SLAIN2’s interactions with multiple different microtubule plus end–tracking proteins stimulate processive microtubule polymerization and ensure proper microtubule organization.
Abstract
The ends of growing microtubules (MTs) accumulate a set of diverse factors known as MT plus end–tracking proteins (+TIPs), which control microtubule dynamics and organization. In this paper, we identify SLAIN2 as a key component of +TIP interaction networks. We showed that the C-terminal part of SLAIN2 bound to end-binding proteins (EBs), cytoplasmic linker proteins (CLIPs), and CLIP-associated proteins and characterized in detail the interaction of SLAIN2 with EB1 and CLIP-170. Furthermore, we found that the N-terminal part of SLAIN2 interacted with ch-TOG, the mammalian homologue of the MT polymerase XMAP215. Through its multiple interactions, SLAIN2 enhanced ch-TOG accumulation at MT plus ends and, as a consequence, strongly stimulated processive MT polymerization in interphase cells. Depletion or disruption of the SLAIN2–ch-TOG complex led to disorganization of the radial MT array. During mitosis, SLAIN2 became highly phosphorylated, and its interaction with EBs and ch-TOG was inhibited. Our study provides new insights into the molecular mechanisms underlying cell cycle–specific regulation of MT polymerization and the organization of the MT network.
Introduction
Microtubules (MTs) are filamentous structures required for various cellular processes, such as intracellular transport, cell division, and locomotion. The remodeling of MT networks depends on MT dynamic instability—spontaneous switching between episodes of growth and shortening (Desai and Mitchison, 1997). Numerous cellular factors control MT polymerization, depolymerization, and pausing or transitions between different states (catastrophes and rescues; Desai and Mitchison, 1997; van der Vaart et al., 2009).
MTs are intrinsically asymmetric, and in cells, only one of the two MT ends, the plus end, can grow. Not surprisingly, it is an important site for the regulation of MT dynamics (Howard and Hyman, 2003). Among MT regulators, MT plus end–tracking proteins (+TIPs) are distinguished by their ability to form cometlike accumulations at the ends of growing MTs (Schuyler and Pellman, 2001). +TIPs can influence MT dynamics in various ways: cytoplasmic linker proteins (CLIPs) and CLIP-associated proteins (CLASPs) stimulate rescues (Komarova et al., 2002; Mimori-Kiyosue et al., 2005), end-binding proteins (EBs) promote MT dynamicity and growth and suppress catastrophes (Tirnauer and Bierer, 2000; Komarova et al., 2009), and the MT depolymerase mitotic centromere-associated kinesin (MCAK) induces catastrophes (Howard and Hyman, 2007).
Although many +TIPs can interact with MTs directly, most of them target growing MT ends by binding to the members of the EB family, which can autonomously localize to growing MT tips (Akhmanova and Steinmetz, 2008). The N-terminal part of the EBs consists of a calponin homology domain, which is the primary determinant of MT tip recognition (Komarova et al., 2009). The C-terminal part of the EBs includes an EB homology (EBH) domain that encompasses a coiled-coil and a four-helix bundle and an acidic tail with a conserved terminal tyrosine residue reminiscent of the ones of α-tubulin and CLIP-170 (Akhmanova and Steinmetz, 2008). To date, two types of interactions between the EBs and their partners have been characterized in detail. Proteins containing cytoskeleton-associated protein (CAP)–Gly domains, such as CLIPs, interact with the EEY/F motifs of the EB tails whereby the C-terminal tyrosine is required for efficient binding (Honnappa et al., 2006; Weisbrich et al., 2007). A large number of other EB partners, including CLASPs and MCAK, associate with a hydrophobic cavity of the EBH domain through basic and serine-rich regions containing the short linear motif SxIP (Honnappa et al., 2009). +TIP interactions with the EBs are transient and competitive, as EB dimers can associate with only two CAP-Gly domains or SxIP motifs at the same time. Additional enrichment of +TIPs at the MT ends can be achieved by binding to other +TIPs (Akhmanova and Steinmetz, 2008). For example, CLASPs associate with the coiled-coil part of CLIPs independently of EB binding (Akhmanova et al., 2001). +TIPs thus form an intricate and dynamic protein network at growing MT plus ends (Akhmanova and Steinmetz, 2008).
A highly conserved and essential +TIP family is represented by XMAP215 in Xenopus laevis and Dis1 in the fission yeast (Slep, 2009). XMAP215 was shown to track MT ends processively and autonomously and to act as an MT polymerase (Brouhard et al., 2008). Experiments in Xenopus egg extracts indicated that XMAP215 is a major MT-stabilizing factor in both mitosis and interphase (Tournebize et al., 2000). In addition to promoting MT polymerization, XMAP215 can also counteract the MT-destabilizing activity of the MT depolymerase XKCM1 (Tournebize et al., 2000; Kinoshita et al., 2001). The mammalian homologue of XMAP215, ch-TOG, also promotes MT assembly in vitro (Charrasse et al., 1998; Bonfils et al., 2007). The cellular function of ch-TOG has been predominantly studied in mitosis, in which it is essential for proper spindle assembly and organization (Gergely et al., 2003; Cassimeris and Morabito, 2004; Holmfeldt et al., 2004; Barr and Gergely, 2008; Cassimeris et al., 2009). However, the role of ch-TOG in interphase cells has not been addressed in detail, and it is unknown whether this protein behaves as an authentic +TIP.
In this study, we identify SLAIN as a new +TIP that associates with EBs, CLASPs, CLIPs, and ch-TOG. We provide evidence that the SLAIN2–ch-TOG complex enriched at MT ends through association with EBs and possibly other +TIPs strongly promotes processive MT growth. During cell division, SLAIN2 is phosphorylated, and thereby, its interaction with the EBs and ch-TOG is inhibited. Therefore, although the disruption of the SLAIN2–ch-TOG complex has a profound effect on MT growth and organization in interphase, it does not affect mitotic progression. Our study provides new insights into the control of MT plus end dynamics during the cell cycle.
Results
Identification of SLAIN1 and SLAIN2 as EB-dependent +TIPs
To identify new EB-interacting partners, we performed GST pull-down assays combined with mass spectrometry using GST-EB1 and different cell extracts (Fig. S1, A and B). Among the new potential EB partners was SLAIN2, which was initially described as a homologue of SLAIN1, a protein named after an amino acid stretch in the C terminus that reads “SLAIN” (Hirst et al., 2006). SLAINs are present in all vertebrates, and two SLAIN homologues exist in mammalian genomes (Hirst et al., 2006). Sequence analysis of SLAIN1 and SLAIN2 predicts a short coiled-coil domain at their N termini (Fig. S1 C). Analysis of a purified SLAIN2 N-terminal fragment (residues 1–43) by circular dichroism spectroscopy, thermal unfolding, and multiangle light scattering showed that it indeed folds into a two-stranded α-helical coiled-coil structure (Fig. S1, D–F). The remaining SLAIN1/2 sequence is rich in serines, prolines, and basic residues, encompasses no conserved sequence regions, and is predicted to be largely unstructured.
Mass spectrometry results were validated by GST-EB pull-down assays with the extracts of cells overexpressing GFP-tagged SLAIN1/2 (Fig. 1 A). Next, we performed GST-EB pull-down assays with different deletion mutants of SLAIN2 (Fig. 1, B and C). The C-terminal part of SLAIN2 contains four SxIP-like sequence motifs as well as a more distantly related motif, RSLP, which are potential EB1 binding sites and MT plus end localization signals (Fig. S1 C; Honnappa et al., 2009). Indeed, two short nonoverlapping C-terminal fragments of SLAIN2 could interact with EB1 in GST pull-down assays, indicating that SLAIN2 has several tandemly arranged EB1 binding sites (Fig. 1, B and C). Both GFP-SLAIN1 and 2 as well as short EB1-binding fragments of SLAIN2 colocalized with EB-positive MT ends (Fig. 1, B and D; Video 1; and not depicted).
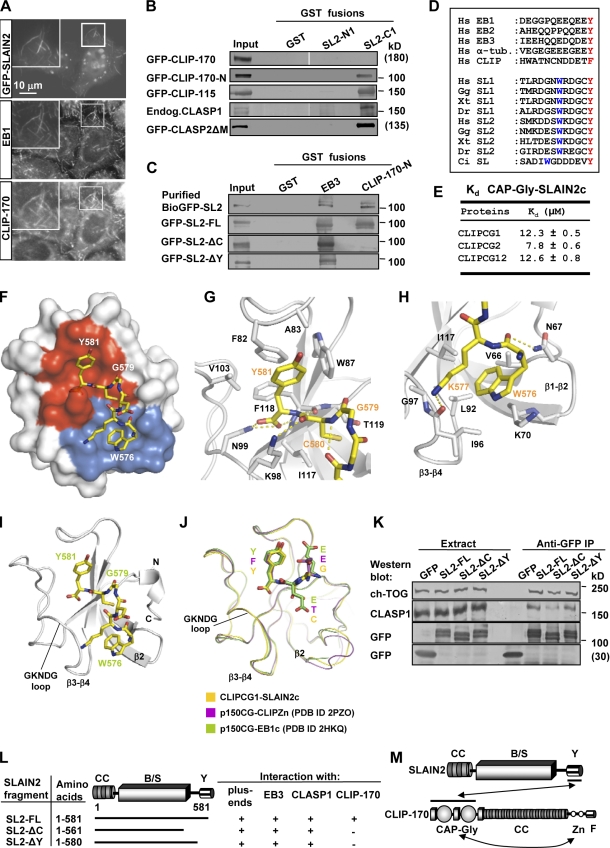
Figure 1.
SLAIN1 and SLAIN2 are EB-dependent +TIPs. (A and C) GST pull-down assays were performed with the indicated GST fusions and lysates of cells expressing different GFP-SLAIN1/2 fusions. Coomassie-stained gels are shown for GST fusions, and Western blots with anti-GFP antibodies are shown for GFP fusions. (B) Mapping of the minimal MT plus end–binding domain of SLAIN2 based on GST-EB1 binding and MT plus end tracking in live cells. CC, coiled coil; B/S, basic and serine rich; Y, C-terminal tyrosine. Asterisks indicate SxIP-like motifs. Mutations in the SxIP-like sites are indicated by red asterisks (see Fig. S1 C for the sequence of SLAIN2). (D) Live-cell imaging of HeLa cells transiently transfected with EB3–monomeric RFP and GFP-SLAIN1/2. Red and green images were collected simultaneously with a beam splitter and a 0.5-s interval; five consecutive frames were averaged. (E) IPs from HeLa cell extracts with rat monoclonal antibodies against HA tag (control) or EB1 and EB3 were analyzed by Western blotting with the indicated antibodies. White lines indicate that intervening lanes have been spliced out. (F and H) 3T3 cells were transfected with the indicated siRNAs, fixed, and stained with the indicated antibodies. The insets show enlargements of the boxed areas. In the overlay, EB1 is shown in green, and SLAIN2 is shown in red. (G) Extracts of 3T3 cells transfected with the indicated siRNAs analyzed by Western blotting with the indicated antibodies. Endog., endogenous; SL2, SLAIN2; WB, Western blot.
To investigate whether the SxIP-like sites in SLAIN2 are required for MT tip tracking, the IP/LP dipeptides within the identified sites were mutated to asparagines (Fig. 1 B and Fig. S1 C). Mutation of the first four sites considerably diminished, but did not abolish, plus end tracking of the C-terminal SLAIN fragment (Video 2). Next, we introduced additional mutations in the fifth, more divergent site, RSLP, as well as two other even more deviant potential EB1 binding sites containing a hydrophobic residue followed by a proline (Fig. S1 C and not depicted). These mutations further reduced plus end tracking (with the RSLP site at position 560 having the strongest contribution to plus end tracking; Fig. S1 C and Video 2), but we still observed a very weak association of the resulting mutants with the growing MT ends as well as binding to EB1 (Video 2 and not depicted). We conclude that SxIP-like sites in SLAIN2 strongly contribute to its plus end–tracking behavior but that additional weak EB1 and MT tip binding sites that do not match the SxIP consensus are present within the SLAIN2 C terminus.
To study endogenous SLAINs, we raised an antibody that recognized both GFP-SLAIN1 and GFP-SLAIN2 (Fig. S1 G). On Western blots of different cell lines, this antibody recognizes protein bands of ∼75–85 kD (Fig. S1 H), which is in agreement with the predicted molecular mass of SLAIN1/2. The presence of multiple bands is likely explained by the existence of alternatively spliced isoforms, phosphorylation, and/or degradation. Endogenous SLAIN1/2 coprecipitates with EB1 (Fig. 1 E) and colocalizes with it at MT plus ends (Fig. 1 F). The accumulation of SLAINs at MT tips depends on EBs, as it is strongly reduced after siRNA-mediated knockdown of EB1 and EB3 (Fig. 1, G and H). Based on these findings, we conclude that SLAIN1/2 accumulate at MT plus ends in an EB-dependent manner.
SLAINs associate with CLIPs
Although at low expression levels, exogenous SLAINs decorated only MT plus ends, at high expression levels, GFP-SLAIN2 bound along MTs and formed bundles of acetylated MTs (Fig. S2 A). The formation of these bundles was dependent on the presence of EB proteins because, in EB1/3-depleted cells, SLAIN2 did not associate with MTs but formed aggregates (Fig. S2 B). In line with these data, we could detect no direct interaction between SLAIN2 and MTs in MT-pelleting assays (unpublished data), indicating that SLAIN2 has no high intrinsic affinity for MTs.
Interestingly, CLASPs and CLIP-170 were also recruited to MT bundles induced by SLAIN overexpression (Fig. 2 A and Fig. S2 A). Previous studies have shown that +TIPs compete for EB binding and that their simultaneous recruitment to MT bundles is observed only when they can interact with each other in an EB-independent manner (Mimori-Kiyosue et al., 2005; Weisbrich et al., 2007). This suggests a direct interaction between SLAIN2 and CLIP-170 and/or CLASPs.
Figure 2.
SLAIN2 interacts with CLIPs and CLASPs. (A) HeLa cells were transiently transfected with GFP-SLAIN2, fixed, and labeled with the indicated antibodies. Insets show enlargements of the boxed areas. (B and C) GST pull-down assays were performed with the indicated GST fusions and lysates of untransfected HeLa cells or cells expressing the indicated GFP fusions (SLAIN2 is abbreviated as SL2). Western blots were performed using the antibodies against GFP or CLASP1. The top lane of C shows a GST pull-down assay with BioGFP-SLAIN2 purified from HEK293 cells. White lines indicate that intervening lanes have been spliced out. (D) Alignment of the C-terminal tails of human EB1, EB2, EB3, α-tubulin (α-tub.), CLIP-170, and SLAIN1 and SLAIN2 from different species. Hs, Homo sapiens; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Ci, Ciona intestinalis. The conserved C-terminal aromatic and the tryptophan residues are highlighted. (E) Equilibrium dissociation constants obtained by ITC for the complexes of the human SLAIN2 peptide (SLAIN2c) with either the first (CLIPCG1), second (CLIPCG2), or both (CLIPCG12) CAP-Gly domains of CLIP-170. (F) Overall view of the heterodimeric complex formed between CLIPCG1 (surface representation) and SLAIN2c (sticks representation). Contact modes A and B are shown in red and blue, respectively. (G and H) Close up views of the interaction network seen in the complex formed between SLAIN2c (yellow carbon atoms) and CLIPCG1 (gray carbon atoms) in the cartoon (main chain) and sticks (contacting residues) representation. Panels G and H depict contact modes A and B, respectively. (I and J) Overall view of the heterodimeric complex formed between CLIPCG1 (ribbon representation) and SLAIN2c (sticks representation; I) and superposition of complexes formed between CAP-Gly domains and C-terminal tyrosine or phenylalanine-containing sequence regions (J). For simplicity, only the last three C-terminal residues of the respective CAP-Gly ligands are shown in sticks representation. (K) IP with anti-GFP antibodies from extracts of HeLa cells expressing GFP or GFP-SLAIN2 fusions were analyzed by Western blotting with the indicated antibodies. (L) Mapping of the SLAIN2 interaction site with CLIP-170, EB3, CLASP1, and MT plus ends. (M) Schematic overview of the SLAIN2–CLIP-170 interaction. CC, coiled-coil; B/S, basic and serine rich; Y, C-terminal tyrosine; Zn, zinc knuckles; F, C-terminal phenylalanine; Endog., endogenous; PDB, Protein Data Bank.
To test this possibility, we performed pull-down assays using GST fusions of SLAIN2 N and C termini but found no significant interaction with full-length CLIP-170 (Fig. 2 B). We hypothesized that this was caused by the formation of an autoinhibitory loop within CLIP-170 blocking the binding of CAP-Gly domains with their partners (Lansbergen et al., 2004). To test this idea, we performed GST pull-down assays with GFP–CLIP-170 N terminus (GFP–CLIP-170-N) or full-length GFP–CLIP-115, two proteins that contain CAP-Gly domains but lack the inhibitory C terminus of CLIP-170 (Lansbergen et al., 2004). We found that both of them associated with the SLAIN2 C terminus (Fig. 2 B). Purified biotinylated and GFP-tagged SLAIN2 (BioGFP-SLAIN2) bound to purified GST–CLIP-170-N as well as GST-EB3 (Fig. 2 C), confirming that the interactions of SLAIN2 with CLIP-170 and EBs are direct and do not depend on the presence of each other or additional +TIPs.
A conspicuous feature of SLAIN sequences is the presence of a highly conserved tyrosine at the outmost C terminus, which is similar to that of α-tubulin, EBs, and CLIP-170 (Fig. 2 D). In the latter proteins, the C-terminal aromatic residue of the EEY/F motif is essential for binding to CAP-Gly domains (Komarova et al., 2005; Honnappa et al., 2006; Mishima et al., 2007; Weisbrich et al., 2007). To determine whether this is also true for the SLAIN2–CLIP-170 interaction, we generated GFP-SLAIN2 deletion mutants lacking the last 20 amino acids (GFP–SLAIN2-ΔC) or only the C-terminal tyrosine (GFP–SLAIN2-ΔY). Both mutants colocalized with endogenous EB1 at MT plus ends and interacted with GST-EB3 in pull-down experiments but displayed no binding to GST–CLIP-170-N (Fig. S2 C and Fig. 2 C). Moreover, MT bundles induced by these SLAIN2 mutants failed to recruit CLIP-170, indicating that the C-terminal tyrosine in SLAIN2 is important for the interaction with CLIP-170 in cells (Fig. S2 C and not depicted).
Biophysical and structural analysis of the SLAIN2–CLIP-170 interaction reveals a new CAP-Gly domain-binding mode
To investigate the SLAIN2–CLIP-170 interaction in more detail, we performed isothermal titration calorimetry (ITC) experiments with a 13–amino acid peptide of SLAIN2 C terminus (SLAIN2c) and with the CAP-Gly domains of CLIP-170 (CLIPCG1 and CLIPCG2). Analysis of the data yielded equilibrium dissociation constants (Kd’s) in the micromolar range (Fig. 2 E and Fig. S2 D). A comparable Kd was obtained for the double CAP-Gly construct CLIPCG12, which bound two SLAIN2c peptides (Fig. 2 E and Fig. S2 D). In contrast, only very weak binding was observed between SLAIN2c and the CAP-Gly domain of the dynactin large subunit p150Glued (p150CG; Fig. S2 D), indicating that the C-terminal regions of SLAINs specifically target the CAP-Gly domains of the CLIPs.
Next, we solved the structure of the SLAIN2c–CLIPCG1 complex by x-ray crystallography (Table S1). As illustrated in Fig. 2 (F–J), the overall structure reveals a heterodimeric complex formed between one CLIPCG1 and one SLAIN2c molecule. The CLIPCG1 subunit displays all features of the CAP-Gly fold (Steinmetz and Akhmanova, 2008). The six C-terminal SLAIN2c residues are tightly packed against the CLIPCG1 fold and assume a looplike conformation (Fig. 2, F–J; and Fig. S2 E).
Analysis of the SLAIN2c–CLIPCG1 binding interface reveals two contact sites, referred to as A and B. Contact A (Fig. 2 G) involves residues from a distinct groove shaped by hydrophobic and polar side chains of CLIPCG1 and the last two C-terminal residues, Cys580 and Tyr581, of SLAIN2c. The side chain of Tyr581, including its α-carboxylate group, is inserted at one end of the CLIPCG1 groove and is specifically recognized by a set of amino acid residues that are highly conserved across CAP-Gly domain homologues (Steinmetz and Akhmanova, 2008). The binding mode of contact A is nearly identical to the ones seen between the CAP-Gly domain of p150Glued and the C-terminal domains of EB1 (Honnappa et al., 2006) and CLIP-170 (Weisbrich et al., 2007) and between the second CAP-Gly domain of CLIP-170 and a peptide derived from the C terminus of α-tubulin (Fig. 2 J; Mishima et al., 2007). The importance of the tyrosine residue for the interaction of the C-terminal SLAIN2 peptide with the two CAP-Gly domains of CLIP-170 was confirmed by ITC (Fig. S2 D).
Contact B (Fig. 2 H) involves residues from a second, distinct groove shaped by hydrophobic and polar side chains of CLIPCG1 and Trp576 and Lys577 of SLAIN2c (Fig. S2 E). The side chain of Trp576, which is highly conserved among SLAIN orthologues (Fig. 2 D), is inserted into this groove and is further buried by the hydrophobic moiety of Lys577. Substitution of Trp576 for alanine abrogates binding of the mutant SLAIN2c peptide (SLAIN2c-W576A) to CLIPCG1 and CLIPCG2 (Fig. S2 D), demonstrating the importance of this conserved residue for binding to the CAP-Gly domains of CLIP-170. Interestingly, the CLIPCG1 residues that contact Trp576 as well as the β1-β2 loop, which establishes one wall of the groove, are only partially conserved in p150CG (Fig. S2 F) and other CAP-Gly domain homologues (Steinmetz and Akhmanova, 2008). Because p150CG binds SLAIN2c more weakly than the two CAP-Gly domains of CLIP-170 (Fig. S2 D), we propose that these residues play an important role in determining the specificity of CAP-Gly domains for tryptophan side chains that precede C-terminal aromatic residues.
SLAINs associate with CLASPs and ch-TOG
Next, we investigated the association of SLAIN2 with CLASPs. Endogenous CLASPs were pulled down by GST–SLAIN2-C1 (Fig. 2 B and not depicted). Although this interaction might be indirect, it was not mediated by EB1, as CLASP2-ΔM, a CLASP2 deletion mutant lacking the EB1 binding site (Komarova et al., 2002; Mimori-Kiyosue et al., 2005), could still bind to GST–SLAIN2-C1 (Fig. 2 B). The CLASP–SLAIN2 interaction was also independent of CLIP-170, as immunoprecipitation (IP) experiments showed that both GFP-SLAIN2 C-terminal mutants that do not bind CLIP-170 could still coprecipitate CLASP1 (Fig. 2 K). Collectively, our results show that the SLAIN2-C1 binds to EBs, CLIPs, and CLASPs (Fig. 2, L and M). Targeting of SLAIN2 to the MT plus ends depends on EBs (Fig. 1, F and H), whereas GFP-SLAIN2 still localizes to MT tips in cells depleted of CLIP-170 and CLASPs (Fig. S2 G). The interactions with CLIPs and CLASPs are likely needed to avoid competition between these proteins at MT tips.
To identify additional SLAIN partners, we performed streptavidin-based pull-down assays of BioGFP-SLAIN1/2 from HeLa cells (Fig. 3 A) and analyzed the resulting proteins by mass spectrometry. The experimental set up was validated by the identification of CLASP1 among the proteins associated with BioGFP-SLAIN2 (Fig. S3 A). Furthermore, on a Coomassie-stained gel, a prominent band of ∼220 kD was visible in both SLAIN1 and SLAIN2 lanes (Fig. 3 A). This protein was identified as ch-TOG (Fig. S3 A).
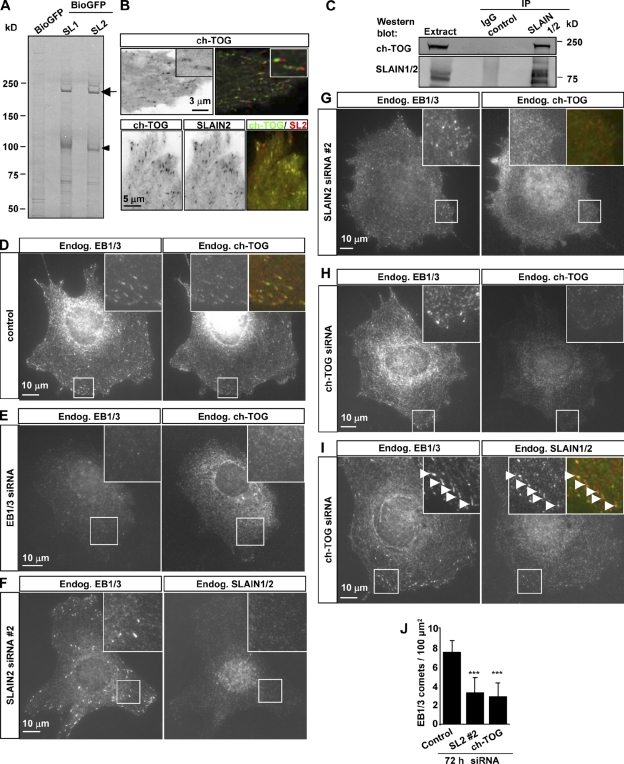
Figure 3.
SLAIN2 interacts with ch-TOG and promotes its MT plus end accumulation. (A) Streptavidin pull-down assay from HeLa cells expressing BioGFP or BioGFP-SLAIN1 (SL1) or -SLAIN2 (SL2) together with BirA. Proteins were analyzed by Coomassie staining. The arrow indicates ch-TOG, and the arrowhead indicates BioGFP-SLAIN1/2. (B) Live imaging of HeLa cells transiently expressing GFP–ch-TOG alone (top) or in combination with mCherry-SLAIN2 (bottom) collected as described for Fig. 1 D. The top right image shows maximum intensity projection of two consecutive averaged frames displayed in green and red. Insets show enlarged views of individual MT plus ends. (C) IPs from HeLa cell extracts with either the IgG control or SLAIN1/2 antibody were analyzed by Western blotting with the indicated antibodies. (D–I) 3T3 cells were transiently transfected with different siRNAs, fixed, and stained with the indicated antibodies. The insets show enlargements of the boxed areas. Arrowheads in I indicate MT plus ends. In the overlay in D, G, and I, EB1/3 is in red, and ch-TOG or SLAIN1/2 is in green. (J) Quantification of the number of EB1/3-positive comets per 100-µm2 surface area in control 3T3 cells or cells depleted of SLAIN2 or ch-TOG (11–14 cells were analyzed for each condition). Error bars show SD. Values significantly different from control are indicated with asterisks; ***, P < 0.001. Endog., endogenous.
Although it is generally accepted that XMAP215 and its homologues act at MT tips (Brouhard et al., 2008), the vertebrate members of the XMAP215/Dis1 family have not been shown to form comet-like accumulations at MT ends in cells. Using live-cell imaging, we could detect GFP-tagged ch-TOG at the growing MT ends, where it colocalized with SLAIN1/2 and EBs (Fig. 3 B, Video 3, and not depicted). Endogenous SLAIN and ch-TOG were coprecipitated with endogenous EB1 and with each other (Fig. 1 E and Fig. 3 C). Endogenous ch-TOG was also coprecipitated with full-length GFP-SLAIN2 and with its mutants deficient in CLIP-170 binding, demonstrating that the SLAIN2–ch-TOG interaction does not require CLIP-170 (Fig. 2 K).
EBs and SLAIN2 promote ch-TOG accumulation at MT tips
In agreement with previous studies, we found that endogenous ch-TOG localized to centrosomes in interphase and to spindle poles and MTs in mitosis (unpublished data; Tournebize et al., 2000; Gergely et al., 2003). In addition, ch-TOG colocalized with endogenous EBs at the MT plus ends of interphase cells (Fig. 3 D). By performing a double knockdown of EB1 and EB3, we found that ch-TOG requires EBs for efficient cometlike accumulation at the MT plus ends but not at the centrosome (Fig. 3 E and not depicted).
Next, we investigated the hierarchy of SLAIN and ch-TOG interactions with MT plus ends. In HeLa cells, all SLAIN-specific bands could be depleted by different siRNAs against SLAIN2, whereas SLAIN1-specific siRNAs had no effect. This result suggests that HeLa cells do not express SLAIN1, a conclusion supported by RT-PCR analysis (Fig. S3, B and C). Using the SLAIN2 siRNA#2, we could efficiently deplete SLAIN2 also from mouse 3T3 cells (Fig. S3 D and Fig. 3 F). Interestingly, in cells depleted of SLAIN2, EB1/3 comets displayed little ch-TOG labeling (Fig. 3 G), indicating that SLAIN2 participates in ch-TOG recruitment to MT ends. In contrast, both CLASPs and CLIPs could still be detected at the plus ends of SLAIN2-depleted cells (Fig. S3, E and F).
Next, we used siRNAs to deplete endogenous ch-TOG. Because of the strong mitotic arrest caused by ch-TOG knockdown (Gergely et al., 2003; Cassimeris and Morabito, 2004), we enriched interphase cells by applying a thymidine block for 2 d. Knockdown of ch-TOG in HeLa and 3T3 cells could be confirmed both by Western blotting (Fig. S3, D and G) and by immunostaining (Fig. 3 H). Although SLAIN2 levels were somewhat reduced in ch-TOG–depleted cells, it could still be detected at the EB1/3-positive MT tips (Fig. S3, D and G; and Fig. 3 I), indicating that SLAIN2 acts upstream of ch-TOG with respect to MT tip localization.
Both SLAIN2 and ch-TOG depletion strongly reduced the number and length of EB1/3-positive comets (Fig. 3 J and not depicted), suggesting a defect in MT growth. An overall disorganization of the MT network was also observed: instead of the radial array typical for 3T3 fibroblasts, MTs often appeared to be circularly arranged and entangled (Fig. S3 H).
SLAIN2 promotes the interaction between EB1 and ch-TOG
To characterize the SLAIN2–ch-TOG interaction in more detail, we performed pull-down experiments with deletion mutants of the two proteins. We found that the whole N-terminal half of SLAIN2 (SLAIN2-N1), but not its shorter fragments, bound to ch-TOG; this interaction depended on the C-terminal domain of ch-TOG (Fig. 4, A–D). Using GST pull-down assays, we found that, as expected, GFP-SLAIN2, but not GFP–SLAIN2-N1, was pulled down by GST-EB1 (Fig. 4 E). Endogenous ch-TOG also interacted with GST-EB1, in line with our mass spectrometry data (Fig. 4 E and Fig. S1 B). This interaction was increased when GFP-SLAIN2 was overexpressed (Fig. 4 E and Fig. S4 A), suggesting that full-length SLAIN2 can stabilize the binding between ch-TOG and EB1. In agreement with this idea, we found that GFP–ch-TOG-C1, which localized at centrosomes and in the cytosol of control cells (Fig. S4 B) was recruited to MT plus ends when SLAIN2 levels were elevated by expressing mCherry-SLAIN2 (Fig. S4 C). On the other hand, when GFP–SLAIN2-N1 was overexpressed, endogenous ch-TOG could no longer be detected at the MT plus ends, and its interaction with GST-EB1 was strongly reduced (Fig. 4, E and F; and Fig. S4 A), indicating that the SLAIN2 N terminus uncouples ch-TOG from EB1.
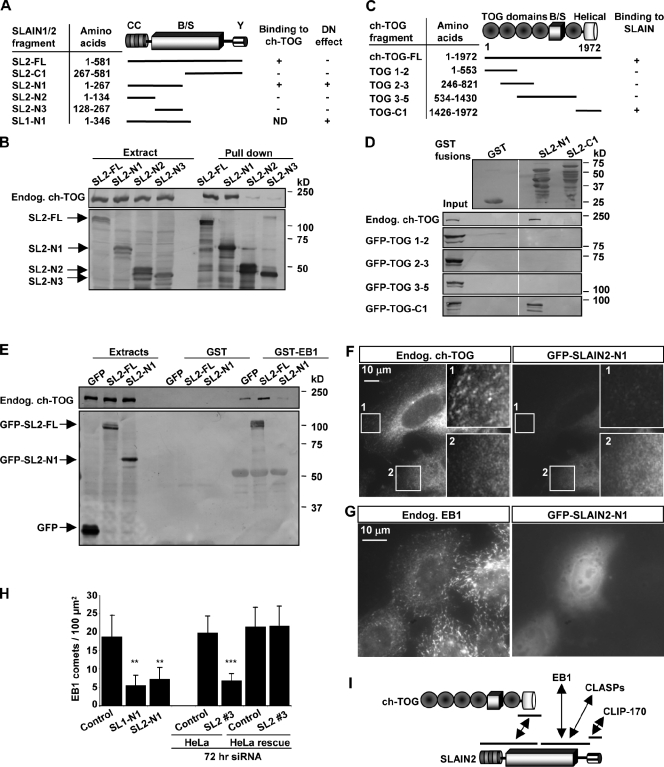
Figure 4.
SLAIN2 links ch-TOG to EB1. (A) Mapping of the ch-TOG–binding domain of SLAIN1 and 2 (SL1 and SL2). CC, coiled coil; B/S, basic and serine rich; Y, C-terminal tyrosine. The ability to exert a dominant-negative (DN) effect on the number of EB1-positive MT tips is indicated. (B) Streptavidin pull-down assays were performed with extracts of HEK293 cells coexpressing BioGFP-SLAIN2 mutants and BirA and analyzed by Western blotting with the indicated antibodies. (C) Mapping of the SLAIN2-binding domain of ch-TOG. (D and E) GST pull-down assays were performed with the indicated GST fusions and lysates of HEK293 cells expressing GFP fusions of ch-TOG or SLAIN2. Coomassie-stained SDS-PAGE is shown for GST fusions. Western blots with anti-GFP antibodies are shown for GFP fusions and with ch-TOG antibodies for endogenous (Endog.) ch-TOG. White lines indicate that intervening lanes have been spliced out. (F and G) 3T3 (F) or HeLa cells (G) were transfected with GFP–SLAIN2-N1, fixed, and labeled with the indicated antibodies. In F, the insets show enlargements of the boxed areas where 1 is an untransfected control cell and 2 is a GFP–SLAIN2-N1–transfected cell. (H) Quantification of the number of EB1-positive comets per 100-µm2 surface area in control or GFP–SLAIN1/2-N1–expressing HeLa cells. Approximately 10–50 cells were analyzed in each experiment. Error bars show SD. Statistically significant differences are indicated (**, P < 0.01; ***, P < 0.001). (I) A scheme of the identified interactions between SLAIN2, ch-TOG, EB1, CLASPs, and CLIP-170.
Next, we examined the functional consequences of the disruption of the ch-TOG–EB1 interaction caused by overexpression of GFP–SLAIN2-N1 and found that it induced a strong reduction in the number of EB1-positive MT tips, whereas MT density remained unchanged (Fig. 4, G and H; and Fig. S4 D). Overexpression of the SLAIN1 N terminus caused an identical phenotype (Fig. 4 H and Fig. S4 D), suggesting that the functions of SLAIN1 and SLAIN2 are similar. Reduction in the number of EB1 comets was also observed in cells overexpressing the ch-TOG C terminus, which is also expected to uncouple endogenous ch-TOG from SLAIN and EB1 (Fig. S4, B and E) as well as after SLAIN2 and ch-TOG depletion (Fig. 3 J, Fig. 4 H, and Fig. S4 D). Importantly, the number of growing MT ends was fully rescued by stable expression of low levels of GFP-SLAIN2 in SLAIN2-depleted cells (Fig. 4 H), confirming the specificity of the SLAIN2 siRNA.
Collectively, these results indicate that SLAIN2 can promote the binding between ch-TOG and EB1 and potentially other +TIPs (Fig. 4 I) and help to recruit ch-TOG to MT plus ends and to maintain the normal number of growing MTs.
SLAIN2–ch-TOG complex is required for persistent MT growth and radial MT organization
To analyze the effect of the loss or disruption of the SLAIN2–ch-TOG complex on MT growth, we used HeLa cells stably expressing fluorescently tagged tubulin. We followed the dynamics of freshly polymerized MTs in the internal cytoplasm after photobleaching of the preexisting MTs (Fig. 5 A). We found that the length of MT stretches polymerized after FRAP was dramatically reduced in cells that were depleted of SLAIN2 and ch-TOG or that were expressing the dominant-negative SLAIN1/2-N1 mutants (Fig. 5, A and B). The mean MT elongation rate, determined using fluorescently tagged tubulin or EB3-GFP without taking into account short pauses or depolymerization events, was reduced approximately twofold (Fig. 5, C and D; and not depicted). This change in rate was primarily caused by a two- to threefold increase in the catastrophe frequency (Fig. S5 A and Table S2). Although in control cells, MT growth episodes were relatively “smooth,” in SLAIN2- and ch-TOG–depleted cells, they were continuously interrupted by short depolymerization events or pauses, an effect that was particularly obvious in kymograph analysis (Fig. 5 E, Table S2, and Fig. S5 A). Interestingly, although the instantaneous rate of MT growth bursts was not significantly changed (Fig. S5 B), EB3-GFP comets became shorter (Fig. 5, F and G; and Video 4), suggesting an altered state of the MT plus end. The MT depolymerization rate was not significantly affected, and the rescue frequency remained high (it was even elevated in SLAIN2-depleted, but not in ch-TOG–depleted, cells (Fig. S5 A and Table S2). The frequent switching between short episodes of growth and depolymerization led to a strong overall decrease in MT elongation (Fig. 5, C–E).
Figure 5.
SLAIN2 and ch-TOG promote MT growth. (A) HeLa cells stably expressing GFP– or mCherry–α-tubulin were transfected with the indicated siRNAs. FRAP assay was performed 72 h later in an internal part of the lamella indicated by a stippled line. Video frames at 33 s after FRAP are shown. Newly polymerized MTs are indicated by arrows. (B) Share of freshly polymerized MT segments longer than 2 µm 30 s after FRAP. Approximately 230–300 growth episodes were analyzed in 15–20 cells for each condition. (C and D) Mean MT elongation rate was measured in internal cytoplasm over periods of 10–50 s from the moment of appearance of the growing MT end until the end of the video or until a catastrophe leading to a processive MT-shortening episode with the length of >1 µm. Analysis was performed in HeLa cells stably expressing EB3-GFP that were transfected with the indicated siRNAs (C) or in HeLa cells stably expressing mCherry–α-tubulin transfected with GFP or GFP–SLAIN1/2-N1 (D). Approximately 70–100 growth episodes in 10–20 cells were analyzed for each condition. (E) Kymographs illustrating MT growth using mCherry–α-tubulin or EB3-GFP after different siRNA treatments or in cells expressing GFP alone (control) or GFP–SLAIN1/2-N1. (F) CHO cells were transiently transfected with EB3-GFP and the indicated shRNAs. Live images were collected with a 0.5-s time interval. Single frames (top) and maximum intensity projections of 100 frames (bottom) are shown. (G) Length of EB3-GFP comets in control, SLAIN2, or ch-TOG–depleted cells determined from live imaging experiments shown in F. Approximately 300 MT tips were analyzed in ∼15 cells per condition. (H and I) Proportion of MT tracks originating from the centrosome with the length exceeding 7.5 µm (H) and MT nucleation frequency from the centrosome in CHO cells transiently transfected with EB3-GFP and the indicated shRNAs (I). In H, ∼100 MT growth episodes were analyzed in ∼15 cells per condition. In I, ∼10 cells were analyzed per condition. (J and K) MT recovery after nocodazole washout. HeLa cells expressing GFP or GFP–SLAIN1/2-N1 for 1 d were treated with 10 µm nocodazole for 2 h. The drug was washed out with fresh medium, and cells were fixed and stained 10 min later (J). The number of MTs per 100 µm2 was counted in 20 cells for each condition (K). (L and M) MT organization in HeLa cells transiently expressing GFP or GFP–SLAIN1/2-N1 1 d after transfection. Cells were stained for β-tubulin (L), and the angles of MT segments in relation to the long axis of the lamella were measured. Angle distributions measured in 10 cells and their Gaussian fits are shown for each condition (M). Transfected cells are indicated by asterisks in J and L. In C, D, G–I, and K, the values significantly different from controls are indicated with asterisks (**, P < 0.01; ***, P < 0.001).
An increased catastrophe frequency was also observed after depletion of SLAIN2 and ch-TOG in 3T3 and CHO cells (Fig. S5 A and Table S2). We have shown previously that in the latter cell type, most of the growing MTs emerging from the centrosome extend persistently to the cell periphery and that ∼80% of the MT growth tracks visualized by EB3-GFP are longer than half of the mean cell radius (7.5 µm; Komarova et al., 2009). Depletion of SLAIN2 and ch-TOG reduced this value to <10% (Fig. 5, F and H), which is consistent with a strong increase in catastrophe frequency.
The depletion of ch-TOG and SLAIN2 might also cause a reduction in the number of growing MT ends by affecting MT nucleation, as XMAP215/Dis1 proteins play an important role at the centrosome (see Cassimeris et al., 2009 and references therein). Indeed, MT nucleation frequency determined with EB3-GFP was significantly reduced after SLAIN2 and ch-TOG knockdown (Fig. 5 I). The involvement of the SLAIN2–ch-TOG complex in MT nucleation was further supported by the fact that its disruption by overexpressing SLAIN1/2 N termini strongly delayed MT recovery after nocodazole treatment: 10 min after the drug washout, the MT network was almost completely restored in control cells but was virtually absent in cells expressing SLAIN1/2 N termini (Fig. 5, J and K).
We further found that in all situations in which the SLAIN2–ch-TOG complex was depleted or disrupted, MTs were severely disorganized: instead of running from the cell center to the cell periphery, MTs were arranged at random angles and strongly entangled (Fig. 5, L and M; Fig. S3 H; and Fig. S5 C). Collectively, these data show that SLAIN2 and ch-TOG act as major MT growth-promoting factors by stimulating MT nucleation and growth and suppressing catastrophes. They further indicate that SLAIN2- and ch-TOG–dependent processive MT polymerization is an important factor for maintaining normal interphase MT array.
Mitotic phosphorylation of SLAIN2 disrupts the EB1–SLAIN2–ch-TOG complex
Depletion of ch-TOG causes severe mitotic defects (Gergely et al., 2003; Cassimeris and Morabito, 2004; Holmfeldt et al., 2004). Indeed, ch-TOG knockdown induced a strong increase in the mitotic index of HeLa cells (Fig. 6 A). In contrast, two of the three SLAIN2 siRNAs that efficiently deplete SLAIN2 (Fig. S3 B) caused no increase in the proportion of mitotic cells (Fig. 6 A) and no apparent abnormalities of the mitotic apparatus (not depicted). Localization experiments using a HeLa cell line stably expressing GFP-SLAIN2 showed that it dissociated from MT plus ends and the centrosome early in prophase and remained cytosolic until late telophase (Fig. 6 B, Video 5, Video 6, and Video 7).
Figure 6.
Mitotic phosphorylation disrupts SLAIN2 interaction with MTs, EB1, and ch-TOG. (A) Proportion of mitotic HeLa cells identified by staining with antibodies against histone H3 phosphorylated at serine 10 after transfection with the indicated siRNAs. (B) HeLa cells stably expressing GFP-SLAIN2 (SL2) were fixed and stained with anti-EB1 antibodies. The prophase cell was distinguished by the strong increase in centrosomal MT nucleation. Insets show enlargements of the boxed areas. (C) IPs with anti-SLAIN1/2 antibodies from control HeLa cells or cells blocked in mitosis with 0.1 µM nocodazole. Where indicated, immunoprecipitated material was treated on beads with λ phosphatase. Western blotting was performed with the indicated antibodies. (D) The same experiment as in C but using a GFP-SLAIN2 stable HeLa cell line and anti-GFP antibodies. (E) GST pull-down assays were performed with GST or GST-EB1, and cell extracts were prepared as in D. (F) The same experiment as in C but using HeLa cells transiently expressing N- and C-terminal fragments of SLAIN2. (G and H) HeLa cells stably expressing GFP-SLAIN2 were blocked in mitosis with 7.5 µM STLC (G) or 0.1 µM nocodazole (H) and released for 1 h into medium containing either the indicated inhibitors, 20 mM NaCl, or 20 mM LiCl. White lines indicate that intervening lanes have been spliced out. S, STLC; N, nocodazole; FP, flavopiridol; Endog., endogenous; Cont., control; Mit., mitotic; Extr., extract.
Western blot analysis of extracts of cells blocked in mitosis showed that endogenous SLAIN2 and GFP-SLAIN2 protein bands are very strongly up-shifted (Fig. 6, C and D). Incubation of immunoprecipitated SLAIN2 with λ phosphatase completely reversed this shift, indicating that it is caused by phosphorylation (Fig. 6, C and D). The presence of multiple phosphorylated sites in SLAIN2 isolated from mitotic cells was confirmed by mass spectrometry (Fig. S1 C).
In agreement with the absence of GFP-SLAIN2 at the MT plus ends in mitosis, hyperphosphorylated GFP-SLAIN2 did not associate with GST-EB1 in a GST pull-down assay (Fig. 6 E). Furthermore, mitotic phosphorylation also inhibited coprecipitation of SLAIN2 and ch-TOG (Fig. 6, C and D). In line with these observations, both SLAIN2 N (the ch-TOG–binding domain) and C termini (the EB1-, CLIP-, and CLASP-binding domains) showed strongly reduced electrophoretic mobility in mitotically blocked cells (Fig. 6 F). These results indicate that mitotic hyperphosphorylation inhibits the interaction of SLAIN2 with EB1 and ch-TOG.
To investigate which kinase is responsible for SLAIN2 phosphorylation, HeLa cells stably expressing GFP-SLAIN2 were blocked in mitosis and subsequently released for 1 h in the presence of different kinase inhibitors. Inhibition of Cdk1 using RO-3306 or flavopiridol caused a strong downward shift of GFP-SLAIN2 protein bands on Western blots (Fig. 6, G and H). This increase in GFP-SLAIN2 electrophoretic mobility was not caused by proteasome activity, as it was also observed in the presence of the proteasome inhibitor MG132 (Fig. 6, G and H). Inhibitors of other kinases, such as Plk1 or GSK3-β, had no effect (Fig. 6 G). Sequence analysis of SLAIN2 predicts multiple major (S/TPxPK/R) and minor (S/TP) Cdk1 consensus sites, and several of them are indeed phosphorylated based on mass spectrometry analysis (Fig. S1 C), suggesting that SLAIN2 might be a direct target of Cdk1. Collectively, these data show that in mitosis, the interaction of SLAIN2 with EB1 and ch-TOG is inhibited by phosphorylation and that SLAIN2-dependent recruitment of ch-TOG to MT tips is, thus, confined to interphase cells.
Discussion
In this study, we identified SLAINs as +TIPs and showed that SLAIN2 plays an important role in regulating MT growth and organization by interacting with multiple +TIPs and targeting ch-TOG to growing MT plus ends. Similar to many +TIPs (Akhmanova and Steinmetz, 2008), SLAINs bind to EBs and depend on them for MT tip accumulation. In addition, the C-terminal part of SLAIN2 interacts with the CLASPs and the CLIPs. The SLAIN2–CLIP interaction involves the C-terminal tail of SLAINs and the CAP-Gly domains of CLIPs. Notably, the conserved terminal tyrosine residue of SLAIN2 is recognized by the CAP-Gly fold in exactly the same way as the ones of the EEY/F motifs of EBs, α-tubulin, and CLIP-170. However, although in EBs, α-tubulin, and CLIP-170, the acidic residues of EEY/F contribute to the stability of the complexes through electrostatic interactions (Mishima et al., 2007), a conserved tryptophan in SLAIN2 interacts with a distinct hydrophobic cavity present in the CAP-Gly domains of CLIP-170. Our structural data therefore extend the specificity portfolio of CAP-Gly domains and explain the functional role of the evolutionarily conserved SLAIN C terminus.
SLAINs combine different and multiple copies of major +TIP elements, such as SxIP-like and EEY/F-like motifs, in one molecule (Fig. 7). This unique property of SLAINs is expected to enable them to associate simultaneously with several different +TIPs and, thus, promote formation of +TIP interaction networks with multiple and partly redundant binding nodes (Fig. 7). The functional significance of each particular interaction might be relatively small; however, their overall effect would be to enable SLAINs to act as adhesive +TIP factors, which have a continuous access to growing MT ends, enhance +TIP interactions, and promote their access to the MT ends.
Figure 7.
A scheme of SLAIN2-linked +TIP network and its biological role. Different types of +TIP interactions are indicated by different colors. The potential direct EB1–ch-TOG interaction based on the data from other species (Rehberg and Gräf, 2002; Wolyniak et al., 2006; Kronja et al., 2009) is not depicted. SLAIN2-dependent interactions enhance ch-TOG accumulation at the MT tip. ch-TOG, in turn, promotes processive MT growth that is required for proper MT organization.
Importantly, we found that SLAINs use their adhesive properties to robustly target ch-TOG to growing MT plus ends. ch-TOG binds to SLAINs through its C-terminal part, which is distinct from its tubulin-binding TOG domains, necessary for MT polymerase activity (Slep, 2009). SLAINs likely promote the activity of ch-TOG by positioning it close to growing MT ends. However, in vitro XMAP215 can autonomously track MT plus ends (Brouhard et al., 2008) and stimulate MT growth in the absence of other +TIPs (Kinoshita et al., 2001). So why would the members of the XMAP215/Dis1 family need an accessory factor in vivo? We propose that in cells, the ability of XMAP215/Dis1 family members to interact with EB proteins and other +TIPs is needed to gain access to growing MT tips that are strongly occluded by EBs and their numerous partners.
Although SLAINs are conserved in vertebrates and tunicates, we could identify no apparent SLAIN counterparts in other taxa, possibly because of a low degree or absence of sequence conservation. Alternatively, lower organisms might not need accessory factors for XMAP215/Dis1 homologues because they are expressed at higher levels or because their +TIP networks are less complex.
Because SLAINs localize to MT tips by interacting with EBs and EB partners (Fig. 7), one could expect that EB depletion should have an MT growth phenotype similar to that of ch-TOG or SLAIN2. Our previous study showed that the depletion of EBs indeed increased the catastrophe frequency (Komarova et al., 2009), but the effect was relatively mild. This difference could be caused by the difficulty to completely deplete all three mammalian EBs (Komarova et al., 2009). Alternatively, EBs can concentrate at MT tips not only in MT-stabilizing proteins, such as CLASPs, but also destabilizing factors, such as MCAK (Montenegro Gouveia et al., 2010). Therefore, when EBs are knocked down, the balance of regulatory activities at MT ends might not be significantly shifted. In contrast, SLAIN2 or ch-TOG depletion affects only growth-promoting, but not destabilizing, activities at MT tips, and MT polymerization is inhibited.
Overall, our functional data are consistent with the view that in interphase cells, SLAIN2 primarily acts as a ch-TOG cofactor at growing MT ends. Remarkably, the SLAIN–ch-TOG as well as SLAIN–EB partnership is disrupted during cell division. As the cell proceeds from interphase into mitosis, MT dynamics drastically changes, and MT turnover strongly increases (Wittmann et al., 2001). Our data suggest that phosphorylation of SLAIN2 at the onset of mitosis is part of this mitosis-specific regulatory switch in MT dynamics. It remains to be investigated whether, in mitosis, ch-TOG targets MT tips autonomously or uses some additional partners. In conclusion, our study identifies SLAIN2 as an important component of a complex MT plus end–targeting mechanism and reveals the function of SLAIN2 in the cell cycle–regulated control of MT dynamics.
Materials and methods
Constructs
GFP-SLAIN1 and -SLAIN2 expression constructs and their deletion mutants were generated using the mouse cDNA IMAGE (integrated molecular analysis of genomes and their expression) clone 6811096 and the human cDNA KIAA1458 (a gift from Kazusa DNA Research Institute) in pEGFP-C1 by PCR-based strategies. In BioGFP fusions, a linker encoding the sequence MASGLNDIFEAQKIEWHEGGG, which is the substrate of biotin ligase BirA is inserted into the NheI and AgeI sites in front of the GFP (pBioGFP-C1). The BirA ligase expression construct, which contains the Escherichia coli BirA open reading frame fused to the triple HA tag and cloned into the cytomegalovirus enhancer and promoter-containing vector pSCT (Driegen et al., 2005) was a gift from D. Meijer (Erasmus MC, Rotterdam, Netherlands). mCherry-SLAIN1/2 were made by recloning SLAIN1/2 into MluI–EcoRI sites of a modified pEGFP vector in which the GFP open reading frame was substituted for that of mCherry (a gift from R. Tsien, University of California, San Diego, San Diego, CA; Shaner et al., 2004). GFP–ch-TOG, based on a human ch-TOG cDNA in pEGFP-C–based vector, was a gift from L. Wordeman (University of Washington, Seattle, WA); this construct was used for generating ch-TOG deletion fragments in pBioGFP-C1 by a PCR-based strategy. We also used the following previously described pEGFP-C1–based expression constructs: GFP–α-tubulin, encoding human α-tubulin (Takara Bio Inc.), GFP–CLIP-115, which encodes full-length rat CLIP-115 (De Zeeuw et al., 1997), GFP–CLIP-170, which encodes full-length rat CLIP-170 (Hoogenraad et al., 2000), GFP–CLIP-170-N, which encodes amino acids 4–309 of the rat CLIP-170 (Komarova et al., 2002) and GFP–CLASP2-ΔM, which encodes amino acids 36–340 fused in frame to amino acids 581–1,294 of CLASP2-γ (Mimori-Kiyosue et al., 2005). mCherry–α-tubulin contained a human α-tubulin coding sequence N-terminally fused to mCherry (Shaner et al., 2004). EB3-GFP, based on the human cDNA, was generated in pEGFP-N1 (Stepanova et al., 2003). Point mutations in the GFP–SLAIN2-C3 fragment were introduced by overlapping PCR. pSuper-based small hairpin RNA (shRNA) vectors (Brummelkamp et al., 2002) were directed against the following target sequences: mouse/rat ch-TOG, 5′-AGAGTCCAGAATGGTCCAA-3′; and mouse/rat/human SLAIN2, 5′-CTCTATAGATAGTGAGTTA-3′.
Cell culture, stable cell lines, and transfection of DNA constructs
HeLa, Swiss 3T3, CHO, and HEK293 were cultured in medium which consisted of 45% DME, 45% Ham’s F10, and 10% fetal calf serum supplemented with penicillin and streptomycin (Akhmanova et al., 2001). HeLa cell lines stably expressing GFP–α-tubulin, EB3-GFP (Mimori-Kiyosue et al., 2005), mCherry–α-tubulin (Splinter et al., 2010), GFP-SLAIN2, and the GFP-SLAIN2 #3 rescue construct and the 3T3 cell line stably expressing EB3-GFP were selected using FACS and cultured in the presence of 0.4 mg/ml G418 (Roche). PolyFect (QIAGEN), FuGENE 6 (Roche), or Lipofectamine 2000 (Invitrogen) reagents were used for plasmid transfection.
siRNAs
siRNAs were synthesized by Invitrogen or Thermo Fisher Scientific. They were directed against the following target sequences: SLAIN1 #1, 5′-GACAUGUAGUGAACAAGAA-3′; SLAIN1 #2, 5′-GUAACAUGCCUUUAUCAAA-3′; SLAIN1 #3, 5′-GCAGCAACAGUAUUAUUC-3′; SLAIN2 #1, 5′-GCGCAGUUCUGGUUCAUCU-3′; SLAIN2 #2, 5′-CUCUAUAGAUAGUGAGUUA-3′; SLAIN2 #3, 5′-GGAACUUGAUGCACAAAGU-3′; control, 5′-GCACUCAUUAUGACUCCAU-3′ (Mimori-Kiyosue et al., 2005); human ch-TOG, 5′-GAGCCCAGAGUGGUCCAAA-3′ (Cassimeris and Morabito, 2004); mouse ch-TOG, ON-TARGETplus SMARTpool L-0470; EB1, 5′-AUUCCAAGCUAAGCUAGAA-3′ (Watson and Stephens, 2006); EB3, 5′-CUAUGAUGGAAAGGAUUAC-3′ (Komarova et al., 2005); CLIP-170, 5′-GGAGAAGCAGCAGCACAUU-3′ (Lansbergen et al., 2004); CLASP1, 5′-GCCAUUAUGCCAACUAUCU-3′; and CLASP2, 5′-GUUCAGAAAGCCCUUGAUG-3′ (Mimori-Kiyosue et al., 2005). Synthetic oligos were transfected using a transfection reagent (HiPerFect; QIAGEN) at a concentration of 5 nM. Cells were analyzed 72 h after transfection.
Drug treatments
In the case of siRNA-mediated ch-TOG and SLAIN2 knockdown, 3T3 and HeLa cells were blocked in interphase 1 d after transfection by adding 2 mM thymidine (Sigma-Aldrich) to culture medium for 2 d. HeLa cells were blocked in mitosis by a 16 h treatment with 0.1 µM nocodazole (Sigma-Aldrich) or with 7.5 µM S-trityl-l-cysteine (STLC; Eg5 inhibitor; Sigma-Aldrich). Cells were released from the mitotic block for 60 min in the presence of the following inhibitors: 10 µM RO-3306 (EMD), 100 nM BI-2536 (Selleck), 20 mM LiCl, 10 µM flavopiridol (Sigma-Aldrich), and 20 µM MG132 (Sigma-Aldrich). Nocodazole washout experiments were performed by applying 10 µM nocodazole for 2 h followed by washout of the drug for 5–20 min.
RT-PCR
Total RNA was isolated from HeLa cells using RNA-Bee (Tel-Test, Inc.) according to the manufacturer’s protocol. cDNA was generated using a reverse transcription system (First-Strand cDNA synthesis SuperScript II RT; Invitrogen). Human brain cDNA was a gift from E. Mientjes (Erasmus MC, Rotterdam, Netherlands). Primers used for amplification of SLAIN2 were as follows: forward, 5′-TAAGTGCTTCAGAATTAGAT-3′; and reverse, 5′-CATCATGCAGTATACCCTG-3′. SLAIN1 primers were previously described (Smith et al., 2010).
Protein purification, peptide preparation, pull-down assays, and protein analysis
GST fusions of the N-terminal and C-terminal fragments of SLAIN2 were generated in pGEX-4T-1. GST-EB1, -EB2, -EB3, –EB1-N (amino acids 1–124), and –EB1-C (amino acids 125–268) were generated in pGEX-3X using full-length mouse EB1 and EB2 (long isoform) and human EB3 (long isoform) cDNAs (Komarova et al., 2005). GST–CLIP-170-N (amino acids 1–310 of rat CLIP-170) was described previously (Lansbergen et al., 2004). GST pull-downs, IPs, and Western blotting were performed according to Komarova et al. (2005) and Lansbergen et al. (2004). Treatments with λ phosphatase (New England Biolabs, Inc.) were performed on beads after GST pull-down assays or IP. All GST fusions were expressed in BL21 E. coli and purified with glutathione–Sepharose 4B (GE Healthcare) according to the manufacturer’s instructions. The His6-tagged SLAIN2 fragment (amino acids 1–43) was inserted in the vector PSTCm1 (Olieric et al., 2010). It was expressed in BL21 (DE3; Agilent Technologies) and purified by immobilized metal affinity chromatography on Ni2+-Sepharose (GE Healthcare) followed by size exclusion chromatography using a separation column (Superdex 200 10/300 GL; GE Healthcare). Identity of the protein was confirmed by liquid chromatography/mass spectrometry. BioGFP-SLAIN2 was purified from HEK293T cells. 70% confluent HEK293T cells were cotransfected with the constructs BioGFP-SLAIN2 and BirA using Lipofectamine 2000. 1 d after transfection, cells were lysed in a buffer containing 20 mM Tris-HCl, 100 mM KCl, 1% Triton X-100, and protease inhibitors (Complete; Roche) and purified with Mutein beads (Roche) according to the manufacturer’s instructions.
Human CLIP-170 and p150Glued fragments CLIPCG1 (residues 56–128), CLIPCG2 (residues 210–282), CLIPCG12 (residues 48–300), and p150CG (residues 18–111) were cloned into pETG20A-MTA using the Gateway cloning system (Invitrogen) and purified as previously described in Weisbrich et al. (2007). In brief, transformed E. coli strains BL21 (DE3; Agilent Technologies; for CLIPCG1, CLIPCG2, and p150CG) and C41 (DE3; Lucigen; for CLIPCG12) were grown at 37°C in Luria-Bertani media to an OD600 of 0.7. Expression was induced with 1mM IPTG and performed overnight at 20°C. The His6-tagged fusion proteins were affinity purified by immobilized metal affinity chromatography on Ni2+-Sepharose at 4°C. Proteolytic cleavage to remove the His6 tag was performed at 4°C using human thrombin (Sigma-Aldrich). Cleaved proteins were subjected to a second Ni2+-Sepharose column and further purified by size exclusion chromatography on Superdex 75 (CLIPCG1, CLIPCG2, and p150CG) or Superdex 200 columns (CLIPCG12; GE Healthcare) equilibrated in PBS (137 mM NaCl, 2.7 mM KCl, 8.3 mM Na2HPO4, and 1.47 mM KH2PO4, pH 7.4). Throughout the CG12 purification, reducing conditions (1 mM β-mercaptoethanol) were maintained. The homogeneity of the recombinant proteins was assessed by SDS-PAGE, and their identity was confirmed by mass spectral analysis.
The SLAIN2c (residues 569–581 of human SLAIN2), SLAIN2c-W576A, and SLAIN2c-ΔY581 peptides were purchased from United Peptide. The purity of the peptides was verified by reversed-phase analytical HPLC, and their identities were assessed by mass spectral analysis.
Size exclusion chromatography coupled to multiangle light scattering was performed on a three-angle detector (DAWN EOS; Wyatt) followed by a refractometer (Optilab Rex; Wyatt). Protein solutions (100 µl of 1–15 mg/ml) were injected on a size exclusion chromatography column (Superdex 200 10/300 GL) equilibrated with PBS. Molecular weights were calculated by using the ASTRA V version 5.3.4.19 software package (Wyatt).
Far-UV circular dichroism spectroscopy was performed on a spectrometer (Chirascan-plus; Applied Photophysics) equipped with a temperature-controlled quartz cell of 0.1-cm path length. A ramping rate of 1°C/min was used to record thermal unfolding profiles. Midpoints of the transitions (Tm’s) were taken as the maximum of the derivative (d): d[θ]222/dT, in which T is the temperature.
ITC
ITC experiments were performed in PBS at 25°C on a calorimeter (iTC200; MicroCal) machine. The sample cell was filled with 160–240 µM CAP-Gly solutions. The syringe was filled with 2.2–2.6 mM SLAIN2c peptide solutions. In the experiment with CLIPCG12, the buffer was supplemented with 1 mM β-mercaptoethanol. 2.6-µl SLAIN2c aliquots from the stirred syringe were injected 14–28 times into the sample cell. To determine the binding stoichiometry and the equilibrium dissociation constant (Kd) of the binding, isotherms were fitted using a nonlinear least squares minimization method provided with the ITC calorimeter. Exact concentrations of protein solutions were determined by absorbance at 280 nm in 6 M GuHCl for the CAP-Gly domains or by quantitative amino acid analysis for the SLAIN2c peptide.
Crystal structure determination
For crystallization, CLIPCG1 and SLAIN2c in PBS were mixed in a 1:1.2 ratio to reach a final complex concentration of 24 mg/ml. Crystals were obtained at 20°C by the hanging-drop vapor diffusion method from a 1:1 mixture of the complex solution and a reservoir composed of 36% PEG 6000 and 100 mM citric acid, pH 4.5.
X-ray diffraction data were collected at 100 K at beamline X06DA of the Swiss Light Source (Villigen PSI). The structure was solved by molecular replacement using the CLIP-170 CAP-Gly structure as a search model (Protein Data Bank accession no. 2E3I). Data processing and refinement statistics are summarized in Table S1. The atomic coordinates of the CLIPCG1–SLAIN2c complex have been deposited in the Protein Data Bank (accession no. 3RDV).
Mass spectrometry
GST pull-down assays followed by mass spectrometry and streptavidin bead pull-down assays from HeLa cells followed by mass spectrometry were performed as previously described by Grigoriev et al. (2007). 1D SDS-PAGE gel lanes were cut into 2-mm slices using an automatic gel slicer and subjected to an in-gel reduction with dithiothreitol, alkylation with iodoacetamide, and digestion with trypsin (sequencing grade; Promega) essentially as previously described by Wilm et al. (1996). Nanoflow liquid chromatography with tandem mass spectrometry was performed on a capillary liquid chromatography system (1100 series; Agilent Technologies) coupled to either an LTQ Orbitrap or an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific) both operating in positive mode and equipped with a nanospray source. Peptide mixtures were trapped on a reversed-phase column (ReproSil C18; Dr. Maisch GmbH; column dimensions were 1.5 cm × 100 µm, packed in house) at a flow rate of 8 µl/min. Peptide separation was performed on another ReproSil C18 reversed-phase column (column dimensions were 15 cm × 50 µm, packed in house) using a linear gradient from 0 to 80% B (A, 0.1% formic acid; B, 80% [vol/vol] acetonitrile and 0.1% formic acid) in 70 min and at a constant flow rate of 200 nl/min using a splitter. The column eluent was directly sprayed into the electrospray ionization source of the mass spectrometer. Mass spectra were acquired in continuum mode; fragmentation of the peptides was performed in the data-dependent mode. Peak lists were automatically created from raw data files using the Mascot Distiller software (version 2.1; Matrix Science). The Mascot search algorithm (version 2.2) was used for searching against the International Protein Index database (release number IPI_mouse_20100507.fasta or IPI_human_20100507.fasta). The peptide tolerance was typically set to 10 ppm for Orbitrap data and to 2 D for ion trap data. The fragment ion tolerance was set to 0.8 D. A maximum number of two missed cleavages by trypsin were allowed, and carbamidomethylated cysteine and oxidized methionine were set as fixed and variable modifications, respectively. The Mascot score cut-off value for a positive protein hit was set to 60. Individual peptide tandem mass spectrometry spectra with Mascot scores <40 were checked manually and either interpreted as valid identifications or discarded. Proteins present in the negative controls (pull-down assays with either GST or bioGFP alone) were omitted from the table.
Phosphorylated peptides were selectively enriched in an offline chromatographic manner using a TiO2 (Titansphere) packed fused silica capillary that is used as a trap, which acts as an 1D separation step in a 2D chromatography system (Pinkse et al., 2004). Phosphorylated peptides were separated from nonphosphorylated peptides by trapping them under acidic conditions on the TiO2 column and ultimately desorbed under alkaline conditions, dried, and dissolved in 0.1 M formic acid. Subsequently, nanoflow liquid chromatography with tandem mass spectrometry was performed on an capillary liquid chromatography system (1100 series) coupled to a mass spectrometer (LTQ Orbitrap) operating in positive mode and equipped with a nanospray source as described in the previous paragraph. The Mascot search algorithm (version 2.2) was used for searching against the International Protein Index database (release number IPI_human_20100507).
Antibodies and immunofluorescent cell staining
Rabbit antibodies against SLAIN1/2 were raised against a bacterially purified GST-SLAIN2 N terminus. We used rabbit polyclonal antibodies against GFP (Abcam), CLASP1 (Mimori-Kiyosue et al., 2005), CLASP2, CLIP-170 (Akhmanova et al., 2001), EB3 (Stepanova et al., 2003), phosphorylated histone H3 (Ser 10; Millipore), cyclin B1 (GNS1; Santa Cruz Biotechnology, Inc.), and ch-TOG (a gift from L. Cassimeris, Lehigh University, Bethelehem, PA; Charrasse et al., 1998); mouse monoclonal antibodies against GFP and HA tag (Roche), EB1 (BD), β-tubulin, acetylated tubulin (Sigma-Aldrich), p150Glued (BD), and actin (Millipore); and the rat monoclonal antibody against EB1/3 (clone 15H11; Absea) and HA tag (Roche). The following secondary antibodies were used: alkaline phosphatase–conjugated anti–rabbit, anti–mouse, or anti–rat antibodies (Sigma-Aldrich); goat anti–rabbit, anti–mouse, and anti–rat IgG (IRDye 800CW; LI-COR Biosciences); and Alexa Fluor 350–, Alexa Fluor 488–, and Alexa Fluor 598–conjugated goat antibodies against rabbit, rat, and mouse IgG (Invitrogen).
Cultured cells were fixed with −20°C methanol for 15 min in the case of EB1/3, ch-TOG, CLASP1/2, and p150Glued labeling. In the case of EB1, SLAIN1/2, CLIP-170, acetylated tubulin, and β-tubulin labeling, cells were fixed with −20°C methanol for 15 min and postfixed in 4% PFA in PBS for 15 min at RT. Cells were rinsed with 0.15% Triton X-100 in PBS; subsequent washing and labeling steps were performed in PBS supplemented with 1% bovine serum albumin and 0.15% Tween 20. At the end, slides were rinsed in 100% ethanol, air dried, and mounted in mounting medium (Vectashield; Vector Laboratories).
Image acquisition and processing
Images of fixed cells were collected with a microscope (DMRBE; Leica) equipped with PL Fluotar 100× 1.3 NA or 40× 1.00–0.50 NA oil objectives with a FITC/EGFP filter 41012 (Chroma Technology Corp.) and Texas red filter 41004 (Chroma Technology Corp.) and a charge-coupled device (CCD) camera (ORCA-ER-1394; Hamamatsu Photonics).
Live-cell imaging was performed on an inverted research microscope (Eclipse Ti-E; Nikon) with a Perfect Focus System (Nikon), equipped with CFI Apo total internal reflection fluorescence (TIRF) 100× 1.49 NA oil objective (Nikon) and an EMCCD camera (QuantEM 512SC; Roper Scientific) and controlled with MetaMorph 7.5 software (Molecular Devices). The 16-bit images were projected onto the CCD chip with intermediate lens 2.5× at a magnification of 0.065 µm/pixel. To keep cells at 37°C, we used a stage-top incubator (model INUG2E-ZILCS; Tokai Hit); cells were imaged in the normal culture medium. The microscope was equipped with a TIRF-E motorized TIRF illuminator modified by Roper Scientific (PICT-IbiSA; Institut Curie). For regular imaging, we used a mercury lamp (HBO-100W/2; Osram) for excitation or 491-nm 50-mW Calypso (Cobolt) and 561-nm 50-mW Jive (Cobolt) lasers. We used an ET-GFP filter set (Chroma Technology Corp.) for imaging of proteins tagged with GFP and an ET-mCherry filter set (Chroma Technology Corp.) for imaging of proteins tagged with mCherry. For simultaneous imaging of green and red fluorescence, we used an ET-mCherry/GFP filter set together with an imaging system (Dual View DV2; Roper Scientific) equipped with dichroic filter (565 DCXR; Chroma Technology Corp.) and an emission filter (HQ530/30m; Chroma Technology Corp.).
The FRAP assay was performed using a FRAP scanning system (I-Las/I-Launch [Roper Scientific]; PICT-IBiSA [Institut Curie]) installed on the same microscope and with the lasers mentioned in the previous paragraph at 100% laser power.
For imaging of mitotic cells, we used a spinning-disc microscope (CSU-X1-A1; Yokogawa) equipped with a 405/491/561 triple band mirror and GFP, mCherry, and GFP/mCherry emission filters (Chroma Technology Corp.) installed on an inverted research microscope (Eclipse Ti-E), which is almost identical to the one described previously in this section. The 16-bit images were projected onto the CCD chip with a 2.0× intermediate lens at a magnification of 0.068 µm/pixel.
Images were prepared for publication using MetaMorph and Photoshop (Adobe). All images were modified by adjustments of levels and contrast; for images of live cells, averaging of several consecutive frames was performed in some cases. In addition to adjustments of levels and contrast, Unsharp Mask and Blur filters (Photoshop) were applied to tubulin images. Maximum intensity projection, kymograph analysis, and various quantifications were performed in MetaMorph. Statistical analysis was performed using a nonparametric Mann-Whitney U test in Statistica for Windows (Microsoft) and SigmaPlot (Systat Software, Inc.).
Measurement of parameters of MT dynamics
Two HeLa cell lines stably expressing GFP– or mCherry–α-tubulin were used for the analysis at 72 h after transfection with SLAIN2 and ch-TOG siRNAs. The data were pooled because no significant differences were observed between the two lines for the measured parameters. Live-cell images were collected with 30, 10, and 2 frames per second. Initial analysis revealed no significant differences in the measured values. Therefore, all the data were averaged to obtain movies with a 0.5-s time interval for the final analysis. Parameters of MT growth were confirmed by independent measurements using HeLa cells stably expressing EB3-GFP. We applied kymograph analysis in order to distinguish very short episodes of growth and shortening, which are relevant for describing the phenotypes of SLAIN2 and ch-TOG depletion. This led to much higher values for transition frequencies than those commonly determined using MT life history plots or particle-tracking algorithms for EB-GFP videos. For comparison, a similar analysis was performed in Swiss 3T3 fibroblasts stably expressing EB3-GFP after siRNA transfection or in CHO cells after transient cotransfection of EB3-GFP and shRNA constructs. As we only focused on MT dynamics in internal cell regions, we did not analyze the frequency and duration of pausing that is mostly associated with region-specific cortical MT stabilization. For measurements of instantaneous growth and shortening rates, the velocity of MT end displacements that were longer than 0.5 μm were taken into account. Statistical analysis was performed using the Mann-Whitney U test.
Online supplemental material
Fig. S1 provides details of the mass spectrometry data on the identification of SLAIN2 and ch-TOG as EB1-binding partners and illustrates dimer formation by the SLAIN2 N terminus and the specificity of anti-SLAIN1/2 antibodies. Fig. S2 provides additional data on the SLAIN2–CLIP-170 interaction. Fig. S3 provides details of the mass spectrometry analysis of SLAIN-binding partners, RT-PCR–based data on expression of SLAIN1 and 2 and characterization of SLAIN2-depleted cells. Fig. S4 provides additional data on SLAIN2-mediated ch-TOG binding to EB1 and to MT tips and the dominant-negative properties of the ch-TOG C terminus. Fig. S5 shows MT density after depletion or disruption of the SLAIN2–ch-TOG complex and MT organization and dynamics in SLAIN2 and ch-TOG–depleted cells. Video 1 shows that GFP-SLAIN2 tracks growing MT ends in interphase HeLa cells. Video 2 shows that SxIP-like motifs of SLAIN2 contribute to its plus end–tracking behavior. Video 3 shows that GFP–ch-TOG colocalizes with mCherry-SLAIN2 at the plus ends of growing MTs. Video 4 shows MT plus end growth visualized with EB3-GFP in control, SLAIN2, and ch-TOG–depleted cells. Video 5 shows that GFP-SLAIN2 does not track MT plus ends in metaphase cells. Video 6 shows that GFP-SLAIN2 does not track MT plus ends in anaphase cells. Video 7 shows that GFP-SLAIN2 reassociates with MT plus ends in late telophase. Table S1 provides information on x-ray data collection and refinement statistics. Table S2 provides the values and statistical analysis of the MT dynamics parameters. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012179/DC1.
Acknowledgments
We are grateful to D. Meijer, E. Mientjes, R. Tsien, L. Wordeman, L. Cassimeris, and Kazusa DNA Research Institute for sharing materials and R. van der Linden and E. Dzierzak for FACS of the stable cell lines.
This study was supported by the Netherlands Organization for Scientific Research ALW (Earth and Life Sciences) open program and ALW-Vici grants to A. Akhmanova, ZonMw (Netherlands Organization for Health Research and Development)-Vidi and European Science Foundation (European Young Investigator) awards to C.C. Hoogenraad, and the Swiss National Science Foundation grants to M.O. Steinmetz.
Footnotes
Abbreviations used in this paper:
- EB
- end-binding protein
- EBH
- EB homology
- CCD
- charge-coupled device
- CLASP
- CLIP-associated protein
- CLIP
- cytoplasmic linker protein
- IP
- immunoprecipitation
- ITC
- isothermal titration calorimetry
- MCAK
- mitotic centromere-associated kinesin
- MT
- microtubule
- shRNA
- small hairpin RNA
- STLC
- S-trityl-l-cysteine
- +TIP
- plus end–tracking protein
- TIRF
- total internal reflection fluorescence
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9:309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935 10.1016/S0092-8674(01)00288-4 [DOI] [PubMed] [Google Scholar]
- Barr A.R., Gergely F. 2008. MCAK-independent functions of ch-Tog/XMAP215 in microtubule plus-end dynamics. Mol. Cell. Biol. 28:7199–7211 10.1128/MCB.01040-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils C., Bec N., Lacroix B., Harricane M.C., Larroque C. 2007. Kinetic analysis of tubulin assembly in the presence of the microtubule-associated protein TOGp. J. Biol. Chem. 282:5570–5581 10.1074/jbc.M605641200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., Hyman A.A. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590 10.1091/mbc.E03-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Becker B., Carney B. 2009. TOGp regulates microtubule assembly and density during mitosis and contributes to chromosome directional instability. Cell Motil. Cytoskeleton. 66:535–545 10.1002/cm.20359 [DOI] [PubMed] [Google Scholar]
- Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D.L., Larroque C. 1998. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 111:1371–1383 [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T.J. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- De Zeeuw C.I., Hoogenraad C.C., Goedknegt E., Hertzberg E., Neubauer A., Grosveld F., Galjart N. 1997. CLIP-115, a novel brain-specific cytoplasmic linker protein, mediates the localization of dendritic lamellar bodies. Neuron. 19:1187–1199 10.1016/S0896-6273(00)80411-0 [DOI] [PubMed] [Google Scholar]
- Driegen S., Ferreira R., van Zon A., Strouboulis J., Jaegle M., Grosveld F., Philipsen S., Meijer D. 2005. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgenic Res. 14:477–482 10.1007/s11248-005-7220-2 [DOI] [PubMed] [Google Scholar]
- Gergely F., Draviam V.M., Raff J.W. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341 10.1101/gad.245603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Splinter D., Keijzer N., Wulf P.S., Demmers J., Ohtsuka T., Modesti M., Maly I.V., Grosveld F., Hoogenraad C.C., Akhmanova A. 2007. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 13:305–314 10.1016/j.devcel.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Hirst C.E., Ng E.S., Azzola L., Voss A.K., Thomas T., Stanley E.G., Elefanty A.G. 2006. Transcriptional profiling of mouse and human ES cells identifies SLAIN1, a novel stem cell gene. Dev. Biol. 293:90–103 10.1016/j.ydbio.2006.01.023 [DOI] [PubMed] [Google Scholar]
- Holmfeldt P., Stenmark S., Gullberg M. 2004. Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J. 23:627–637 10.1038/sj.emboj.7600076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F.K., Steinmetz M.O. 2006. Key interaction modes of dynamic +TIP networks. Mol. Cell. 23:663–671 10.1016/j.molcel.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S.M., Weisbrich A., Damberger F.F., Bhavesh N.S., Jawhari H., Grigoriev I., van Rijssel F.J., Buey R.M., Lawera A., et al. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 138:366–376 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Akhmanova A., Grosveld F., De Zeeuw C.I., Galjart N. 2000. Functional analysis of CLIP-115 and its binding to microtubules. J. Cell Sci. 113:2285–2297 [DOI] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758 10.1038/nature01600 [DOI] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. 2007. Microtubule polymerases and depolymerases. Curr. Opin. Cell Biol. 19:31–35 10.1016/j.ceb.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Arnal I., Desai A., Drechsel D.N., Hyman A.A. 2001. Reconstitution of physiological microtubule dynamics using purified components. Science. 294:1340–1343 10.1126/science.1064629 [DOI] [PubMed] [Google Scholar]
- Komarova Y.A., Akhmanova A.S., Kojima S., Galjart N., Borisy G.G. 2002. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159:589–599 10.1083/jcb.200208058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G.G., Akhmanova A. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol. Biol. Cell. 16:5334–5345 10.1091/mbc.E05-07-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y., De Groot C.O., Grigoriev I., Gouveia S.M., Munteanu E.L., Schober J.M., Honnappa S., Buey R.M., Hoogenraad C.C., Dogterom M., et al. 2009. Mammalian end binding proteins control persistent microtubule growth. J. Cell Biol. 184:691–706 10.1083/jcb.200807179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I., Kruljac-Letunic A., Caudron-Herger M., Bieling P., Karsenti E. 2009. XMAP215-EB1 interaction is required for proper spindle assembly and chromosome segregation in Xenopus egg extract. Mol. Biol. Cell. 20:2684–2696 10.1091/mbc.E08-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Komarova Y., Modesti M., Wyman C., Hoogenraad C.C., Goodson H.V., Lemaitre R.P., Drechsel D.N., van Munster E., Gadella T.W.J., Jr, et al. 2004. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166:1003–1014 10.1083/jcb.200402082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168:141–153 10.1083/jcb.200405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M., Maesaki R., Kasa M., Watanabe T., Fukata M., Kaibuchi K., Hakoshima T. 2007. Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proc. Natl. Acad. Sci. USA. 104:10346–10351 10.1073/pnas.0703876104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia S., Leslie K., Kapitein L.C., Buey R.M., Grigoriev I., Wagenbach M., Smal I., Meijering E., Hoogenraad C.C., Wordeman L., et al. 2010. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr. Biol. 20:1717–1722 10.1016/j.cub.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Olieric N., Kuchen M., Wagen S., Sauter M., Crone S., Edmondson S., Frey D., Ostermeier C., Steinmetz M.O., Jaussi R. 2010. Automated seamless DNA co-transformation cloning with direct expression vectors applying positive or negative insert selection. BMC Biotechnol. 10:56 10.1186/1472-6750-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkse M.W., Uitto P.M., Hilhorst M.J., Ooms B., Heck A.J. 2004. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76:3935–3943 10.1021/ac0498617 [DOI] [PubMed] [Google Scholar]
- Rehberg M., Gräf R. 2002. Dictyostelium EB1 is a genuine centrosomal component required for proper spindle formation. Mol. Biol. Cell. 13:2301–2310 10.1091/mbc.E02-01-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C., Pellman D. 2001. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 105:421–424 10.1016/S0092-8674(01)00364-6 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Slep K.C. 2009. The role of TOG domains in microtubule plus end dynamics. Biochem. Soc. Trans. 37:1002–1006 10.1042/BST0371002 [DOI] [PubMed] [Google Scholar]
- Smith B., Treadwell J., Zhang D., Ly D., McKinnell I., Walker P.R., Sikorska M. 2010. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS ONE. 5:e11109 10.1371/journal.pone.0011109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350 10.1371/journal.pbio.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M.O., Akhmanova A. 2008. Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33:535–545 10.1016/j.tibs.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C.C., Lansbergen G., Dortland B., De Zeeuw C.I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Bierer B.E. 2000. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149:761–766 10.1083/jcb.149.4.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A.J., Rybina S., Pozniakovsky A., Mayer T.U., Walczak C.E., Karsenti E., Hyman A.A. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2:13–19 10.1038/71330 [DOI] [PubMed] [Google Scholar]
- van der Vaart B., Akhmanova A., Straube A. 2009. Regulation of microtubule dynamic instability. Biochem. Soc. Trans. 37:1007–1013 10.1042/BST0371007 [DOI] [PubMed] [Google Scholar]
- Watson P., Stephens D.J. 2006. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J. Cell Sci. 119:2758–2767 10.1242/jcs.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A., Steinmetz M.O. 2007. Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 14:959–967 10.1038/nsmb1291 [DOI] [PubMed] [Google Scholar]
- Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 379:466–469 10.1038/379466a0 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Hyman A., Desai A. 2001. The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3:E28–E34 10.1038/35050669 [DOI] [PubMed] [Google Scholar]
- Wolyniak M.J., Blake-Hodek K., Kosco K., Hwang E., You L., Huffaker T.C. 2006. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 17:2789–2798 10.1091/mbc.E05-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]