Ubiquitin-dependent degradation of PDZ-RhoGEF restricts RhoA activity to facilitate neurite outgrowth.
Abstract
The induction of neurite outgrowth and arborization is critical for developmental and regenerative processes. In this paper, we report that the BTB-kelch protein KLHL20 promoted neurite outgrowth and arborization in hippocampal and cortical neurons through its interaction with Cullin3 to form a ubiquitin ligase complex. This complex targeted PDZ–Rho guanine nucleotide exchange factor (RhoGEF), a protein abundantly expressed in the brain, for ubiquitin-dependent proteolysis, thereby restricting RhoA activity and facilitating growth cone spreading and neurite outgrowth. Importantly, targeting PDZ-RhoGEF to KLHL20 required PDZ-RhoGEF phosphorylation by p38 mitogen-activated protein kinase. In response to p38-activating neurotrophins, such as brain-derived neurotrophic factor and neurotrophin-3, KLHL20-mediated PDZ-RhoGEF destruction was potentiated, leading to neurotrophin-induced neurite outgrowth. Our study identified a ubiquitin-dependent pathway that targets PDZ-RhoGEF destruction to facilitate neurite outgrowth and indicates a key role of this pathway in neurotrophin-induced neuronal morphogenesis.
Introduction
Neurite outgrowth and arborization are crucial for establishing anatomical connections during the development and remodeling of the nervous system. The Rho family GTPases, such as Rho, Rac, and Cdc42, regulate various aspects of neuronal differentiation, including neurite outgrowth and arborization, by activating multiple effector pathways that affect actin and microtubule dynamics (Luo, 2000; Govek et al., 2005; Hall and Lalli, 2010). Rho GTPases are activated by a large family of Rho guanine nucleotide exchange factors (RhoGEFs; Schmidt and Hall, 2002), and several RhoGEFs are known to regulate neuronal morphogenesis in response to a variety of extracellular cues (Rossman et al., 2005). Among them, the brain-enriched PDZ-RhoGEF (Kuner et al., 2002) is activated by interacting with the activated Gα12/13 (Fukuhara et al., 1999), thereby mediating the neurite retraction effect of lysophosphatidic acid (Togashi et al., 2000). PDZ-RhoGEF also plays a role in axon guidance by interacting with plexin-B1, thereby mediating semaphorin 4D–induced Rho activation, growth cone collapse, and axon retraction (Perrot et al., 2002; Swiercz et al., 2002). Although PDZ-RhoGEF is known to induce neurite or axon retraction, it is unclear whether and how its activity is restrained under conditions that permit neurite outgrowth.
The ubiquitin-proteasome system (UPS) plays crucial roles in various aspects of neuronal development, such as axon formation, elongation, and pruning, and synapse formation and elimination (Yi and Ehlers, 2007; Tai and Schuman, 2008; Segref and Hoppe, 2009). The Cullin3 (Cul3)-based ubiquitin E3 ligases use BTB domain–containing proteins as substrate adaptors (Petroski and Deshaies, 2005). We recently identified KLHL20, a protein possessing a BTB domain and six kelch repeats, as such an adaptor (Lee et al., 2010). Based on the expression pattern illustrated in the Genepaint database, KLHL20 mRNA is abundantly expressed in the brain of an embryonic day 14.5 (E14.5) mouse embryo, implying its role in neural development. In the adult mouse brain (Allen Brain Atlas), KLHL20 mRNA is highly expressed in the hippocampus, especially in the dentate gyrus, where a lifelong neurogenesis occurs (Ming and Song, 2005). Here, we show that the KLHL20-based E3 ligase targets PDZ-RhoGEF for ubiquitination and degradation, thereby participating in neurotrophin-induced neurite growth.
Results and discussion
KLHL20 promotes neurite outgrowth/arborization
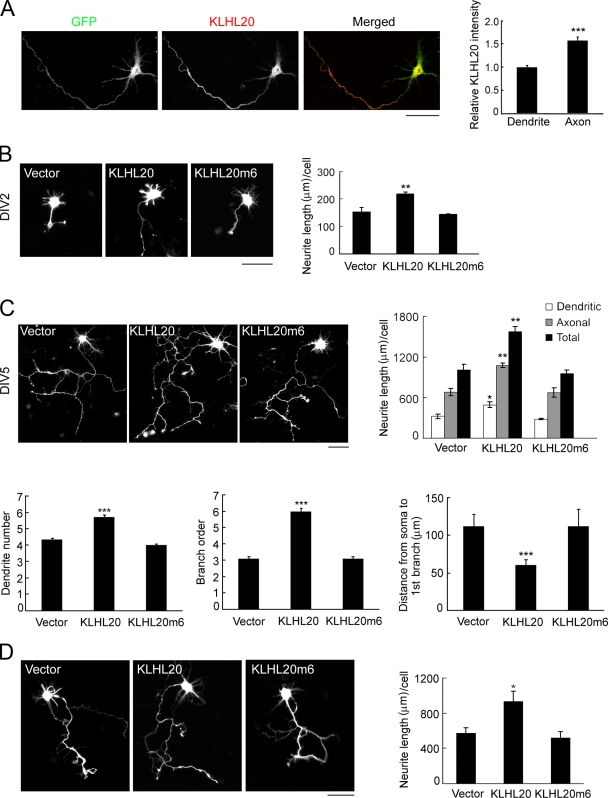
We first determined the expression of the KLHL20 protein and its subcellular distribution in primary rat hippocampal neurons by immunofluorescent analysis, and antibody specificity was confirmed by the reduction of staining intensity in neurons expressing KLHL20 siRNA (Fig. S1 A). KLHL20 was distributed throughout the cell body, axon, and dendrites, albeit at a lower expression level in dendrites than the axon (Fig. 1 A). To investigate the effect of KLHL20 on neuronal morphogenesis, hippocampal neurons at day in vitro (DIV) 0 were transfected with KLHL20 and analyzed at DIV2 or DIV5. At both time points, we detected an increase of neurite lengths in cells overexpressing KLHL20 (Figs. 1, B and C; and S1 B). Furthermore, this effect was more significantly observed in axon than dendrites (Fig. 1 C, top). KLHL20 also increased the complexity of the neurite arbor. This was reflected by the increase of dendrite number and axon branching, and the latter was inferred by a higher branch order and shorter distance from soma to the first branch point (Fig. 1 C, bottom). KLHL20, however, did not affect neuronal polarity (Fig. S1 C). Overexpression of KLHL20 in primary cortical neurons also stimulated neurite outgrowth (Figs. 1 D and S1 D). However, introducing KLHL20m6, a mutant defective in Cul3 binding (Lee et al., 2010), to hippocampal or cortical neurons failed to elicit any effect (Fig. 1, B–D). We further showed that KLHL20-induced neurite outgrowth was abrogated by Cul3 siRNA (Fig. S1 E). Our data suggest that KLHL20 promotes neurite outgrowth/arborization through the formation of a KLHL20–Cul3 complex.
Figure 1.
KLHL20 promotes neurite outgrowth/arborization. (A) Confocal image of GFP-transfected DIV4 neurons stained with the anti-KLHL20 antibody (left). The relative intensities of the KLHL20 signal in axon and dendrites were quantified (see Materials and methods) and normalized by the intensity of GFP (right). Data represent means ± SEM from three independent experiments (***, P < 0.0005; n = 15). (B and C) Hippocampal neurons at DIV0 were transfected with the indicated plasmids together with GFP at a ratio of 4:1, and the morphologies of GFP-positive neurons were examined at DIV2 (B) or DIV5 (C). Representative morphologies of neurons are shown. Neurite length and various parameters of neuronal morphology (see Materials and methods) were quantified and plotted. (D) Cortical neurons were transfected as in B and monitored for neurite outgrowth at DIV4. Data in B–D represent means ± SEM from three independent experiments (*, P < 0.05; **, P < 0.005; ***, P < 0.0005; n ≥ 35). Bars, 50 µm.
KLHL20 interacts with PDZ-RhoGEF
To identify the substrate of Cul3–KLHL20 mediating the neuronal morphogenesis function of KLHL20, we performed a yeast two-hybrid screen using the substrate-binding, kelch repeat domain as bait. Several positive clones identified in this screen encoded for C-terminal fragments of PDZ-RhoGEF. Notably, KLHL20 interacted specifically with PDZ-RhoGEF(951–1,562) but not with a corresponding fragment of a related protein, LARG (Fig. 2 A). Immunoprecipitation analysis demonstrated the interaction of KLHL20-Myc with PDZ-RhoGEF–Flag in transfected cells (Fig. 2 B). This interaction was abolished by deleting the kelch repeat domain of KLHL20 (KLHL20ΔK), whereas the kelch repeat domain alone was sufficient for interaction. An in vitro binding assay demonstrated that GST-KLHL20 could specifically pull down baculovirally purified PDZ-RhoGEF (Fig. 2 D). Furthermore, using in vitro binding and immunoprecipitation analyses, we found that the C-terminal segment (amino acids 960–1,309) of PDZ-RhoGEF was required for KLHL20 binding (Fig. 2, C and D). The binding of endogenous KLHL20 to endogenous PDZ-RhoGEF was observed using mouse brain extract (Fig. 2 E). We further detected an interaction of endogenous PDZ-RhoGEF with endogenous Cul3, and this interaction was diminished by depletion of KLHL20 (Fig. 2 F). Likewise, coimmunoprecipitation of Cul3 with both KLHL20 and PDZ-RhoGEF was observed using primary cortical neurons (Fig. 2 G). Together, these data indicate PDZ-RhoGEF as a KLHL20-interacting protein and reveal the formation of a Cul3–KLHL20–PDZ-RhoGEF tripartite complex.
Figure 2.
KLHL20 interacts with PDZ-RhoGEF. (A) Yeast cotransformed with the indicated constructs was assayed for the His3 phenotype (−His) and β-galactosidase activity (β-gal). (B and D) Interaction of PDZ-RhoGEF–Flag with KLHL20-Myc and mapping of their interaction domains. 293T cells transfected with various constructs were analyzed by immunoprecipitation (IP) and/or Western blotting (WB). The various deletion mutants of PDZ-RhoGEF are shown on the top of C. (C) GST pull-down analysis of the KLHL20 interaction with baculovirally purified PDZ-RhoGEF and its deletion mutants. The equal input of the GST fusion protein and the expression levels of various PDZ-RhoGEF mutants are shown on the bottom and right, respectively. (E) Coimmunoprecipitation analysis of the interaction between endogenous PDZ-RhoGEF and endogenous KLHL20 in the mouse brain. The dotted line indicates that unrelated lanes were removed. (F) KLHL20 mediates the interaction between Cul3 and PDZ-RhoGEF. HeLa cells infected with a lentivirus carrying KLHL20 siRNA or control siRNA were assayed by immunoprecipitation and/or Western blotting. (G) Coimmunoprecipitation analysis of the interaction of Cul3 with KLHL20 and PDZ-RhoGEF in cortical neurons.
KLHL20 promotes PDZ-RhoGEF ubiquitination and destruction to inactivate RhoA
We next examined the effect of KLHL20-based ligase on PDZ-RhoGEF ubiquitination. The ubiquitination level of PDZ-RhoGEF was greatly elevated by coexpression of Cul3, KLHL20, and Roc1 (a subunit of the Cul3 complex; Fig. 3 A). Replacement of KLHL20 with KLHL20m6 or Cul3 with Cul3H2/5, a mutant defective in binding BTB proteins (Xu et al., 2003), failed to promote PDZ-RhoGEF ubiquitination. Depletion of KLHL20 by two independent siRNAs suppressed PDZ-RhoGEF ubiquitination (Fig. 3 B). To demonstrate PDZ-RhoGEF as a direct substrate of the Roc1–Cul3–KLHL20 ligase, in vitro ubiquitination was performed. Incubation of the KLHL20–Cul3–Roc1 complex purified from transfected cells with baculovirally purified PDZ-RhoGEF in the in vitro ubiquitination reaction resulted in PDZ-RhoGEF polyubiquitination. Omission of the ubiquitin, E1, E2, or E3 complex abrogated this ubiquitination (Fig. 3 C). PDZ-RhoGEF ubiquitination was also impaired by the replacement of KLHL20 with KLHL20m6 or KLHL20ΔK or Cul3 with Cul3ΔC (a mutant defective in Roc1 binding). These results indicate PDZ-RhoGEF as a bona fide substrate of the Roc1–Cul3–KLHL20 ligase.
Figure 3.
KLHL20 promotes PDZ-RhoGEF ubiquitination to inactivate RhoA. (A and B) KLHL20 promotes PDZ-RhoGEF ubiquitination in vivo. 293T cells (A) or HeLa cells stably expressing control or KLHL20 siRNA (B) were transfected with the indicated constructs and treated with MG132. Cells were lysed for immunoprecipitation (IP) and/or Western blot (WB) with the indicated antibodies. (C) PDZ-RhoGEF purified from baculovirus was subject to in vitro ubiquitination reaction in the presence of the E1, E2, and E3 complex and/or ubiquitin and then analyzed by Western blotting with the anti-Flag antibody. (D) Western blot analysis of the ectopic (left) or endogenous (right) level of PDZ-RhoGEF in 293T cells transfected with the indicated constructs and treated with or without MG132. (E) KLHL20 promotes PDZ-RhoGEF turnover. Western blot analysis of endogenous PDZ-RhoGEF level in 293T cells transfected with indicated constructs and treated with cycloheximide (CHX) for various time points. (F) Western blot analysis of endogenous PDZ-RhoGEF level in HeLa cells stably expressing indicated siRNA. (G–I) 293T cells (G), HeLa cells (H), or hippocampal neurons (I) were transfected with various constructs and/or siRNAs and then were lysed for assaying GTP-bound RhoA by pull-down (G and H) or G-LISA (I) or the expression of various proteins. Data in I represent means ± SEM (**, P < 0.005; n = 3). The efficacy of PDZ-RhoGEF siRNA to down-regulate the endogenous protein in HeLa cells is shown in Fig. S2 E. In D–F, the relative amounts of PDZ-RhoGEF were quantified, normalized to the amounts of tubulin, and marked below the blot. si, siRNA; Ub, ubiquitin.
Most cullin-based ligases catalyze K48-linked polyubiquitination to promote substrate degradation. Accordingly, overexpression of KLHL20, but not KLHL20ΔK or KLHL20m6, decreased the levels of ectopic and endogenous PDZ-RhoGEF, and this effect was reversed by the proteasome inhibitor MG132 (Fig. 3 D). A cycloheximide chase experiment revealed that KLHL20 enhanced PDZ-RhoGEF turnover (Fig. 3 E). Furthermore, endogenous PDZ-RhoGEF level was elevated in cells expressing KLHL20 siRNA (Fig. 3 F). These data support a role of KLHL20-associated ligase in promoting PDZ-RhoGEF proteasomal degradation. Next, we investigated the effect of KLHL20 on regulating RhoA. As expected, overexpression of PDZ-RhoGEF stimulated RhoA activity. This effect, however, was markedly attenuated by coexpression of KLHL20 but not KLHL20m6 (Fig. 3 G). Conversely, depletion of KLHL20 enhanced RhoA activity, which was reversed by overexpressing an siRNA-resistant KLHL20 (KLHL20r) or a PDZ-RhoGEF–specific siRNA (Fig. 3 H). Notably, in these two experiments, the alteration of RhoA activity correlated with the level of PDZ-RhoGEF. Finally, we showed that depletion of KLHL20 in hippocampal neurons also potentiated RhoA activity without affecting RhoA expression (Fig. 3 I). Our data support that KLHL20 promotes PDZ-RhoGEF degradation to inactivate RhoA.
KLHL20 promotes neurite outgrowth and growth cone spreading through PDZ-RhoGEF degradation
Given the well-documented role of RhoA in suppressing neurite outgrowth (Luo, 2000; Govek et al., 2005; Hall and Lalli, 2010), our findings suggest a contribution of PDZ-RhoGEF degradation to the neuronal morphogenesis function of KLHL20. In support of this notion, overexpression of PDZ-RhoGEF abolished the effect of KLHL20 on neurite outgrowth in hippocampal neurons (Figs. 4 A and S2 A). In the next set of experiments, KLHL20 siRNA was introduced to hippocampal neurons, and its efficacy to deplete KLHL20 was confirmed (Fig. S2 B). We showed that neurons expressing KLHL20 siRNA displayed shorter neurites. This effect was rescued by cotransfection of KLHL20r but not its m6 mutant (Fig. 4 B). More importantly, depletion of PDZ-RhoGEF (Figs. 4 B and S2 C) or blockage of the RhoA downstream effector Rho-associated protein kinase (ROCK) by Y27632 or myosin II by blebbistatin (Fig. S2 D) completely reversed the inhibitory effect of KLHL20 siRNA on neurite outgrowth. Notably, the KLHL20 siRNA failed to elicit any effect on Y27632- or blebbistatin-treated neurons (Fig. S2 D), suggesting that KLHL20, ROCK, and myosin II act in the same pathway to suppress neurite outgrowth.
Figure 4.
KLHL20 promotes neurite outgrowth and growth cone spreading through down-regulating PDZ-RhoGEF. (A and B) Hippocampal neurons at DIV0 were transfected with the indicated plasmids and/or siRNAs together with GFP. Neurons were imaged at DIV5 (A) or at DIV3, 4, and 5 (B), and neurite lengths of GFP-positive neurons were quantified and plotted. The efficacy of PDZ-RhoGEF siRNA to down-regulate endogenous PDZ-RhoGEF in hippocampal neurons is shown in Fig. S2 E. (C and D) Hippocampal neurons transfected with the indicated constructs and/or siRNAs were fixed at DIV3 and then stained by rhodamine-conjugated phalloidin and Tau-1 (not depicted). The percentage of axons showing collapsed growth cones was quantified and plotted (C, left). Representative growth cone morphologies are shown on the right. Data in all panels represent means ± SEM (*, P < 0.05; **, P < 0.005; ***, P < 0.0005; n ≥ 50). Bars, 10 µm. WT, wild type.
The growth cones undergo dynamic changes of their morphology in response to extracellular cues, thereby determining the growth or retraction of neurites (Lowery and Van Vactor, 2009). Consistent with a previous study (Swiercz et al., 2002), PDZ-RhoGEF overexpression markedly increased the percentage of hippocampal neurons displaying growth cone collapse. This effect was abolished by coexpression of KLHL20 (Fig. 4 C). Expression of KLHL20 alone also attenuated growth cone collapse, presumably mediated by endogenous PDZ-RhoGEF. Conversely, KLHL20 siRNA induced growth cone collapse, and this effect was rescued by overexpression of KLHL20r but not its m6 mutant (Fig. 4 D). More importantly, the effect of KLHL20 siRNA was also reversed by PDZ-RhoGEF silencing. Together, our data indicate that KLHL20-mediated PDZ-RhoGEF degradation is important to maintain growth cone spreading, thereby facilitating neurite outgrowth.
KLHL20-mediated PDZ-RhoGEF destruction is potentiated by p38 MAPK and participates in neurotrophin-induced morphogenesis
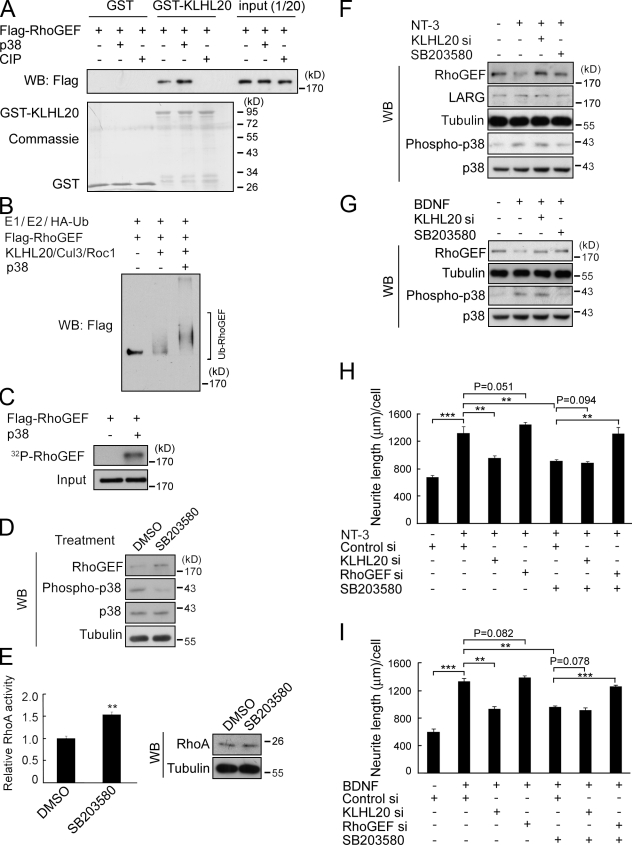
Next, we investigated whether the KLHL20-mediated PDZ-RhoGEF ubiquitination could be regulated by a physiological signal. Intriguingly, treatment of baculovirally purified PDZ-RhoGEF with calf intestine phosphatase abrogated its binding to KLHL20 (Fig. 5 A) and ubiquitination by KLHL20–Cul3–Roc1 in vitro (Fig. S3 A). Preincubation of PDZ-RhoGEF with p38 MAPK, but not extracellular signal–related kinase (ERK), in a kinase reaction potentiated the subsequent PDZ-RhoGEF ubiquitination catalyzed by the same E3 ligase (Figs. 5 B and S3 B). We further demonstrated that p38 directly phosphorylated PDZ-RhoGEF (Fig. 5 C) and that this phosphorylation promoted the PDZ-RhoGEF interaction with KLHL20 (Fig. 5 A). Thus, p38-induced phosphorylation is crucial for targeting PDZ-RhoGEF to KLHL20-based E3 ligase. In line with this notion, pharmacological inhibition of p38 MAPK in hippocampal neurons increased PDZ-RhoGEF level and RhoA activity without affecting RhoA expression (Fig. 5, D and E). Consistent with a previous study (Ying et al., 2002), treatment of hippocampal neurons with neurotrophin-3 (NT-3) increased p38 activity. Importantly, this p38 activation was accompanied by a down-regulation of PDZ-RhoGEF, which was reversed by p38 inhibitor or KLHL20 siRNA (Fig. 5 F). The expression of LARG, however, was not affected. PDZ-RhoGEF down-regulation was also induced by brain-derived neurotrophic factor (BDNF) in a p38- and KLHL20-dependent manner (Fig. 5 G). These data suggest that these p38-activiating neurotrophins up-regulate the KLHL20-dependent PDZ-RhoGEF degradation pathway. Next, we determined the contribution of the p38–KLHL20–PDZ-RhoGEF signaling axis to neurotrophin-induced neurite outgrowth. The BDNF- and NT-3–induced neurite outgrowth was significantly suppressed by KLHL20 depletion or p38 inhibition (Fig. 5, H and I, compare second bar with third and fifth bars). Cotreatment of neurons with KLHL20 siRNA and p38 inhibitor, however, did not cause an additional inhibitory effect (Fig. 5, H and I, sixth bar), suggesting that p38 and KLHL20 act in the same pathway. To address whether the suppressive effect of p38 inhibitor on neurotrophin-induced neurite outgrowth was caused by PDZ-RhoGEF up-regulation, PDZ-RhoGEF siRNA was introduced. PDZ-RhoGEF siRNA reversed the effect of p38 inhibitor on neurotrophin-treated neurons (Fig. 5, H and I, compare seventh bar with fifth bar). Notably, in neurotrophin-treated neurons without exposure to p38 inhibitor, PDZ-RhoGEF siRNA did not significantly affect neurite outgrowth (Fig. 5, H and I, compare second bar with fourth bar), which is consistent with a persistent degradation of PDZ-RhoGEF under such conditions. These results collectively support a critical role of the p38–KLHL20–PDZ-RhoGEF pathway in neurotrophin-induced neuronal morphogenesis.
Figure 5.
KLHL20-mediated PDZ-RhoGEF ubiquitination is stimulated by p38 MAPK and contributes in part to neurotrophin-induced morphogenesis. (A) GST pull-down analysis of the KLHL20 interaction with p38-phosphorylated or calf intestine phosphatase (CIP)–treated PDZ-RhoGEF. (B) Baculovirally purified PDZ-RhoGEF was phosphorylated by p38 and then subject to in vitro ubiquitination assay as in Fig. 3 C. (C) In vitro phosphorylation of PDZ-RhoGEF by p38. Substrate phosphorylation was analyzed by autoradiograph. (D and E) p38 inhibitor elevates PDZ-RhoGEF level (D) and RhoA activity (E) in hippocampal neurons. Hippocampal neurons at DIV1 were treated with 20 µM SB203580 for 3 h and then analyzed by Western blotting (WB) for protein expression (D) or G-LISA for RhoA activity (E, left). The expression of RhoA was determined by Western blotting (E, right). Data represent means ± SEM (**, P < 0.005; n = 3). (F and G) NT-3 (F) and BDNF (G) down-regulate PDZ-RhoGEF through p38 and KLHL20. Hippocampal neurons were transfected with control or KLHL20 siRNA at DIV0, treated at DIV2 with SB203580 and/or 200 ng/ml NT-3 or BDNF, and analyzed by Western blotting. (H and I) p38- and KLHL20-mediated PDZ-RhoGEF destruction participates in BDNF- and NT-3–induced differentiation. Hippocampal neurons were transfected with the indicated siRNAs and treated as in F. Neurite lengths were assayed at DIV4. Data represent means ± SEM (**, P < 0.005; ***, P < 0.0005; n ≥ 35). si, siRNA; Ub, ubiquitin.
In summary, our study reveals a PDZ-RhoGEF degradation pathway mediated by p38 MAPK and KLHL20-based E3 ligase, thereby participating in neurotrophin-induced neurite outgrowth. RhoA has been well recognized as a negative regulator of neurite outgrowth, but how extracellular cues restrict RhoA activity to promote neuritogenesis is largely unknown. Our study provides important insights into the mechanism of RhoA regulation during physiological neuritogenesis and also implies a previously unrecognized role of KLHL20 in the proper development of neural circuits. In addition to neurotrophin-induced morphogenesis, PDZ-RhoGEF also participates in semaphorin 4D–induced axon guidance by interacting with the receptor plexin-B1 through its PDZ domain (Swiercz et al., 2002). However, PDZ-RhoGEF is constitutively associated with plexin-B1, and the interaction domain does not overlap with the KLHL20-binding region. Thus, it is unlikely that KLHL20 could affect semaphorin signaling by competing with plexin-B1 for PDZ-RhoGEF binding. Notably, BDNF was recently reported to induce UPS-dependent degradation of RhoA via a PKA/Smurf1-dependent pathway, thereby facilitating axon formation (Cheng et al., 2011). The UPS is also known to promote axonal outgrowth by targeting the RhoA downstream molecule LIM kinase 1 via the E3 ligase Rnf6 localized in an axonal growth cone (Tursun et al., 2005). Thus, the UPS targets multiple proteins along the RhoA signaling pathway via distinct ubiquitin ligases to regulate diverse aspects of neuronal development.
Materials and methods
Cell culture and transfection
293T and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Primary hippocampal and cortical neurons were prepared as previously described (Goslin et al., 1998; Meberg and Miller, 2003). In brief, hippocampi and cortex were dissected from E18 rat fetuses in ice-cold Dulbecco’s modified Eagle’s medium. The cells were dissociated with papain for 10 min at 37°C and then triturated gently. The dissociated cells were then plated on poly-d-lysine–coated glass coverslips and maintained in neurobasal medium (Invitrogen) supplemented with 2% B27 (Invitrogen) and 0.5 mM l-glutamine.
Transfection of HeLa and 293T cells was performed by the calcium phosphate method as previously described (Chen et al., 2005). Transfection of hippocampal and cortical neurons was performed using nucleofection. In brief, freshly isolated neurons were suspended in 100 µl transfection solution (Lonza) containing 4 µg cDNA or 150 pmol siRNA oligonucleotides. Nucleofection was performed using the eletroporator (Lonza) setting in the program G-13. After transfection, the neurons were plated on poly-d-lysine–coated dishes and then refed with neurobasal medium supplemented with B27 and l-glutamine at 1 h after plating. For siRNA rescue experiments, neurons were cotransfected with 150 pmol siRNA oligonucleotides and 2 µg of appropriate plasmid constructs.
Plasmids
Constructs for KLHL20, KLHL20m6, KLHL20ΔK, KLHL20-kelch domain, Roc1, Cul3, and Cul3ΔC were described previously (Lee et al., 2010). The full-length PDZ-RhoGEF clone (BC057394) was purchased from Thermo Fisher Scientific and then amplified by PCR using a 5′ primer containing an EcoRI site and a 3′ primer containing an XhoI site. The PCR product was digested with EcoRI and XhoI and then inserted into pRK5-Flag. The deletion mutants of PDZ-RhoGEF were generated by PCR and then subcloned to pRK5-Flag. The full-length LARG clone (BC152900) was purchased from Thermo Fisher Scientific. A fragment corresponding to amino acids 965–1,544 was amplified by PCR using a 5′ primer containing a BamHI site and a 3′ primer containing an XhoI site. The PCR product was digested with BamHI and XhoI and then inserted into the pACT2 vector. pCDNA3-Myc-hCul3H2/5 was provided by Y. Xiong (University of North Carolina, Chapel Hill, NC). KLHL20r-Myc and KLHL20m6r-Myc that contain two silent mutations introduced in the 21-bp KLHL20 siRNA-targeting sequence were generated by site-directed mutagenesis.
RNA interference
Knockdown of KLHL20 in HeLa cells was performed by lentiviral transduction as previously described (Lee et al., 2010). The sequences of human KLHL20 siRNAs are KLHL20 siRNA#1, 5′-GGTGGCGTAGGAGTTATTAA-3′ (corresponding to KLHL20 mRNA 1,959–1,977), and KLHL20 siRNA#2, 5′-GCCTGCTGTGAATTCTTAA-3′ (corresponding to KLHL20 mRNA 645–663). Knockdown of PDZ-RhoGEF, KLHL20, or Cul3 in hippocampal neurons was performed by transfection with an siRNA oligonucleotide specific to rat PDZ-RhoGEF, 5′-CCUCAUCUUCUACCAGCGCAU-3′ (corresponding to PDZ-RhoGEF mRNA 2,295–2,315); rat KLHL20, 5′-GCCUGCUGUGAAUUCUUAA-3′ (corresponding to KLHL20 mRNA 645–663); or rat Cul3, 5′-UGACAGAAAACACGAGAUA-3′ (corresponding to Cul3 mRNA 2,031–2,049). All siRNA oligonucleotides (including control siRNA) were purchased from Thermo Fisher Scientific. Lentiviruses were provided by the National RNAi Core Facility.
Yeast two-hybrid screen
The DNA fragment encoding the kelch repeat domain of KLHL20 (amino acids 267–609) was cloned to pBTM116 to produce the LexA-kelch for screening a human fetal brain cDNA library fused to the Gal4 activation domain (Takara Bio Inc.). Yeast two-hybrid screen was performed as previously described (Chen et al., 2005). In brief, the yeast strain L40 was sequentially transformed with the pBTM116-based construct and library constructs. The double transformants were plated on selective medium lacking uracil, histidine, tryptophan, lysine, and leucine followed by β-galactosidase assays and then verified by one-on-one transformation. The positive clones were isolated and then rescued into Escherichia coli DH5-α and sequenced.
Antibodies and reagents
Polyclonal anti-KLHL20 antibody was described previously (Lee et al., 2010). Anti-Myc (9E10), anti-RhoA (26C4), and anti–PDZ-RhoGEF (R-295 and H300) antibodies were obtained from Santa Cruz Biotechnology, Inc. Anti-Flag (M2) and anti-HA antibodies and cycloheximide were purchased from Sigma-Aldrich. Antibodies to p38 MAPK and phospho-p38 were purchased from Cell Signaling Technology. The anti–α-tubulin antibody and the anti–Tau-1 antibody (MAB2340) were obtained from Millipore. The anti-LARG antibody was purchased from Santa Cruz Biotechnology, Inc., and anti-Cul3 antibodies were purchased from Santa Cruz Biotechnology, Inc. and Gene Tex. MG132, SB203580, blebbistatin, and Y27632 were purchased from EMD. NT-3 and BDNF were obtained from Cell Signaling Technology.
Immunofluorescence
Neurons were fixed by PBS containing 4% formaldehyde and 4% sucrose for 10 min at room temperature and then permeabilized with 0.5% Triton X-100 in PBS for 15 min. Neurons were blocked with PBS containing 0.1% Triton X-100 and 5% donkey serum for 1 h. Neurons were then incubated with various primary antibodies diluted in PBS containing 0.2% BSA and 5% donkey serum at 4°C overnight, washed with PBS containing 0.1% Triton X-100, and labeled with a rhodamine- or Cy5-conjugated secondary antibody. Neurons were washed, mounted, and then examined at room temperature with a laser-scanning confocal microscope (510 Meta; Carl Zeiss) equipped with a 40× 1.2 NA water immersion (for monitoring neurons). Fluorescent images were analyzed by the laser-scanning microscope software (LSM510; Carl Zeiss). The images were processed using Photoshop software (Adobe).
To quantify the KLHL20 immunointensity, neurons were transfected, cultured, and immunostained in parallel under identical conditions. Images were obtained on the same day using identical laser power and acquisition settings by confocal microscopy, and the middle-most image among serial confocal sections was chosen for analysis. The region showing the GFP signal at the GFP channel was selected by ImageJ software (National Institutes of Health), and the mean pixel intensity (MPI) in the corresponding region at the KLHL20 channel was measured. For quantifying the KLHL20 signal at axon and dendrites, the MPI of KLHL20 in axon and dendrites was calculated separately and normalized by the MPI of GFP in the corresponding regions.
Analyses of neurite outgrowth, neuronal morphology, and growth cone morphology
For monitoring neurite outgrowth and neuronal morphology, primary hippocampal or cortical neurons were fixed with 4% formaldehyde in PBS for 10 min and then examined at room temperature by an epifluorescent microscope (IX71; Olympus) equipped with a 10× 0.25 NA objective lens and a camera (F-View II; Olympus) using a controller software (analySIS LS Research; Olympus). Axonal and dendritic processes were identified by their morphological characteristics. For counting dendrite numbers, only processes longer than 20 µm were considered. Axon and dendrite lengths together with the distance from soma to the first axonal branch were measured using the NeuronJ neurite tracing program (Meijering et al., 2004). Axonal branch order was determined by counting the number of branching points encountered from the axon’s first branch point to the tip, and only processes longer than 20 µm were counted.
To visualize growth cones, neurons were fixed at DIV3, stained with rhodamine-conjugated phalloidin and Tau-1, and examined at room temperature with a laser-scanning confocal microscope (510 Meta) equipped with a 100× 1.3 NA oil immersion objective lens. Growth cones were counted as collapsed if they contain no lamellipodia and two or fewer filopodia.
Production of baculovirus
PDZ-RhoGEF–Flag was cloned to pVL1393. This plasmid, together with the linearized baculovirus DNA (BaculoGold; BD), was used to infect monolayers of Sf9 cells cultured in Grace’s medium (Life Technologies) supplemented with 10% fetal calf serum. The recombinant virus was harvested, amplified, and then used to infect the monolayer of Sf9 cells in Trichoplusia ni Medium–Formulation Hink medium. After a 4-d incubation, PDZ-RhoGEF–Flag was purified using the anti-Flag M2 agarose (Sigma-Aldrich).
In vitro phosphorylation
100 ng baculovirally purified PDZ-RhoGEF was incubated with 50 ng p38-α MAPK (Millipore) at 37°C for 15 min in the kinase reaction mixture containing 50 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM DTT, 2 mM NaF, 12.5 µM ATP, and 10 µCi γ-[32P]ATP. The reaction was stopped by boiling in SDS sample buffer. Protein phosphorylation was detected by autoradiography.
Immunoprecipitation and GST pull-down
For immunoprecipitation, cells were lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1% sodium deoxycholate) supplemented with 1 mM PMSF, 10 µg/ml aprotinin, and 10 µg/ml leupeptin. The lysate was incubated with anti-Flag or anti-KLHL20 antibody and then with protein A–Sepharose conjugated on rabbit anti–mouse antibody. The recovered immunocomplex was washed with lysis buffer and then analyzed by Western blotting. For GST pull-down, PDZ-RhoGEF purified from baculovirus was phosphorylated by p38 MAPK and then incubated with GST or GST-KLHL20 immobilized on glutathione–Sepharose beads in binding buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, and 1% NP-40. The beads were washed, and bound protein was analyzed by Western blotting.
In vitro ubiquitination
293T cells were transfected with GST-Cul3, KLHL20-Myc, and Roc1-Myc or appropriate controls and lysed with NP-40 lysis buffer containing 50 mM Tris, pH 7.5, 1% NP-40, and 150 mM NaCl. The Cul3 complexes were isolated by glutathione–Sepharose beads, and then, the beads were washed with reaction buffer containing 50 mM Tris, pH 7.4, 2 mM NaF, and 5 mM MgCl2. The beads were incubated with 0.5 µg purified Flag–PDZ-RhoGEF pretreated with or without 0.5 U calf intestine phosphatase or various kinases in a 20-µl reaction mixture containing 50 mM Tris, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.6 mM DTT, 2 µM ubiquitin aldehyde, 10 µM MG132, 1 µg HA-ubiquitin, 40 ng E1, and 300 ng E2 (UbcH5a) at 37°C for 60 min. The reaction was terminated by boiling for 5 min in sample buffer and analyzed by Western blotting.
RhoA activity assay
The level of GTP-bound RhoA was detected with the GST-rhotekin pull-down assay as previously described (Shen et al., 2008). In brief, cells was lysed with buffer containing 25 mM Hepes, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 10% glycerol, 1 mM EDTA, 2 mM PMSF, 10 µg/ml leupeptin, and 10 µg/ml aprotinin. The lysates were incubated with 50 µg GST-rhotekin beads for 40 min at 4°C. The beads were washed and analyzed by Western blotting to detect the bound RhoA. For assaying RhoA activity in hippocampal neurons, G-LISA assay (Cytoskeleton) was performed according to the manufacturer’s instructions.
Online supplemental material
Fig. S1 shows the specificity of the KLHL20 antibody and the effect of KLHL20 on neuronal polarity and neurite outgrowth. Fig. S2 shows the effect of the KLHL20/PDZ-RhoGEF/Rho/ROCK/MLCII signaling axis on neuron morphogenesis. Fig. S3 shows the effect of p38 MAPK and ERK on PDZ-RhoGEF ubiquitination catalyzed by the KLHL20–Cul3–Roc1 ligase. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103015/DC1.
Acknowledgments
We thank Yue Xiong and the National RNAi Core Facility for reagents and Chin-Chun Hung for confocal analysis.
This work was supported by the National Science Council Frontier grant 99-2321-B-001-081 and the Academia Sinica Investigator Award.
Footnotes
Abbreviations used in this paper:
- BDNF
- brain-derived neurotrophic factor
- Cul3
- Cullin3
- DIV
- day in vitro
- ERK
- extracellular signal–related kinase
- MPI
- mean pixel intensity
- NT-3
- neurotrophin-3
- RhoGEF
- Rho guanine nucleotide exchange factor
- ROCK
- Rho-associated protein kinase
- UPS
- ubiquitin-proteasome system
References
- Chen C.H., Wang W.J., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H. 2005. Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J. 24:294–304 10.1038/sj.emboj.7600510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.L., Lu H., Shelly M., Gao H., Poo M.M. 2011. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 69:231–243 10.1016/j.neuron.2010.12.021 [DOI] [PubMed] [Google Scholar]
- Fukuhara S., Murga C., Zohar M., Igishi T., Gutkind J.S. 1999. A novel PDZ domain containing guanine nucleotide exchange factor links heterotrimeric G proteins to Rho. J. Biol. Chem. 274:5868–5879 10.1074/jbc.274.9.5868 [DOI] [PubMed] [Google Scholar]
- Goslin K., Asmussen H., Banker G. 1998. Rat hippocampal neurons in low-density culture. Culturing Nerve Cells. Second edition Banker G., Goslin K., MIT Press, Cambridge, MA: 339–370 [Google Scholar]
- Govek E.E., Newey S.E., Van Aelst L. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49 10.1101/gad.1256405 [DOI] [PubMed] [Google Scholar]
- Hall A., Lalli G. 2010. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2:a001818 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner R., Swiercz J.M., Zywietz A., Tappe A., Offermanns S. 2002. Characterization of the expression of PDZ-RhoGEF, LARG and G(alpha)12/G(alpha)13 proteins in the murine nervous system. Eur. J. Neurosci. 16:2333–2341 10.1046/j.1460-9568.2002.02402.x [DOI] [PubMed] [Google Scholar]
- Lee Y.R., Yuan W.C., Ho H.C., Chen C.H., Shih H.M., Chen R.H. 2010. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 29:1748–1761 10.1038/emboj.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L.A., Van Vactor D. 2009. The trip of the tip: understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10:332–343 10.1038/nrm2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173–180 10.1038/35044547 [DOI] [PubMed] [Google Scholar]
- Meberg P.J., Miller M.W. 2003. Culturing hippocampal and cortical neurons. Methods Cell Biol. 71:111–127 10.1016/S0091-679X(03)01007-0 [DOI] [PubMed] [Google Scholar]
- Meijering E., Jacob M., Sarria J.C., Steiner P., Hirling H., Unser M. 2004. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 58A:167–176 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]
- Ming G.L., Song H. 2005. Adult neurogenesis in the mammalian central nervous system. Annu. Rev. Neurosci. 28:223–250 10.1146/annurev.neuro.28.051804.101459 [DOI] [PubMed] [Google Scholar]
- Perrot V., Vazquez-Prado J., Gutkind J.S. 2002. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J. Biol. Chem. 277:43115–43120 10.1074/jbc.M206005200 [DOI] [PubMed] [Google Scholar]
- Petroski M.D., Deshaies R.J. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6:167–180 10.1038/nrm1587 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Hall A. 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16:1587–1609 10.1101/gad.1003302 [DOI] [PubMed] [Google Scholar]
- Segref A., Hoppe T. 2009. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 10:44–50 10.1038/embor.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.H., Chen H.Y., Lin M.S., Li F.Y., Chang C.C., Kuo M.L., Settleman J., Chen R.H. 2008. Breast tumor kinase phosphorylates p190RhoGAP to regulate rho and ras and promote breast carcinoma growth, migration, and invasion. Cancer Res. 68:7779–7787 10.1158/0008-5472.CAN-08-0997 [DOI] [PubMed] [Google Scholar]
- Swiercz J.M., Kuner R., Behrens J., Offermanns S. 2002. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 35:51–63 10.1016/S0896-6273(02)00750-X [DOI] [PubMed] [Google Scholar]
- Tai H.C., Schuman E.M. 2008. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9:826–838 10.1038/nrn2499 [DOI] [PubMed] [Google Scholar]
- Togashi H., Nagata K., Takagishi M., Saitoh N., Inagaki M. 2000. Functions of a rho-specific guanine nucleotide exchange factor in neurite retraction. Possible role of a proline-rich motif of KIAA0380 in localization. J. Biol. Chem. 275:29570–29578 10.1074/jbc.M003726200 [DOI] [PubMed] [Google Scholar]
- Tursun B., Schlüter A., Peters M.A., Viehweger B., Ostendorff H.P., Soosairajah J., Drung A., Bossenz M., Johnsen S.A., Schweizer M., et al. 2005. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 19:2307–2319 10.1101/gad.1340605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., Harper J.W. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 425:316–321 10.1038/nature01985 [DOI] [PubMed] [Google Scholar]
- Yi J.J., Ehlers M.D. 2007. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol. Rev. 59:14–39 10.1124/pr.59.1.4 [DOI] [PubMed] [Google Scholar]
- Ying S.W., Futter M., Rosenblum K., Webber M.J., Hunt S.P., Bliss T.V., Bramham C.R. 2002. Brain-derived neurotrophic factor induces long-term potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and upregulation of Arc synthesis. J. Neurosci. 22:1532–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]