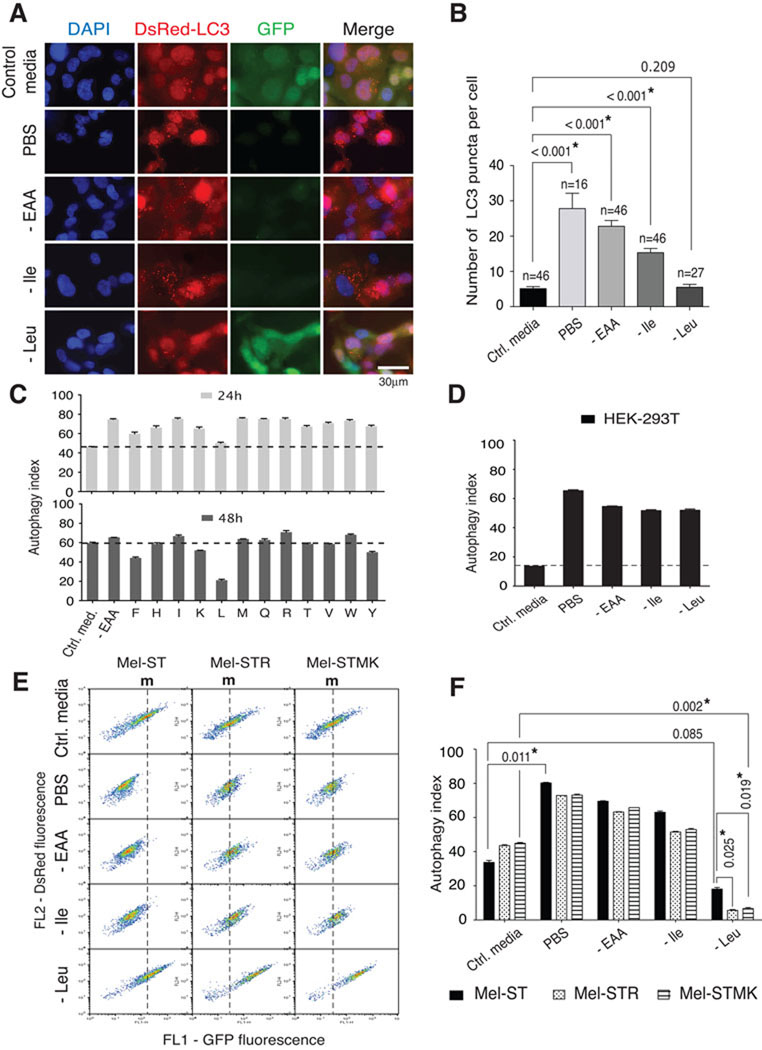
Figure 3. Deprivation of leucine does not activate autophagy in melanoma cells with activated Ras-MEK signaling.
(A) Fluorescent micrographs showing autophagy markers. Control, complete RPMI-1640 media control; PBS, phosphate buffered saline; -EAA, deprivation of all essential amino acids, -Ile, deprivation of isoleucine; -Leu, deprivation of leucine. DAPI, cell nuclei; DsRed-LC3, red fluorescence from DsRed-LC3 puncta; GFP, green fluorescence from the uncleaved DsRed-LC3-GFP reporter; Merge, merged image of DAPI, DsRed, and GFP signals. (B) Quantitation of DsRed-LC3 puncta. Bar graphs display the mean ± s.d. of DsRed-LC3 puncta per cell following each type of nutrient starvation. The numbers of cells examined are indicated. (C) Flow cytometric analyses of autophagic activity. The bar graphs show mean ± s.d. of autophagy indexes obtained after deprivation of single essential amino acids for 24 or 48 hours (n = 3). Dotted lines indicate the autophagy index of cells incubated in control media. (D) Autophagy index in HEK-293T cells following PBS incubation or indicated amino acid deprivations. (E) Flow cytometric quantitation of the autophagy activity. m, marks line indicating median fluorescence intensity of FL1 (GFP fluorescence) in cells in the control media. (F) Bar graphs show mean ± s.d. of the autophagy index (n = 3) and * indicates values that are significantly different from controls. See also Figure S2.
Mel-ST cells displayed a steady state level of autophagic activity with a ~40% autophagy index when growing in control media. As expected, the autophagy index increased (to 70 – 80%) following deprivation of all amino acids or most single amino acids. In contrast, leucine deprivation failed to significantly activate autophagy (Figures 3A – 3C). Immunoblot analyses also showed that deprivation of just leucine did not reduce the level of the full-length DsRed-LC3-GFP reporter while deprivation of all amino acids or just isoleucine did (Figure S2B), indicating a defect in the regulation of autophagy upon leucine deprivation. This difference is unlikely due to a change in proteasomal activity because deprivation of all amino acids, isoleucine, or leucine equally affected the levels of Cyclin D1, a short-lived protein whose turnover is regulated by the ubiquitin-proteasome pathway (Figure S1A and Figure S2B)(Alao, 2007; Diehl et al., 1997). In HEK-293T cells, the capacity of leucine deprivation to induce autophagy was indistinguishable from that of isoleucine, methionine, or all amino acids (Figure 3D). Thus, in a melanocyte-derived cell line, leucine is exceptional amongst the amino acids in that its deprivation does not activate autophagy.
It also quickly became apparent that, compared to the parental Mel-ST cells, the Mel-STR and Mel-STMK cells have a significant defect in autophagy upon nutrient withdrawal (Figures 3E and 3F). In these engineered melanoma cells, PBS incubation, complete amino acid deprivation, and isoleucine deprivation activated autophagy to smaller degrees than the same treatments did in parental Mel-ST cells (Figures 3E and 3F). Importantly, upon leucine deprivation, autophagy levels were even more greatly suppressed in the engineered melanoma cells, indicating that leucine deprivation has a profound impact on autophagy in the melanocyte-derived lines with activated RAS-MEK signaling (Figures 3E and 3F).
Constitutive activation of MEK renders the mTORC1 pathway resistant to leucine deprivation
Because mTORC1 suppresses autophagy (Kamada et al., 2000; Noda and Ohsumi, 1998) and amino acids activate mTORC1 (Hara et al., 1998), we asked if inappropriate regulation of mTORC1 might explain why leucine deprivation does not stimulate autophagy in cells with activated RAS-MEK signaling. We monitored mTORC1 activity by measuring the phosphorylation of S6K1 at Thr-389, a site which mTORC1 directly phosphorylates (Burnett et al., 1998). In the same cells we also monitored autophagy activity by examining endogenous LC3-I to LC3-II conversion and the eventual degradation of LC3 (Figure 4A). Deprivation of all amino acids acutely suppressed mTORC1 and activated autophagy in Mel-ST cells. Mel-STMK cells behaved similarly except that these cells maintained some residual mTORC1 activity even after a long period of essential amino acid deprivation (Figure 4A). The more interesting findings, however, were obtained upon leucine deprivation. In contrast to all amino acid deprivation, leucine deprivation was a much poorer inhibitor of mTORC1 and consequently inducer of autophagy (Figure 4A). Strikingly, in Mel-STMK cells, leucine deprivation barely inhibited mTORC1 activity. Consistent with the results obtained with the autophagy reporter, leucine deprivation did not cause substantial LC3-I to LC3-II conversion or loss of endogenous LC3 in Mel-STMK cells (Figure 4A).
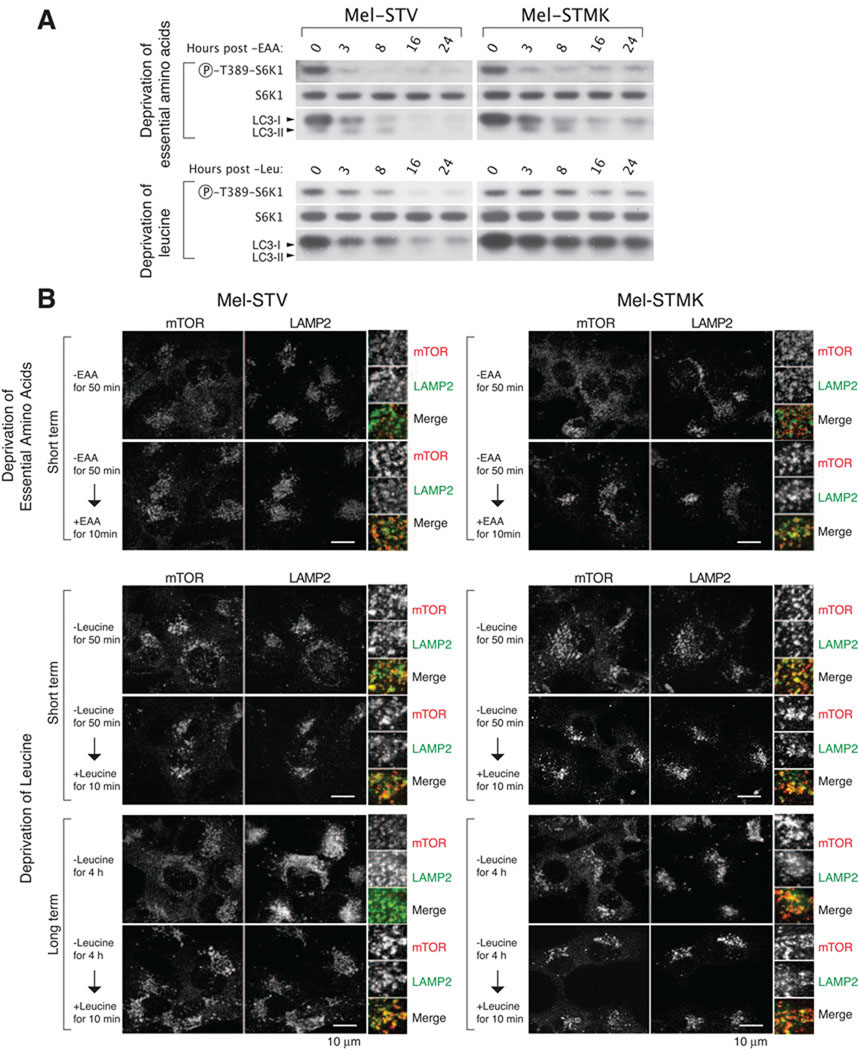
Figure 4. Deregulated activation of the mTORC1 pathway by constitutively active MEK correlates with the inappropriate localization of mTOR to the lysosomal surface.
(A) Immunoblot analyses showing time-dependent changes in mTORC1 and autophagy activity in indicated cell types following deprivation for all essential amino acids or leucine. (B) Immunofluorescence analyses showing mTOR localization upon the deprivation of all essential amino acids (-EAA) or leucine (-Leu). Cells were deprived of indicated amino acids for short (50 minutes) or long (4 hours) periods of time, and re-fed with the amino acids for 10 minutes before processing for co-immunostaining for mTOR (red) and LAMP2 (green), and imaging. In all images, insets show selected fields that were magnified five times and their overlay. Scale bar = 10 µm. See also Figure S3.
To further investigate why leucine deprivation fails to suppress mTORC1 signaling in cells that have constitutively active MEK, we examined the amino acid-sensitive translocation of mTORC1 to the lysosomal surface (Figure 4B and Figure S3 for high resolution images and quantitation). Recent work indicates that the key event in amino acid signaling to mTORC1 is the amino acid-induced movement of mTORC1 to lysosomal membranes where it can interact with its activator, Rheb, a small GTPase (Sancak et al., 2010). Constitutive targeting of mTORC1 to the lysosomal surface is sufficient to render the mTORC1 pathway insensitive to amino acid levels (Sancak et al., 2010). As expected, mTORC1 did not co-localize with the lysosomal marker LAMP2 in Mel-ST and Mel-STMK cells deprived of all amino acids for 50 minutes (Figure 4B). In contrast, in both lines, the deprivation of just leucine for 50 minutes did not greatly affect the co-localization of mTORC1 with lysosomes. However, after depriving the cells of leucine for a longer period of time (4 hours), the two lines diverged in their behavior: in Mel-ST cells, mTORC1 no longer co-localized with lysosomes while in Mel-STMK cells mTORC1 remained lysosome-associated (Figure 4B). These findings are consistent with the RAS-MEK pathway impacting mTORC1 upstream of the leucine-sensitive machinery that regulates the subcellular localization of mTORC1.
Inappropriate activation of the mTORC1 and RAS-MEK pathways confers sensitivity to apoptosis upon leucine deprivation
To determine if the failure to suppress the mTORC1 pathway in melanoma cells with activated RAS-MEK signaling causes cell death, we examined the effects of small molecule inhibitors of the signaling pathways. Mel-STR cells treated with rapamycin or U-0126 not only reactivated autophagy (Figure 5A) but also suppressed caspase-3 activation (Figure 5B). Moreover, across several patient-derived melanoma lines, rapamycin and U-0126 were equally effective at suppressing the cleavage of caspase-3 caused by leucine deprivation (Figures 5C, 5D, and 5E). Importantly, the mTORC1 or MEK inhibitor significantly increased the survival of leucine-deprived melanoma cells (Figures 5F, 5G, and 5H).
Figure 5. The inhibition of autophagy synergizes with low leucine concentrations in inducing apoptosis in melanoma cells.
(A) Bar graphs displaying the autophagy index in the presence or absence of rapamycin (RAPA) or U-0126. (B–E) Immunoblot analyses for cleavage and activation of caspase-3 and cleavage of PARP. (F–H) Cell survival assay. (I) Immunoblot analyses showing validation of shRNA-mediated knockdowns of ATG1. (J) Knockdown of ATG1 mimics effects of expressing Ras-G12V or MEK1-Q56P in sensitizing Mel-ST cells to caspase-3 activation upon leucine deprivation. (K) Immunoblots show cleavage of caspase-3 and PARP in cells incubated with decreasing amounts of leucine in the presence or absence of chloroquine. (L) Chloroquine (CQ) sensitizes A-2058 melanoma cells to partial leucine deprivation. Immunoblots show and graph quantitates activation of caspase-3 in relation to leucine concentration in media with or without chloroquine. (M) Percent survival of A-2058 cells cultured under indicated conditions for 2 days. Data are represented as mean ± s.d. and * indicates values that are significantly different from controls. See also Figure S4.
Autophagy inhibition mimics activated RAS-MEK signaling in conferring sensitivity to leucine deprivation
Upon leucine deprivation, autophagy is more strongly inhibited in Mel-STR than Mel-ST cells (Figures 3E and 3F). To determine if this difference is sufficient to confer on Mel-STR cells the capacity to trigger caspase-3 cleavage upon leucine withdrawal, we asked if the suppression of autophagy sensitizes, like RAS-MEK pathway activation, Mel-ST cells to leucine deprivation. To inhibit autophagy we employed two distinct shRNAs targeting ATG1 (autophagy related gene 1, also known as ULK1) that greatly reduce ATG1 protein expression (Figure 5I). ATG1 is an evolutionarily conserved, Ser/Thr protein kinase that plays an essential role in the early stages of autophagy (Chan et al., 2007; Matsuura et al., 1997). Indeed, the knockdown of ATG1 was as effective as the expression of Ras-G12V or MEK1-Q56P in sensitizing Mel-ST cells to leucine deprivation (Figure 5J). Similar results were obtained by knocking down VPS34, the class III phosphoinositide 3-kinase (PI3K) (Figure S4A). These results confirm that a particular level of autophagy is necessary for cells to survive essential amino acid deprivation, and suggest that, when deprived of leucine, melanoma cells with activated RAS-MEK fall below this threshold.
A small molecule inhibitor of autophagy sensitizes melanoma cells to partial leucine deprivation in vitro
To explore the potential therapeutic implications of the finding that melanoma cells die upon complete leucine deprivation, we needed to identify a way to sensitize melanoma cells to partial leucine deprivation as it is currently not possible to completely deprive, in vivo, cancer cells of extracellular leucine. As all cells have a basal level of autophagy that likely contributes to the recycling of essential amino acids, we reasoned that if we partially suppressed autophagy with a small molecule inhibitor the melanoma cells might induce apoptosis even if some leucine remained in the extracellular environment.
The feasibility of this idea was investigated using chloroquine, a small molecule inhibitor of autophagy. Chloroquine is a lysosomotrophic drug that inhibits the late stages of the autophagy pathway (Boya et al., 2005) and is currently in many clinical trials as an anti-cancer agent (clinicaltrials.gov). As expected, in A-2058 cells, chloroquine increased the levels of p62/SQSTM1/Sequestosome-1 and prevented loss of LC3-II (Figure S5A). By directly binding to LC3, p62 is incorporated onto autophagosomes and degraded (Bjorkoy et al., 2005) so that the level of p62 inversely correlates with autophagic activity (Mizushima et al., 2010).
To ask if chloroquine can induce apoptosis of melanoma cells upon a partial depletion of leucine, we first determined the greatest concentration of extracellular leucine that is still low enough to trigger apoptosis of the melanoma cells in culture (Figures 5K and 5L). We examined a range of leucine concentrations: 380 µM (the concentration of leucine in RPMI media); 120 µM (approximately the plasma leucine concentration of mice fed a normal diet); and 60 µM (approximately the plasma leucine concentration of mice fed an isocaloric, leucine-free synthetic diet (Anthony et al., 2004)). In the absence of chloroquine, only an extracellular leucine concentration of 30 µM or below was able to activate caspase-3 cleavage (Figures 5K and 5L). In contrast, when also treated with a moderate amount of chloroquine, A-2058 and Mel-STR cells triggered caspase-3 activation even when cultured in media containing 60 µM leucine (Figures 5K and 5L). When completely deprived of leucine and treated with chloroquine, Mel-ST cells did cleave some caspase-3 (Figures 5K), consistent with the results obtained upon the knockdowns of ATG1 and VPS34 (Figure 5J and Figure S4A). Interestingly, the combination of chloroquine treatment with the deprivation of all amino acids or just methionine also promoted apoptosis, albeit to a smaller extent than that caused by chloroquine and leucine deprivation (Figure S4B). Most importantly, the combination of media containing 60 µM leucine and chloroquine synergistically decreased the survival of A-2058 cells (Figure 5M). Collectively, these results demonstrate that chloroquine-mediated suppression of autophagy sensitizes melanoma cells to the levels of plasma leucine that can be achieved by feeding animals a leucine-deficient diet (Anthony et al., 2004).
Dietary leucine deprivation and autophagy inhibition synergistically target melanoma xenografts in vivo
To assess the potential of the combination strategy in vivo, A-2058 cells were injected subcutaneously into immunocompromised mice to establish tumor xenografts. When the tumors were about ~100 mm3 in volume, the host animals were fed: (1) a control diet, which consisted of a leucine-free diet supplemented with leucine; (2) an isocaloric leucine-free diet; (3) the control diet and treated with chloroquine; or (4) the leucine-free diet and treated with chloroquine (Figures 6). The amount of chloroquine used was 60 mg/kg body weight, which is similar to the dose employed by others (Amaravadi et al., 2007) and which in our hands caused no obvious toxicity to the animal. Chloroquine treatment did inhibit autophagy in vivo because an immunohistochemical assay revealed the expected increase in p62 levels in the tumors of chloroquine-treated mice (Figure S5B).
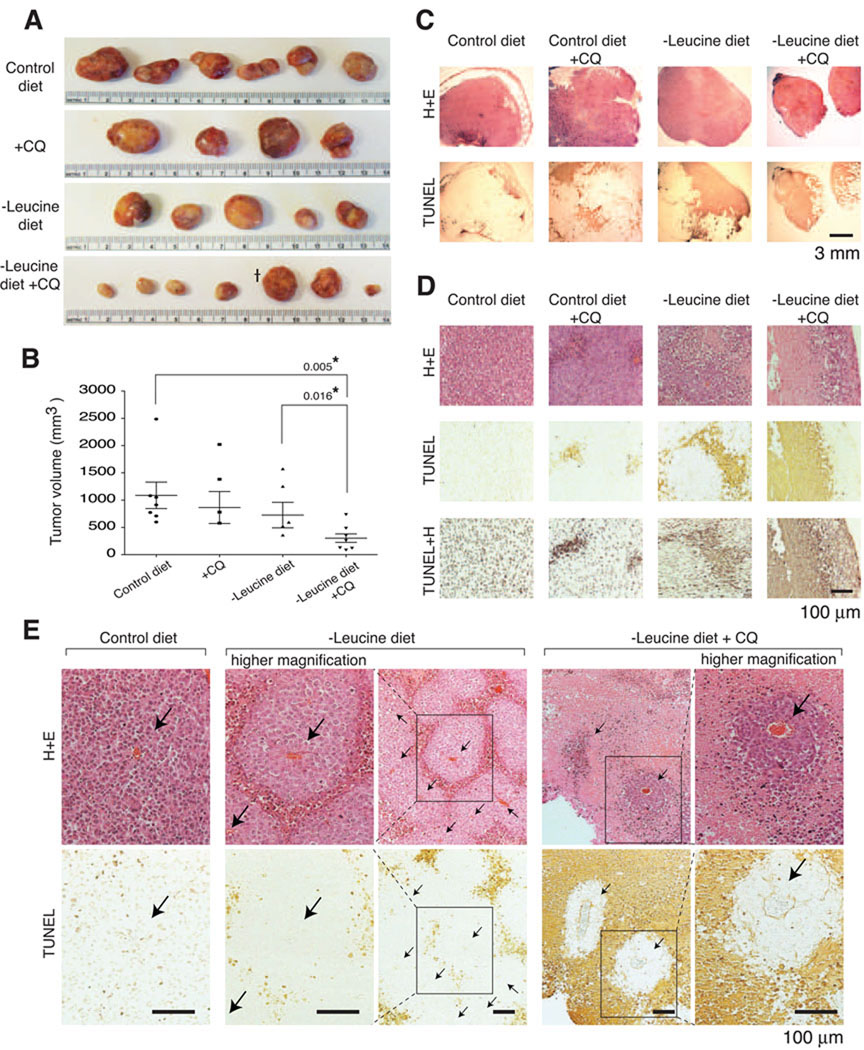
Figure 6. Synergistic inhibition of melanoma tumor growth in mice deprived of dietary leucine and treated with an autophagy inhibitor.
(A) Photographs of excised tumor xenografts following feeding for 14 days with an isocaloric control diet with added leucine (Control diet), control diet plus chloroquine (+CQ), leucine-free diet (-Leucine diet), or leucine-free diet plus chloroquine (-Leucine diet +CQ). (B) Column scatter dot graph displays the mean ± s.e.m. volume of the tumors. * indicates volumes that are significantly different from controls. Note: tumor that is third from the right in the − Leucine diet + CQ group had a flattened disc shape rather than the spherical shape of the large tumors obtained in the other groups. Thus, it appears deceptively large in the photograph. (C) In situ TUNEL assay. H+E, representative micrograph images of tumor sections stained with hematoxylin and eosin; TUNEL, representative images of tumor sections processed in the TUNEL assay; TUNEL+H, representative images of TUNEL results counterstained with hematoxylin. Scale bar = 3 mm. (D) Representative high magnification micrographs of tumor sections showing TUNEL-positive, apoptotic regions. Scale bar = 100 µm. (E) Apoptosis of the melanoma cells inside tumors correlates with the distance from tumor capillaries, and inhibition of autophagy significantly shrinks the viable cuffs surrounding tumor capillaries. Micrographs show corresponding high and low magnification images of tumor sections with capillaries indicated (with arrows) and TUNEL-positive, apoptotic regions. Scale bar =100 µm.
On its own, dietary leucine deprivation did not significantly affect tumor size. In contrast, the tumors in the mice treated with a combination of dietary leucine deprivation and chloroquine were significantly smaller than those in mice in the control groups (Figures 6A and 6B). The inability of a leucine-free diet to reduce tumor growth on its own likely reflects the fact that a leucine-free diet decreases plasma leucine levels from ~133 to ~76 µM (Anthony et al., 2004), which our in vitro results show is not low enough on its own to induce significant death of melanoma cells (Figures 5L and 5M).
To determine if the synergistic effects on tumor size of dietary leucine deprivation and chloroquine treatment reflect the increased death of the melanoma cells, we stained the tumor sections with an in situ TUNEL assay (Figures 6C – 6E). Analogous to the results in culture (Figures 5L and 5M), the combination treatment had a strong pro-death effect in vivo, so that extensive TUNEL-positive staining was observed in nearly all areas of the tumors (Figure 6C). Only cells in the outer shell of the tumors and in the immediate vicinity to the microcapillaries appeared to be spared (Figures 6D and 6E). This pattern of survival likely reflects that melanoma cells in cuffs surrounding the tumor vessels have access to greater amounts of leucine than cells farther away from the blood supply. Dietary leucine deprivation alone showed a significant, but less effective induction of death of the melanoma cells in vivo, and chloroquine treatment alone promoted the death of the cells in only a few isolated areas of the tumor (Figures 6C and 6D).
Immunohistochemical analyses of the tumors with a site-specific (D175) cleaved caspase-3 antibody revealed that, like in vitro, partial leucine deprivation in combination with chloroquine treatment caused caspase-3 cleavage in vivo (Figures 7A and 7B). The cleaved caspase-3 signal was highest at the border between the viable cuffs surrounding capillaries and the large areas of strongly TUNEL-positive dead cells (Figure 7B). This pattern suggests that caspases likely initiate cell death but as the apoptotic program progresses the amount of cleaved caspase-3 drops while the DNA fragments in the apoptotic bodies persist.
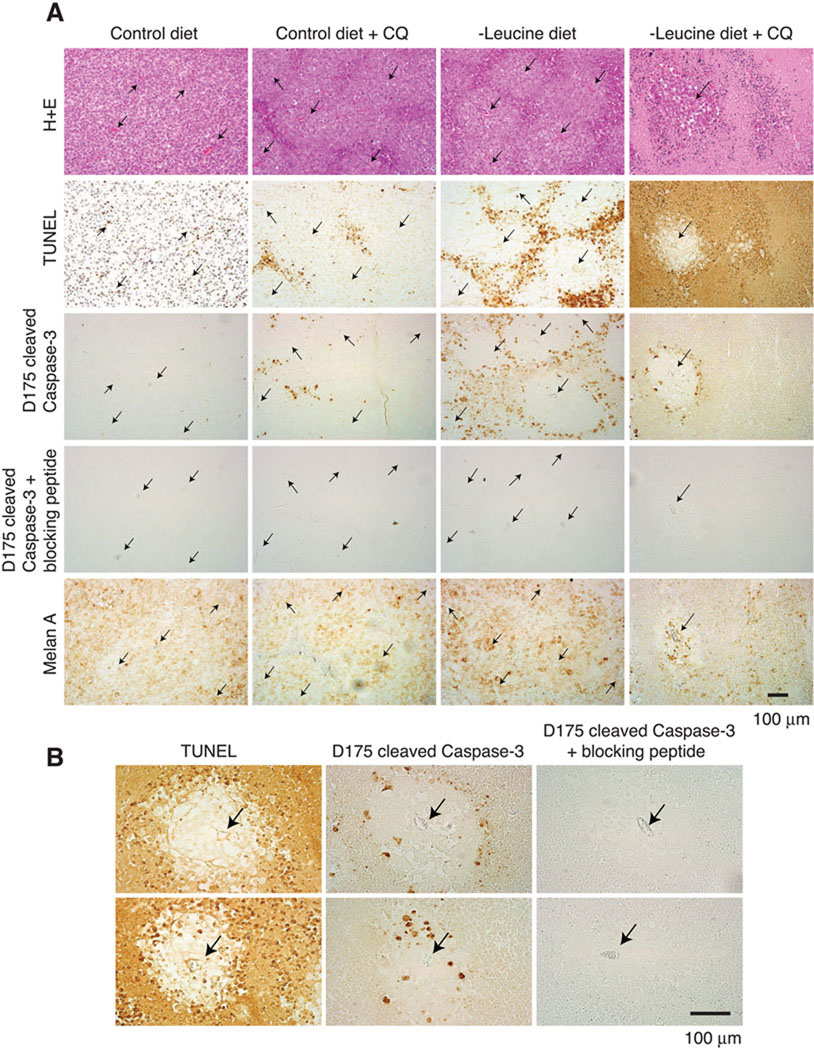
Figure 7. Combination of dietary leucine deprivation and autophagy inhibition induces activation of caspase-3 in melanoma tumors in vivo.
(A) Immunohistochemical analyses showing caspase-3 cleavage in vivo. H+E, images of tumor sections stained with hematoxylin and eosin where capillaries are denoted with arrows; TUNEL, images of tumor sections processed for the TUNEL assay; D175 cleaved Caspase-3, images of tumor sections stained for active caspase-3 with the anti-Asp-175 site-specific cleaved caspase-3 antibody; D175 cleaved Caspase-3 + blocking peptide, images of tumor sections stained for active caspase-3 with the anti-Asp-175 site-specific cleaved caspase-3 antibody that was pre-incubated with the epitope blocking peptide; Melan A, images of tumor sections stained with anti-Melan A, a human melanocyte specific marker, antibodies. Scale bar = 100 µm. (B) Representative high magnification micrographs of tumor tissues showing geographic correlation between the TUNEL-positive signals and the D175 cleaved caspase-3 positive signals where capillaries are denoted with arrows. Scale bar =100 µm. See also Figure S5.
Most of the live cells within the tumors stained for the human-specific melanocyte marker, Melan-A, except for the murine endothelial cells of the capillaries and a thin layer of cells on the tumor surfaces (Figure 7A and Figure S5C). These Melan-A-negative murine cells did not stain for cleaved caspase-3, indicating that the combination of dietary leucine deprivation and chloroquine treatment did not affect the non-transformed cells of the tumors (Figure S5C).
DISCUSSION
There is mounting interest in the roles nutrient-sensing and metabolic pathways play in tumorigenesis and in the potential of these pathways to harbor targets for cancer therapies. Autophagy, for example, is increasingly recognized as important for eukaryotic cells and organisms to survive periods of nutrient withdrawal (Boya et al., 2005; Degenhardt et al., 2006; Kuma et al., 2004) and small molecule inhibitors of autophagy, such as chloroquine, are of interest for anti-cancer uses (reviewed in (Rubinsztein et al., 2007)).
In examining how cancer cells respond to deprivation of single essential amino acids, we made the observation that the deprivation of leucine, but not other essential amino acids, induces apoptosis in all the human melanoma lines studied. Substantial evidence suggests that leucine deprivation triggers apoptosis because, unlike in other cell types, it does not inhibit the mTORC1 pathway and thus does not activate autophagy. In fact, the mTORC1 inhibitor rapamycin—normally thought of as an anti-cancer agent— reactivates autophagy in leucine-deprived melanoma cells and promotes their survival. It is odd that the mTORC1 pathway is so resistant to leucine withdrawal in melanoma cells because this amino acid is a canonical activator of the pathway and its deprivation inhibits mTORC1 signaling in a wide-variety of normal and transformed cells (Guertin and Sabatini, 2007). Amino acid signaling to mTORC1 is a subject of intense study, but the amino acid sensing mechanism remains a mystery. It is possible that once the mechanism is understood it will be found to be different in melanoma cells than in other cell types. The hyperactivation of the RAS-MAPK pathway that is a common occurrence in melanoma cells clearly contributes to the insensitivity of mTORC1 to leucine deprivation. So far, our work suggests that RAS-MEK signaling perturbs the normally leucine-sensitive localization of mTORC1 to the lysosomal surface. Kinases that are part of the MAPK pathway, such ERK and p90 RSK1, phosphorylate and repress the function of TSC2, a tumor suppressor that is a negative regulator of mTORC1 (Inoki et al., 2003; Ma et al., 2005; Roux et al., 2004; Tee et al., 2003). However, TSC2 does not appear to play a major role in amino acid signaling to mTORC1 (Byfield et al., 2005; Nobukuni et al., 2005; Smith et al., 2005) so it is likely that the MAPK pathway affects the leucine sensitivity of mTORC1 in melanoma cells through TSC2-independent mechanisms. Interestingly, the RAS pathway negatively regulates autophagy in budding yeast and in flies, but it is unclear if the TORC1 pathway is involved in autophagy repression in these organisms (Berry and Baehrecke, 2007; Budovskaya et al., 2004).
Autophagy inhibition alone did not trigger apoptosis in melanoma cells in vitro, and chloroquine treatment failed to show anti-tumor effects in mice fed a control diet in vivo. This is likely because most cells inside tumors have access to more than the minimal level of extracellular leucine required for survival. On the other hand, in animals fed a leucine free diet, the inhibition of autophagy likely results in little leucine being liberated from internal sources so that cellular levels of this essential amino acid may fall below the threshold needed for survival. To implement this combination strategy in the current study we used chloroquine to inhibit autophagy. One caveat of chloroquine however is that as a lysosomotrophic compound it may not only inhibit the autophagic process but also non-autophagy related functions of lysosomes. Inhibitors to proteins essential for autophagy, such as the protease, ATG4, and the kinases, ATG1/ULK1 and VPS34, are likely to be developed in the future. Using RNAi we have shown that ATG1 and VPS34 are important for determining the sensitivity of melanoma cells to leucine deprivation.
To deprive the melanoma xenografts of leucine we fed mice a leucine-free diet that is known to reduce the plasma leucine concentration in rodents and humans without greatly affecting blood insulin levels (Anthony et al., 2004; Guo and Cavener, 2007; Hambraeus et al., 1976). A leucine-free diet is unlikely to be the ideal approach to deprive tumors of leucine in a clinical setting. In the future it may be possible to deliver, intravenously, enzymes that specifically degrade leucine or small molecule inhibitors of leucine uptake. As a model for the former, asparaginase (L-asparagine amidohydrolase) is an FDA-approved enzyme that hydrolyzes asparagine to aspartic acid and is a successful therapy for acute lymphocytic leukemia (ALL). Enzymes in the leucine catabolic pathway, such as BCAT (branched chain aminotransferase) (Berg, 2007), could be used in an analogous way if their substrate specificity could be engineered to be limited to leucine (Conway et al., 2003; Onuffer and Kirsch, 1995). There appear to be many transporters that mediate leucine uptake as well (Broer, 2008), and some of the better characterized ones, such as LAT1, may be druggable. Alternatively, the easiest strategy to obtain synergistic effects with autophagy inhibition and nutrient deprivation on tumor cell survival may be to co-administer a drug that can deprive tumor cells of many nutrients, such as an angiogenesis blocker, along with a specific autophagy inhibitor. Our finding that in the presence of an autophagy inhibitor melanoma cells trigger the activation of caspase-3 when deprived not only of leucine but also of all essential amino acids, supports the feasibility of this idea.
EXPERIMENTAL PROCEDURES
Materials
Reagents were obtained from the following sources: Adriamycin, Rapamycin, and U-0126 from Calbiochem and LC Laboratories; Cell culture grade pure amino acids, glucose, and chloroquine from Sigma; JC-1 dye from Invitrogen; Immunohistochemistry kits from Vector Laboratories and Dako; cDNA clones for MEK1, ATG1, and LC3 from Open Biosystems; cDNA clone for H-RAS-G12V from the Laboratory of Dr. Robert Weinberg (Whitehead Institute); cDNA clones for BRAF-WT, BRAF-Δ3, BRAF-V600E, and BRAF-Δ3-V600E from Dr. David Tuveson (Cancer Research UK) (Karreth et al., 2009); Lentiviral shRNA constructs from The RNAi Consortium (Broad Institute); antibodies to SQSTM1/p62 from American Research Products; antibodies to ATG1/ULK1, Bcl-xL, caspase-3, cyclin D1, LC3, Melan-A, PARP, phospho-T202/Y204-ERK, ERK, phopho-T389 S6K1, S6K1, VPS34, as well as HRP-conjugated anti-mouse, anti-rabbit secondary antibodies from Santa Cruz Biotechnology Inc. and Cell Signaling Technology.
Cell lines and Tissue culture
Cell lines were obtained from the American Type Culture Collection. Cell culture media powder and sera were purchased from the following sources: Dulbecco’s MEM (DMEM), RPMI-1640, fetal bovine serum (FBS), dialyzed fetal bovine serum (dFBS), heat-inactivated fetal bovine serum (IFS) from Invitrogen; Amino acid-free, glucose-free RPMI-1640 from US Biological, Inc. Cells were cultured in the following media: HEK-293T cells in DMEM with 10% IFS; A-2058, SK-MEL-3, SK-MEL-5, SK-MEL-28, Mel-ST, and Mel-ST-derivatives in DMEM with 10% FBS. For survival assay, we treated cells with 20 µM Q-VD-OPH or 100 µM Z-VAD-fmk when depriving cells of essential amino acids.
Essential amino acid deprivation protocol
To produce cell culture media deficient of single essential amino acids, we reconstituted the amino acid-free, glucose-free RPMI-1640 media by supplementing it with glucose and individual amino acids except the amino acid to be omitted. Cells were plated in the complete culture media one day prior to the amino acid deprivation experiments so that the plated cells reached ~80% of confluency at the day of experiment. To deprive cells of single amino acids, we replaced culture media twice with target amino acid-free RPMI-1640 media and incubated cells until sampling for analysis.
Autophagy assay using the DsRed-LC3-GFP reporter
To develop a dual-color autophagy reporter, we inserted rat LC3 (also known as ATG8) cDNA in between the cDNAs for DsRed and EGFP so that the DsRed protein is fused with N-terminus of LC3 protein and C-terminus of the protein is connected to EGFP. When indicated, we introduced a deletion of five amino acids (TALAV) at the ATG4-recognition site near C-terminus of LC3 to make a chimeric protein resistant to ATG4-mediatd cleavage. To produce stable cell lines continuously reporting autophagy activity, recombinant retroviruses expressing the DsRed-LC3-GFP reporter were generated and used to infect target cells. The autophagy index is a measure of the relative change in median fluorescence intensity of GFP to that of DsRed and was calculated with the formula: autophagy index = 100 − (100 X (FL1/FL2)), where FL1 is Fluorescence 1 (the median fluorescence intensity of GFP fluorescence) and FL2 is Fluorescence 2 (the median fluorescence intensity of DsRed fluorescence). See Figure S2 for details on the development and validation of the autophagy reporter.
Flow cytometric analyses
Analysis of apoptosis induction using Annexin-V-fluorescein was carried out according to the assay kit manufacturer’s instructions (Roche). Briefly, cultured cells were harvested and washed once in PBS, then incubated with the Ready-to-Use solution of Annexin-V-Fluorescein in a HEPES buffer containing Propidium Iodide. Samples were analyzed using a flow cytometer. Changes in mitochondrial outer membrane permeabilization (MOMP) were measured using the MOMP-sensitive cationic JC-1 dye according to the supplier’s instruction (Roche). JC-1 exhibits membrane potential-dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green (monomeric form in cytosol) to red (aggregates in mitochondria). Briefly, cultured cells were stained with 2 µM JC-1 for 15 minutes at the growth condition, washed with PBS, and analyzed using flow cytometry. When necessary, the concentration of JC-1 was optimized for different cell types using oligomycin as a control.
Immunofluorescence Assay
Mel-ST and Mel-STMK cells were plated on fibronectin-coated glass cover slips in 12-well tissue culture plates. Twenty-four hours later, the slides were rinsed with PBS once and fixed for 15 min with 4% paraformaldehyde in PBS warmed to 37°C. The slides were rinsed twice with PBS and cells were permeabilized with 0.05% Triton X-100 in PBS for 5 minutes. After rinsing twice with PBS, the slides were incubated with primary antibody in 5% normal donkey serum for 2 hours at room temperature, rinsed four times with PBS, and incubated with secondary antibodies produced in donkey (1:400 in 5% normal donkey serum) for 1 hour at room temperature in the dark, washed four times with PBS. Slides were finally mounted on glass cover slips using Vectashield (Vector Laboratories) and images were collected and analyzed on a Perkin Elmer spinning disk confocal microscopy system.
Human xenograft tumor model and dietary leucine deprivation
Immunodeficient mice (NCR nude, nu/nu; Taconic Laboratories) were maintained in a pathogen-free facility and were given autoclaved food and water ad libitum, if not otherwise specified. A-2058 melanoma cells were xenografted into six-week old immunodeficient mice. Briefly, 1 × 106 melanoma cells were resuspended in 200 µl of media and injected subcutaneously in the upper flank region of mice that had been anaesthetized with isoflurane. Tumors were allowed to grow to ~100 mm3 in size and the mice were then initiated on the dietary leucine restriction using an isocaloric leucine free, synthetic diet alone or along with chloroquine treatment. Both the isocaloric control diet with added leucine and the leucine-free synthetic diet were obtained from Research Diet, Inc. Based on a series of preliminary experiments, chloroquine was injected intraperitonealy at 60 mg/kg body weight two times per week. Tumor volumes were estimated with the formula: volume = (2a × b)/2, where a = shortest and b = longest tumor lengths, respectively, in millimeters. When necessary, animals were sacrificed and tumors were harvested and analyzed using standard histology and immunohistochemistry methods. Animal research protocols were approved by the MIT Committee on Animal Care, and all experiments were performed according to the official guidelines of the MIT Committee on Animal Care and the American Association of Laboratory Animal Care.
Lentiviral shRNA-mediated RNAi
For the gene knockdown experiments, we obtained lentiviral shRNA constructs from The RNAi Consortium (TRC) at the Broad Institute and produced recombinant lentiviruses using a transient transfection protocol. Briefly, we transfected HEK-293T cells with the lentiviral shRNA plasmids and the packaging plasmids (pdeltaVPR and pVSVG) according to the TRC standard protocols and used lentiviral supernatants to infect target cells (Moffat et al., 2006).
Statistical analyses
Experimental results were analyzed with a Student’s t-test and graphed using Prism software (GraphPad Software, Inc.). In vitro data are expressed as means ± s.d. and in vivo data as mean ± s.e.m. P < 0.05 was considered statistically significant.
HIGHLIGHTS.
Defective autophagy upon leucine deprivation reveals a liability of melanoma cells.
Leucine deprivation triggers caspase activation and apoptosis of melanoma cells.
mTORC1 and MAPK pathways determine the sensitivity to leucine deprivation.
Dietary leucine deprivation and chloroquine synergistically induce apoptosis in vivo.
SIGNIFICANCE.
Melanoma is a highly aggressive cancer for which additional therapies are needed. Autophagy is a cytoprotective mechanism that may help cancer cells survive in nutrient limiting conditions and is a potential target for anti-cancer therapies. However, in pre-clinical tumor models inhibition of autophagy alone has so far had relatively modest anti-tumor effects. Here, we show that the combination of leucine deprivation and autophagy inhibition induces the caspase-dependent death, in vitro and in vivo, of human melanoma cells driven by the RAS-MEK pathway, but not of non-transformed cells. The results represent a starting point for developing combination therapies involving autophagy inhibitors to target melanoma.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the NIH (R01 CA103866 and CA1299105) to D.M.S., as well as post-doctoral fellowships from the US Department of Defense (W81XWH-04-1-0496) to J.H.S, from the Jane Coffin Childs Memorial Fund for Medical Research to R.Z., and from the American Brain Tumor Association to D.K. We thank members of the Sabatini lab for helpful suggestions, Dr. Robert Weinberg (Whitehead Institute) for reagents and experimental protocols, Dr. David Tuveson (Cancer Research UK) for reagents, and the US Biological, Inc., Whitehead Institute FACS facility, MIT Division of Comparative Medicine, and Histology core facility at the MIT Koch Institute for Integrative Cancer Research for experimental advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TG, McDaniel BJ, Byerley RL, McGrath BC, Cavener DR, McNurlan MA, Wek RC. Preservation of liver protein synthesis during dietary leucine deprivation occurs at the expense of skeletal muscle mass in mice deleted for eIF2 kinase GCN2. J Biol Chem. 2004;279:36553–36561. doi: 10.1074/jbc.M404559200. [DOI] [PubMed] [Google Scholar]
- Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, Flanagan A, Teague J, Futreal PA, Stratton MR, Wooster R. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th ed. W.H.Freeman and Company; 2007. [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottorff D, Stang S, Agellon S, Stone JC. RAS signalling is abnormal in a c-raf1 MEK1 double mutant. Mol Cell Biol. 1995;15:5113–5122. doi: 10.1128/mcb.15.9.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol Rev. 2008;88:249–286. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- Caserta TM, Smith AN, Gultice AD, Reedy MA, Brown TL. Q-VD-OPh, a broad spectrum caspase inhibitor with potent antiapoptotic properties. Apoptosis. 2003;8:345–352. doi: 10.1023/a:1024116916932. [DOI] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chauvier D, Ankri S, Charriaut-Marlangue C, Casimir R, Jacotot E. Broad-spectrum caspase inhibitors: from myth to reality? Cell Death Differ. 2007;14:387–391. doi: 10.1038/sj.cdd.4402044. [DOI] [PubMed] [Google Scholar]
- Conway ME, Yennawar N, Wallin R, Poole LB, Hutson SM. Human mitochondrial branched chain aminotransferase: structural basis for substrate specificity and role of redox active cysteines. Biochim Biophys Acta. 2003;1647:61–65. doi: 10.1016/s1570-9639(03)00051-7. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Eagle H. Amino acid metabolism in mammalian cell cultures. Science. 1959;130:432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, Bazan NG, Blagosklonny MV, Blomgren K, Borner C, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Gong H, Zolzer F, von Recklinghausen G, Havers W, Schweigerer L. Arginine deiminase inhibits proliferation of human leukemia cells more potently than asparaginase by inducing cell cycle arrest and apoptosis. Leukemia. 2000;14:826–829. doi: 10.1038/sj.leu.2401763. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Guo F, Cavener DR. The GCN2 eIF2alpha kinase regulates fatty-acid homeostasis in the liver during deprivation of an essential amino acid. Cell Metab. 2007;5:103–114. doi: 10.1016/j.cmet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambraeus L, Bilmazes C, Dippel C, Scrimshaw N, Young VR. Regulatory role of dietary leucine on plasma branched-chain amino acid levels in young men. J Nutr. 1976;106:230–240. doi: 10.1093/jn/106.2.230. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, DeNicola GM, Winter SP, Tuveson DA. C-Raf inhibits MAPK activation and transformation by B-Raf(V600E) Mol Cell. 2009;36:477–486. doi: 10.1016/j.molcel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Kreis W, Baker A, Ryan V, Bertasso A. Effect of nutritional and enzymatic methionine deprivation upon human normal and malignant cells in tissue culture. Cancer Res. 1980;40:634–641. [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, Ding L, Mardis ER, Wilson RK, Solit D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res. 2008;68:5524–5528. doi: 10.1158/0008-5472.CAN-08-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Ohtawa K, Ueno T, Mitsui K, Kodera Y, Hiroto M, Matsushima A, Nishimura H, Inada Y. Apoptosis of leukemia cells induced by valine-deficient medium. Leukemia. 1998;12:1651–1652. doi: 10.1038/sj.leu.2401139. [DOI] [PubMed] [Google Scholar]
- Onuffer JJ, Kirsch JF. Redesign of the substrate specificity of Escherichia coli aspartate aminotransferase to that of Escherichia coli tyrosine aminotransferase by homology modeling and site-directed mutagenesis. Protein Sci. 1995;4:1750–1757. doi: 10.1002/pro.5560040910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schworer CM, Shiffer KA, Mortimore GE. Quantitative relationship between autophagy and proteolysis during graded amino acid deprivation in perfused rat liver. J Biol Chem. 1981;256:7652–7658. [PubMed] [Google Scholar]
- Scott L, Lamb J, Smith S, Wheatley DN. Single amino acid (arginine) deprivation: rapid and selective death of cultured transformed and malignant cells. Br J Cancer. 2000;83:800–810. doi: 10.1054/bjoc.2000.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee EA, Zhu H, Chow SC, MacFarlane M, Nicholson DW, Cohen GM. Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem J. 1996;315(Pt 1):21–24. doi: 10.1042/bj3150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–217. doi: 10.1016/j.ceb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley PV, Dion RL, Bono VH., Jr Effects of tryptophan deprivation on L1210 cells in culture. Cancer Res. 1974;34:1010–1014. [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.