Abstract
Purpose
Congenital cataracts are a clinically and genetically heterogeneous lens disorder. The purpose of this study was to identify the genetic mutation and the molecular phenotype responsible for the presence of an autosomal dominant congenital nuclear cataract disease in a Chinese family.
Methods
Patients were given a physical examination and their blood samples were collected for DNA extraction. Genotyping was performed by microsatellite markers, and a logarithm of odds (LOD) score was calculated using the LINKAGE programs. Mutation detection was performed by direct sequencing.
Results
Linkage to the crystallin beta A1 (CRYBA1) locus was identified. DNA sequencing of the gene revealed a c.279–281delGAG mutation in exon 4, which resulted in a glycine residue deletion at position 91 (ΔG91). This mutation was identified in all of the affected individuals but was not found in the 100 control chromosomes.
Conclusions
Our results identify that the c.279–281delGAG mutation in CRYBA1 is responsible for the autosomal dominant congenital nuclear cataract disease in this Chinese family.
Introduction
Congenital cataracts are common and are the cause of approximately one third of infant blindness. They are found in approximately 1–6/10,000 live births. One quarter of congenital cataract diseases are hereditary [1-4]. Congenital cataracts are a clinically and genetically heterogeneous lens disorder. Cataracts that are phenotypically identical can result from mutations at different genetic loci and can have different inheritance patterns. Conversely, cataracts with dissimilar phenotypes may result from mutations in a single gene or gene family. It is believed that the type of genetic mutation is related to the morphology of the cataract [5]. To date, approximately 40 genetic loci have been linked to congenital cataracts, and 26 genes have been cloned and sequenced, including crystallins, connexins, heat shock transcription factor-4, aquaporin-0, and the beaded filament structural protein-2 [5].
Recently, a Chinese family in Northeast China has been identified with five generations of congenital nuclear cataracts. This pedigree is further characterized and identified in relation to the recurrent mutation c.279–281delGAG of crystallin, beta A1 (CRYBA1), which is associated with this eye phenotype.
Methods
Patients and clinical data
The five-generation family that enrolled in this study was located in Northeast China. A clinical examination, peripheral blood collection, and DNA extraction were performed on the participating family members, in the Department of Ophthalmology, at the Peking Union Medical College Hospital. The study was performed in accordance with the Declaration of Helsinki and approved by the Institutional Review Board and Ethics Committee of Peking City, and informed consent was obtained from all participants. The recruited family members included fourteen patients that were confirmed with congenital nuclear cataracts. Blood was collected from eight patients and one unaffected family member. The clinical data of these nine subjects was ascertained by detailed ocular examinations.
Genotyping and linkage analysis
DNA was isolated from the blood samples of the nine subjects using the method of phenol-chloroform to extract the DNA. Fluorescently labeled microsatellite markers (Sangon Biotech Co., Ltd., Shanghai, China) were used for linkage analysis of 26 candidate gene regions. A two-point linkage analysis was performed with MLINK, from the LINKAGE program package.
Mutation analysis
The coding exons of CRYBA1 were amplified by a polymerase chain reaction (PCR) and a set of six pairs of primers (Table 1) [6]. Briefly, PCR amplification conditions were: Reaction Mixture Set Up (50 μl); 2 ml (40 ng/ml) of DNA, 1.5 ml (10 μM)) of each exon primer, 20 ml of water , and 25 ml of PCR mix (2× EasyTaq PCR SuperMix; TransGen, Beijing, China) in a final reaction volume of 50 ml. Thermal cycling conditions were: an initial denaturation step at 95 °C for 5 min, 38 cycles (30 min at 95 °C, 30 min at 52 °C, and 30 min at 72 °C) and a final 10 min 72 °C extension.The PCR products were sequenced on an ABI3730 automated sequencer (PE Biosystems, Foster City, CA).
Table 1. Primers used for CRYBA1 amplification.
| Exon | Primer (5′-3′) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|
| CRYBA1-1F |
GGCAGAGGGAGAGCAGAGTG |
|
|
| CRYBA1-1R |
CACTAGGCAGGAGAACTGGG |
207 |
52 |
| CRYBA1-2F |
AGTGAGCAGCAGAGCCAGAA |
|
|
| CRYBA1-2R |
GGTCAGTCACTGCCTTATGG |
293 |
52 |
| CRYBA1-3F |
AAGCACAGAGTCAGACTGAAGT |
|
|
| CRYBA1-3R |
CCCCTGTCTGAAGGGACCTG |
269 |
52 |
| CRYBA1-4F |
GTACAGCTCTACTGGGATTG |
|
|
| CRYBA1-4R |
ACTGATGATAAATAGCATGAACT |
357 |
52 |
| CRYBA1-5F |
GAATGATAGCCATAGCACTAG |
|
|
| CRYBA1-5R |
TACCGATACGTATGAAATCTGA |
290 |
52 |
| CRYBA1-6F |
CATCTCATACCATTGTGTTGAG |
|
|
| CRYBA1-6R | GCAAGGTCTCATGCTTGAGG | 295 | 52 |
Cited from [6]
Results
Clinical findings
We had verified a five-generation family with fourteen confirmed individuals that had congenital cataracts (Figure 1 and Figure 2). Blood samples were collected from eight of the patients. All of the patients that participated have had no other clinically related ophthalmic syndromes.
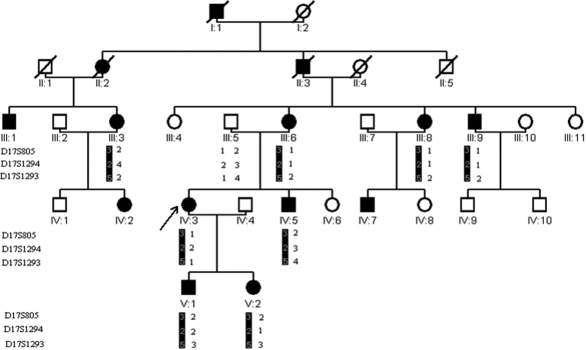
Figure 1.
The pedigree and haplotype of the family. A five-generation pedigree with nine subject family members is shown. Three markers: D17S805, D17S1294, and D17S1293 that are adjacent to CRYBA1 were selected to be analyzed. There was a cosegregation of the diseased haplotype (represented by the black bar) in eight patients of the family. The arrow indicates the proband.
Figure 2.
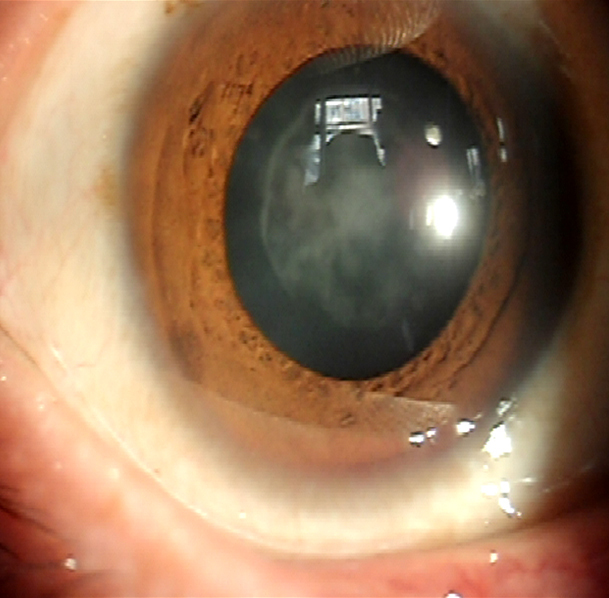
A slit lamp photograph showing the nuclear cataract of a patient III:8 (from Figure 1).
Linkage and haplotype analysis
Positive logarithm of odds (LOD) scores at 17q were obtained (Table 2). A maximum positive LOD score of 1.50 at θ=0.00 was obtained by marker D17S1293. There was a cosegregation of the haplotype in all eight of the affected subjects that were analyzed (in Figure 1).
Table 2. Result of linkage analysis.
|
|
LOD scores at θ= |
|||||
|---|---|---|---|---|---|---|
| Marker | 0.00 | 0.01 | 0.10 | 0.20 | 0.30 | 0.40 |
| D17S805 |
1.38 |
1.35 |
1.08 |
0.78 |
0.50 |
0.23 |
| D17S1294 |
0.94 |
0.91 |
0.69 |
0.45 |
0.25 |
0.09 |
| D17S1293 | 1.50 | 1.46 | 0.69 | 0.80 | 0.48 | 0.21 |
Mutation analysis
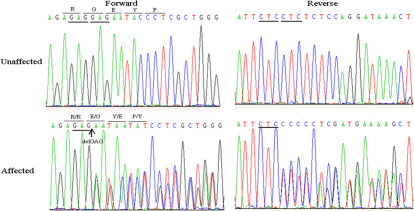
By direct sequencing of the CRYBA1coding region, a c.279–281delGAG mutation was detected (Figure 3). The mutation results in a glycine residue deletion at position 91(ΔG91). The cosegregation of the mutation was only found in the subjects and not in other members of the family, or in any of the 100 control chromosomes that were analyzed from the same ethnic background.
Figure 3.
DNA sequences of CRYBA1 in unaffected and affected individuals.
Discussion
This study identified a recurrent mutation (c.279–281delGAG) in the CRYBA1 gene of a five-generation Chinese pedigree with autosomal dominant nuclear cataract disease. CRYBA1comprises 6 exons that are alternatively spliced to produce 2 proteins (βA1- and βA3-crystallins). The βA1- and βA3-crystallins consist of seven protein domains: four homologous Greek key motifs, a connecting peptide, and NH2- and COOH-terminal extensions. These two proteins are identical except for 17 additional amino acid residues found on the NH2-terminal arm of βA3-crystallin.
In previous reports, three mutations have been identified in CRYBA1 in multiple family and ethnic backgrounds, and have resulted in eye disease [6-15]. One previously identified mutation (c.279–281delGAG) is a 3 bp (GAG) deletion that occurs at the nucleotide positions 279–281, and results in an in-frame deletion of a glycine residue at position 91 (ΔG91). This mutation has been identified in an English family with lamellar cataracts [8], a Chinese family with nuclear cataracts [7], a Swiss family with suture-sparing nuclear cataracts [6], and two Chinese families with pulverulent nuclear cataracts and pulverulent lamellar cataracts [9]. Reddy et al. [8] studied the mechanism of the mutation of p.G91del in CRYBA1, and found that the mutant protein created defective folding and a reduction in solubility.
Two different mutations were identified that altered the same position in the third IVS (IVS3+1G>A and IVS3+1G>C), which caused the G residue to alter into either an A or C. The IVS3+1G>A mutation was detected in an Indian family with zonular sutural cataracts [10], an Australian family with nucleus and Y-sutural opacities [12], a Chinese family with posterior polar cataracts [14], a Chinese family with progressive cataracts [15],and two Indian families with zonular lamellar opacification [13]. The IVS3+1G>C mutation was present in a Brazilian family with pulverulent cataracts [11].
Since the mutations c.279–281delGAG and IVS3+1 have been found in multiple families, it suggests that these two locations are hot-point mutation sites.
In conclusion, the c.279–281delGAG mutation in CRYBA1 is responsible for the pedigree of this Chinese family. It can be further argued that CRYBA1is responsible for autosomal dominant congenital nuclear cataract disease.
References
- 1.Rahi JS, Dezateux C, British Congenital Cataract Interest Group Measuring and interpreting the incidence of congenital ocular anomalies: lessons from a national study of congenital cataract in the UK. Invest Ophthalmol Vis Sci. 2001;42:1444–8. [PubMed] [Google Scholar]
- 2.Wirth MG, Russell-Eggitt IM, Craig JE, Elder JE, Mackey DA. Aetiology of congenital and paediatric cataract in an Australian population. Br J Ophthalmol. 2002;86:782–6. doi: 10.1136/bjo.86.7.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert SR, Drack AV. Infantile cataracts. Surv Ophthalmol. 1996;40:427–58. doi: 10.1016/s0039-6257(96)82011-x. [DOI] [PubMed] [Google Scholar]
- 4.Rahi JS, Dezateux C. Congenital and infantile cataract in the United Kingdom: underlying or associated factors. Invest Ophthalmol Vis Sci. 2000;41:2108–14. [PubMed] [Google Scholar]
- 5.Hejtmancik JF. Cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–49. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrini W, Schorderet DF, Othenin-Girard P, Héon E, Munier FL. CRYBA3/A1 gene mutation associated with suturesparing autosomal dominant congenital nuclear cataract: a novel phenotype. Invest Ophthalmol Vis Sci. 2004;45:1436–41. doi: 10.1167/iovs.03-0760. [DOI] [PubMed] [Google Scholar]
- 7.Qi Y, Jia H, Huang S, Lin H, Gu J, Su H, Zhang T, Gao Y, Qu L, Li D, Li Y. A deletion mutation in the betaA1/A3 crystallin gene (CRYBA1/A3) is associated with autosomal dominant congenital nuclear cataract in a Chinese family. Hum Genet. 2004;114:192–7. doi: 10.1007/s00439-003-1049-7. [DOI] [PubMed] [Google Scholar]
- 8.Reddy MA, Bateman OA, Chakarova C, Ferris J, Berry V, Lomas E, Sarra R, Smith MA, Moore AT, Bhattacharya SS, Slingsby C. Characterization of the G91del CRYBA1/3-crystallin protein: a cause of human inherited cataract. Hum Mol Genet. 2004;13:945–53. doi: 10.1093/hmg/ddh110. [DOI] [PubMed] [Google Scholar]
- 9.Lu S, Zhao C, Jiao H, Kere J, Tang X, Zhao F, Zhang X, Zhao K, Larsson C. Two Chinese families with pulverulent congenital cataracts and deltaG91 CRYBA1 mutations. Mol Vis. 2007;13:1154–60. [PubMed] [Google Scholar]
- 10.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the BA1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 11.Bateman JB, Geyer DD, Flodman P, Johannes M, Sikela J, Walter N, Moreira AT, Clancy K, Spence MA. A new BA1-crystallin splice junction mutation in autosomal dominant cataract. Invest Ophthalmol Vis Sci. 2000;41:3278–85. [PubMed] [Google Scholar]
- 12.Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE, Elder JE, Dickinson JL, Sale MM. Investigation of crystallin genes in familial cataract, and report of two disease associated mutations. Br J Ophthalmol. 2004;88:79–83. doi: 10.1136/bjo.88.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devi RR, Yao W, Vijayalakshmi P, Sergeev YV, Sundaresan P, Hejtmancik JF. Crystallin gene mutations in Indian families with inherited pediatric cataract. Mol Vis. 2008;14:1157–70. [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Z, Ji B, Wan C, He G, Zhang J, Zhang M, Feng G, He L, Gao L. A splice site mutation in CRYBA1/A3 causing autosomal dominant posterior polar cataract in a Chinese pedigree. Mol Vis. 2010;16:154–60. [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Shentu X, Wang W, Li J, Jin C, Yao K. A Chinese family with progressive childhood cataracts and IVS3+1G>A CRYBA3/A1 mutations. Mol Vis. 2010;16:2347–53. [PMC free article] [PubMed] [Google Scholar]