A complex specifically required for the biogenesis of the respiratory chain component cytochrome b binds to the tunnel exit of yeast mitochondrial ribosomes to coordinate protein synthesis and assembly.
Abstract
Mitochondria contain their own genetic system to express a small number of hydrophobic polypeptides, including cytochrome b, an essential subunit of the bc1 complex of the respiratory chain. In this paper, we show in yeast that Cbp3, a bc1 complex assembly factor, and Cbp6, a regulator of cytochrome b translation, form a complex that associates with the polypeptide tunnel exit of mitochondrial ribosomes and that exhibits two important functions in the biogenesis of cytochrome b. On the one hand, the interaction of Cbp3 and Cbp6 with mitochondrial ribosomes is necessary for efficient translation of cytochrome b transcript. On the other hand, the Cbp3–Cbp6 complex interacts directly with newly synthesized cytochrome b in an assembly intermediate that is not ribosome bound and that contains the assembly factor Cbp4. Our results suggest that synthesis of cytochrome b occurs preferentially on those ribosomes that have the Cbp3–Cbp6 complex bound to their tunnel exit, an arrangement that may ensure tight coordination of cytochrome b synthesis and assembly.
Introduction
The membrane-embedded complexes driving oxidative phosphorylation in mitochondria consist of subunits that are encoded in either the nuclear or the organellar DNA. Most of these proteins are nuclear encoded; they are synthesized in the cytosol and imported posttranslationally into the organelle (Neupert and Herrmann, 2007; Chacinska et al., 2009). In yeast mitochondria, the genetic system contributes seven hydrophobic translation products that represent the catalytic cores of the complexes (Borst and Grivell, 1978). Their assembly with the nuclear-encoded subunits necessitates complex biogenesis pathways. Over 30 genes have been identified that are obligatory for the formation of cytochrome c oxidase (Mick et al., 2011). These genes encode factors required for maturation and translation of COX mRNAs as well as proteins that help to insert the redox-active prosthetic groups. Similarly, numerous genes have been identified as being important for biogenesis of ATP synthase and the bc1 complex. In the case of the bc1 complex, these factors include three translational activators (Cbs1, Cbs2, and Cbp6) that are mitochondria-specific proteins required for translation of the cytochrome b mRNA (COB mRNA) and four assembly factors (Cbp3, Cbp4, Bcs1, and Bca1). However, not much is known about how they mediate the formation of a functional bc1 complex.
Protein synthesis in mitochondria is performed by organelle-specific ribosomes. These ribosomes developed from the translation system of the bacterial ancestor of mitochondria. Surprisingly little is known about how these ribosomes mediate protein synthesis and how they are organized in the context of respiratory chain assembly. Newly synthesized proteins emerge from the ribosome at the polypeptide tunnel exit, which serves as a docking site for a variety of biogenesis factors. This is well documented in bacteria (Kramer et al., 2009). The interactors of the tunnel exit of bacterial ribosomes can be classified into three different groups, namely (1) processing enzymes like peptide deformylase, (2) chaperones like trigger factor, and (3) targeting and membrane insertion components like the signal recognition particle and the SecYEG complex (Ménétret et al., 2000; Kramer et al., 2002; Gu et al., 2003; Bingel-Erlenmeyer et al., 2008). These factors interact with the rim of the polypeptide tunnel exit that is formed by RNA moieties and four conserved proteins, namely L22, L23, L24, and L29. The exact composition and structure of the mitochondrial ribosomal tunnel exit is not known. Cryo-EM reconstruction of mitochondrial ribosomes indicated that their tunnel exits differ substantially from those of their bacterial counterparts (Sharma et al., 2003, 2009). Although homologues of the bacterial ribosomal proteins found at the tunnel exit are conserved in mitochondria, this site also contains proteins found exclusively in these organelles (Gruschke et al., 2010). Hence, this important structure of the ribosome was considerably modified in the course of evolution, implying novel ways to organize translation in mitochondria.
Here, we report that Cbp3, an assembly factor of cytochrome b (Wu and Tzagoloff, 1989), can be cross-linked in isolated mitochondria to Mrpl4, a homologue of the conserved tunnel exit protein L29. This positioning of Cbp3 at the tunnel exit supports an early interaction of Cbp3 with the newly synthesized cytochrome b. Interestingly, Cbp3 forms a complex with Cbp6, a yet ill-defined translational activator of the COB mRNA (Dieckmann and Tzagoloff, 1985). By using a combination of yeast genetics and biochemistry, we reveal that the Cbp3–Cbp6 complex has two different functions for biogenesis of cytochrome b. On the one hand, the complex interacts with mitochondrial ribosomes to allow efficient translation of mRNAs containing the 5′ untranslated region (UTR) of the COB mRNA. On the other hand, the Cbp3–Cbp6 complex is part of a non–ribosome-bound assembly intermediate of the bc1 complex that contains newly synthesized cytochrome b and the assembly factor Cbp4. Our data suggest that these two functions of the Cbp3–Cbp6 complex allow coupled synthesis and assembly of cytochrome b, thereby facilitating biogenesis of this central subunit of the bc1 complex.
Results
Cbp3 binds to mitochondrial ribosomes in proximity to the polypeptide tunnel exit
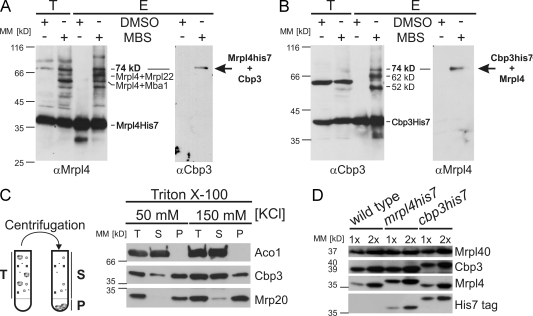
We recently analyzed the composition of the yeast mitochondrial ribosomal tunnel exit by a chemical cross-linking approach using isolated mitochondria that carried His7-tagged versions of the conserved proteins located at the tunnel exit, namely Mrpl22, Mrp20, Mrpl4, and Mrpl40 (Gruschke et al., 2010). The resulting cross-linking products of these proteins were purified by metal affinity chromatography and analyzed by mass spectrometry. Fig. 1 A shows the formation of several cross-linking products to Mrpl4His7 upon incubation with m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS). Two of these cross-linking products could be clearly identified (Mrpl4-Mba1 and Mrpl4–Mrpl22; Gruschke et al., 2010). Interestingly, in the 74-kD band, we recovered, in addition to fragments of Mrpl4, three different peptides of Cbp3, a protein described to be essential for the assembly of the bc1 complex of the respiratory chain (Wu and Tzagoloff, 1989).
Figure 1.
Cbp3 binds to mitochondrial ribosomes in proximity to the polypeptide tunnel exit. (A) Identification of Cbp3 as a cross-linking partner of Mrpl4. Mitochondria containing Mrpl4His7 were incubated with the cross-linker MBS or mock treated (DMSO). Mrpl4His7 and cross-linked proteins were purified on Ni-NTA beads and analyzed by Western blotting with antibodies against Mrpl4 (left) and Cbp3 (right). E, elution after Ni-NTA purification; T, 10% total before Ni-NTA purification. (B) Verification of the cross-link between Mrpl4 and Cbp3. The analysis depicted in A was repeated with mitochondria containing Cbp3His7. Fractions were analyzed by Western blotting using antibodies against Cbp3 (left) and Mrpl4 (right). (C) Cbp3 binds to mitochondrial ribosomes in a salt-sensitive manner. (left) Experimental setup. Ribosomes and co-migrating proteins (P, pellet) are separated from the soluble fraction (S, supernatant) by high-speed centrifugation through a sucrose cushion. (right) Triton X-100 lysates of wild-type mitochondria were fractionated in the presence of 50 or 150 mM KCl. The fractions were analyzed by Western blotting using antibodies against Aco1 (a soluble protein), Mrp20 (a ribosomal component), and Cbp3. T, 100% total before high-speed centrifugation. (D) Cbp3 and ribosomes are present in similar quantities in mitochondria. Increasing amounts of proteins of wild-type, mrpl4his7, and cbp3his7 cells were analyzed by Western blotting using the indicated antibodies. MM, molecular mass.
To confirm that Cbp3 forms a cross-link to Mrpl4, we repeated the experiment using Western blotting for analysis. While the band at 74 kD was absent from mock-treated mitochondria, it appeared after incubation with MBS and could be enriched by metal affinity chromatography from lysates of mitochondria containing Mrpl4His7 (Fig. 1 A). The same band was detected by a Cbp3-specific antibody, confirming that it represents a cross-linking product of Mrpl4 and Cbp3 (Fig. 1 A). To corroborate this result, we performed the cross-linking experiment with mitochondria containing Cbp3His7. Western blotting using antibodies against Mrpl4 proved that the 74-kD band is a cross-linking product of Mrpl4 and Cbp3 (Fig. 1 B).
We next tested whether an interaction of Cbp3 with mitochondrial ribosomes can also be observed by an independent approach. We therefore lysed mitochondria with Triton X-100 and fractionated these lysates by centrifugation through a high density sucrose cushion into supernatant and a ribosome-containing pellet (Fig. 1 C, left). Although the soluble protein aconitase remained in the supernatant fraction, Cbp3 cosedimented in a salt-sensitive fashion with ribosomes (Fig. 1 C, right). The finding that Cbp3 binds almost quantitatively to ribosomes under low salt conditions inspired us to test whether all or only a subset of ribosomes would contain Cbp3. We therefore estimated the relative abundance of Cbp3 over ribosomes by comparing the amounts of Cbp3His7 and Mrpl4His7. The His7 tag did not influence the level of either Cbp3 or Mrpl4 (Fig. 1 D). Western blotting using antibodies against the His7 tag revealed that both proteins are present in similar quantities (Fig. 1 D), suggesting that the levels of Cbp3 are high enough to allow each ribosome to bind one Cbp3. In conclusion, Cbp3 is a novel interactor of the mitochondrial ribosome that binds to the tunnel exit in proximity to Mrpl4.
Cbp3 stabilizes newly synthesized cytochrome b
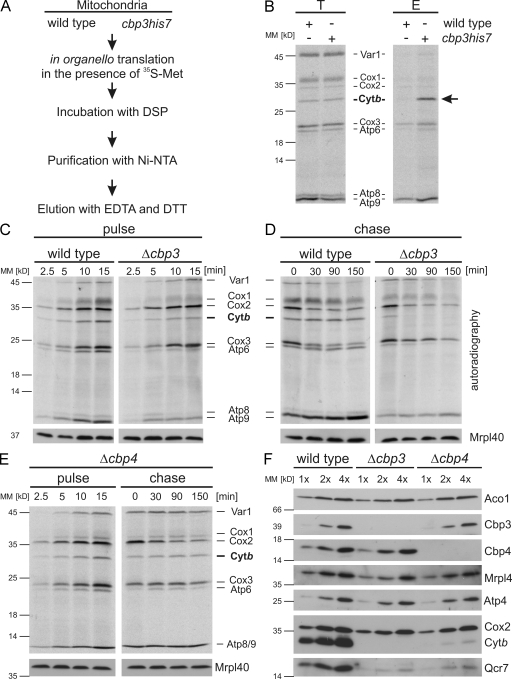
Cbp3, together with Cbp4, Bcs1, and Bca1, is one of four known assembly factors of the bc1 complex (Nobrega et al., 1992; Crivellone, 1994; Zara et al., 2009; Mathieu et al., 2011), but its molecular function is still unclear. Cbp3 is a highly conserved factor and is present in those bacteria and mitochondria that contain a bc1 complex. Because of the binding of Cbp3 in proximity to the polypeptide tunnel exit, we asked whether Cbp3 interacts with newly synthesized proteins. Translation products of mitochondria isolated from either a wild-type strain or a strain expressing Cbp3His7 were labeled with [35S]methionine in the presence of the cleavable cross-linker dithiobis(succinimidyl propionate) (DSP; Fig. 2 A). Next, Cbp3His7 was purified, and the radioactive proteins cross-linked to Cbp3His7 were released by incubation with a reducing agent. The newly synthesized cytochrome b was enriched on Cbp3His7 (Fig. 2 B, arrow), whereas only background signals were obtained when mitochondria from wild-type cells were used. This indicates that Cbp3 interacts directly with newly synthesized cytochrome b.
Figure 2.
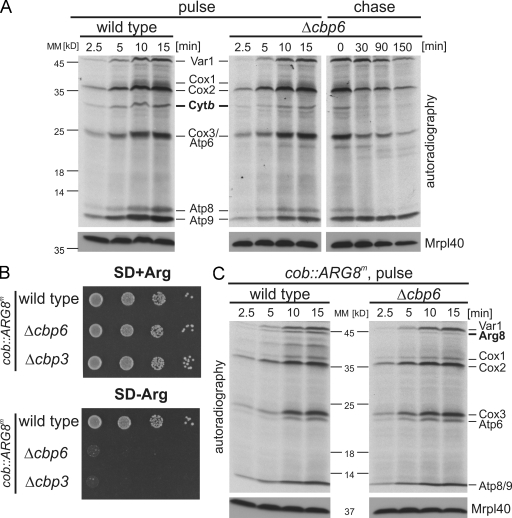
Cbp3 stabilizes newly synthesized cytochrome b. (A) An experimental procedure to analyze the contact of Cbp3 to mitochondrial translation products. (B) Cbp3 can be cross-linked to newly synthesized cytochrome b. Translation products of mitochondria isolated from wild-type or cbp3his7 cells were labeled with [35S]methionine in the presence of the cleavable cross-linking reagent DSP and purified on Ni-NTA beads. The samples were analyzed by autoradiography. The black arrow indicates the newly synthesized cytochrome b that is copurified with Cbp3His7. E, elution after Ni-NTA purification; T, 1% total before Ni-NTA purification. (C) Mitochondrial translation products of wild-type or Δcbp3 cells were labeled with [35S]methionine in the presence of cycloheximide to block cytosolic protein synthesis. Samples were taken after the indicated time points and subjected to alkaline lysis followed by SDS-PAGE and autoradiography. A Western blot using anti-Mrpl40 antibody served as a loading control. (D) Newly synthesized cytochrome b is destabilized in the absence of Cbp3. Radiolabeled translation products of the indicated strains were followed after the labeling had been stopped by addition of unlabeled methionine. (E) Cytochrome b is less efficiently labeled in Δcbp4 cells but is stable for up to 2.5 h. The experiments described in C and D were repeated with Δcbp4 cells. (F) The absence of Cbp3 and Cbp4 reduces the levels of components of the bc1 complex. Increasing amounts of proteins of wild-type, Δcbp3, and Δcbp4 cells were analyzed by Western blotting using the indicated antibodies. MM, molecular mass.
To analyze the fate of newly synthesized cytochrome b in the absence of Cbp3, we followed the synthesis and stability of mitochondrially encoded proteins in whole cells (Fig. 2 B). To avoid any interference with the complex splicing of mitochondrial mRNAs, we used strains carrying intronless mitochondrial genomes. Thus, the mRNAs are present in constant quantities, allowing us to assess their translation independent of the splicing processes. Mitochondrial translation products of wild type or cells lacking CBP3 were pulse labeled with [35S]methionine. After 15 min, labeling was stopped, and incubation was continued for up to 2.5 h. This approach revealed that both cell types can synthesize all mitochondrial translation products (Fig. 2 C), but cytochrome b was less efficiently labeled in Δcbp3 cells (Fig. 2 C, right). All newly synthesized proteins in wild-type cells were stable for at least 2.5 h (Fig. 2 D, left). In contrast, newly synthesized cytochrome b of the Δcbp3 strain was completely degraded after 30 min (Fig. 2 D, right). Atp6 and Cox1 were also affected in this strain, but this did not impair accumulation of ATPase and cytochrome c oxidase (Fig. 2 F), indicating that their biogenesis does not depend on Cbp3.
The reduced stability of newly synthesized cytochrome b could be either a result of a specific defect caused by the absence of Cbp3 or a result of a general defect in the assembly of cytochrome b. We therefore analyzed the fate of mitochondrial translation products in cells impaired in the assembly of the bc1 complex. We chose to characterize a Δcbp4 strain. Cbp4 is an assembly factor of the bc1 complex and interacts with Cbp3 (Kronekova and Rödel, 2005). In contrast to the situation in the absence of Cbp3, the newly synthesized cytochrome b is rather stable in Δcbp4 cells (Fig. 2 E, right). This indicates that defects in the assembly of the bc1 complex do not generally provoke a rapid degradation of newly synthesized cytochrome b. Therefore, it appears that Cbp3 is particularly required to protect newly synthesized cytochrome b from proteolytic degradation.
We next assessed the levels of mitochondrial proteins that accumulate in cells lacking Cbp3 or Cbp4. Subunits of cytochrome c oxidase (Cox2) and ATPase (Atp4) were not affected in both mutants (Fig. 2 F). Importantly, both mutants failed to accumulate bc1 complex subunits (Cytb and Qcr7) to wild-type levels (Fig. 2 F). This indicates a defect in the assembly of the bc1 complex that results in degradation of the nonassembled subunits. However, in light of the rapid proteolysis of cytochrome b observed only in Δcbp3 cells, it appears that Cbp3 and Cbp4 have different functions during the assembly of cytochrome b.
Cbp3, but not Cbp4, is critical for efficient translation of the cob::ARG8m mRNA
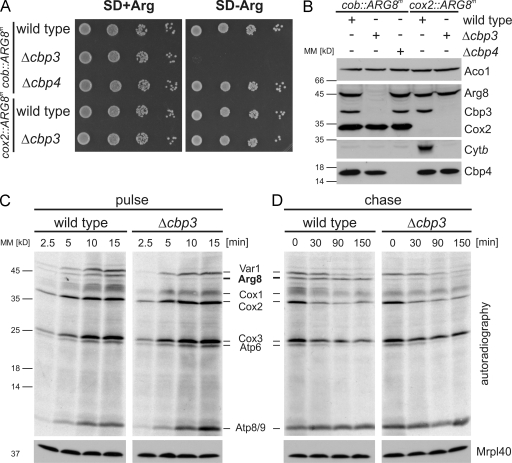
The low level of labeling of cytochrome b during the pulse kinetics in the Δcbp3 mutant (Fig. 2 C) could be explained by either a rapid turnover of the newly synthesized protein or by a direct impairment of translation of its messenger, the COB mRNA. To differentiate between both possibilities, we used a system that allows following the synthesis of a reporter protein that does not require a functional bc1 complex assembly machinery. Translational control in yeast mitochondria is mainly exerted on the 5′ UTRs of the mRNA (Costanzo and Fox, 1988). To analyze whether the absence of Cbp3 or Cbp4 has a direct influence on COB mRNA translation, we used a strain with a mitochondrial genome in which the coding sequence of cytochrome b is replaced by the coding sequence of Arg8. Hence, Arg8 is synthesized on mitochondrial ribosomes from an mRNA that contains the 5′ and 3′ UTRs of cytochrome b (COB-ARG8m-COB). Arg8 is a soluble protein of the mitochondrial matrix that is required for biosynthesis of arginine. Because the nuclear version of ARG8 is deleted in this strain, growth on media lacking arginine (SD-arginine) can be used to score for the ability to translate the COB-ARG8m-COB mRNA. Deletion of CBP4 in this background did not impair Arg8 synthesis because the cells could grow similar to the wild type in media with or without arginine (Fig. 3 A). In contrast, the absence of CBP3 impaired the growth on media lacking arginine (Fig. 3 A), indicating that Cbp3, but not Cbp4, is required for efficient synthesis and accumulation of this reporter (Fig. 3 B).
Figure 3.
Cbp3, but not Cbp4, is critical for efficient translation of the cob::ARG8m mRNA. (A) Growth test of wild-type, Δcbp3, or Δcbp4 yeast harboring mitochondrial genomes where a recoded version of Arg8 was inserted into the mitochondrial genome replacing the ORF of Cytb (cob::ARG8m) or Cox2 (cox2::ARG8m). Cells were plated in serial 10-fold dilutions onto media containing or lacking arginine and incubated for 2 d. SD, synthetic medium supplemented with glucose. (B) Western blot analysis of steady-state levels of proteins from strains described in A. (C and D) The absence of Cbp3 in cells harboring the cob::ARG8m mitochondrial genome leads to a strong decrease in the synthesis of Arg8 (C) but does not affect its stability (D). Mitochondrial translation products from cob::ARG8m wild-type or Δcbp3 cells were labeled with [35S]methionine as described in Fig. 2 C. Western blotting using anti-Mrpl40 antibody served as a loading control. MM, molecular mass.
To check whether Cbp3 is also required for the synthesis of Arg8 when translated from an mRNA containing COB-unrelated flanking regions, we deleted CBP3 in a strain in which the coding sequence of ARG8m was inserted in place of the coding sequence of COX2 (Bonnefoy and Fox, 2000). Here, deletion of CBP3 had no influence on growth requiring expression of this gene (Fig. 3 A). Consistent with the growth phenotypes, deletion of CBP3 in cells with a cob::ARG8m mitochondrial genome provoked a substantial decrease in the steady-state levels of Arg8, whereas there was no effect when cells contained the cox2::ARG8m mitochondrial genome (Fig. 3 B).
To further substantiate this finding, we analyzed mitochondrial protein synthesis in cells containing the cob::ARG8m mitochondrial genome (Fig. 3 C). In wild-type cells, Arg8 is efficiently made, whereas the absence of Cbp3 leads to a strong decrease in Arg8 production (Fig. 3 C). In contrast to newly synthesized cytochrome b that is unstable in the absence of Cbp3 (Fig. 2 D), the small quantities of Arg8 that are synthesized are stable (Fig. 3 D). Collectively, these data indicate that Cbp3 plays an unexpected role in promoting translation of mRNAs that contain the 5′ and 3′ UTRs of COB mRNA.
Cbp3 forms a complex with Cbp6
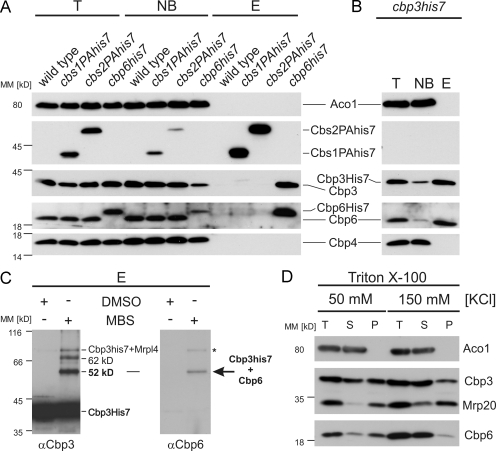
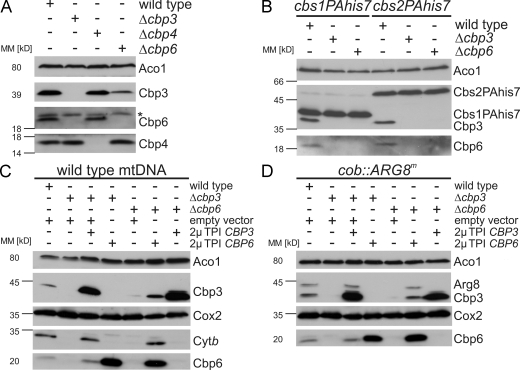
Because a role of Cbp3 in cytochrome b synthesis was not shown before, we asked whether Cbp3 might cooperate with the already identified translational activators of the COB mRNA, namely Cbs1, Cbs2, and Cbp6 (Rödel, 1997). To investigate whether one of them would interact with Cbp3, we purified Cbs1, Cbs2, and Cbp6 under native conditions using His7-tagged variants of the proteins (Fig. 4 A). While no Cbp3 was enriched on Cbs1PAHis7 or Cbs2PAHis7, it was efficiently copurified with Cbp6His7. To confirm this interaction, we repeated the experiment using mitochondria from a strain expressing Cbp3His7 and found that Cbp6 was efficiently copurified (Fig. 4 B). In contrast, Cbp4 did not copurify with the Cbp3–Cbp6 complex under these conditions (Fig. 4, A and B).
Figure 4.
Cbp3 forms a complex with Cbp6. (A) Native purification of Cbs1, Cbs2, and Cbp6 complexes. Cbs1 and Cbs2 were equipped with C-terminal ProteinAHis7 tags to allow sensitive detection as well as purification of the proteins via metal affinity chromatography, whereas Cbp6 was expressed with a C-terminal His7 tag. Triton X-100 lysates of the indicated mitochondria were subjected to Ni-NTA chromatography, and the resulting fractions were analyzed by Western blotting. E, elution after Ni-NTA purification; NB, 20% of unbound material after Ni-NTA purification; PAHis7 tag, ProteinA-heptahistidine tag; T, 20% total before Ni-NTA purification. (B) Cbp6 is efficiently purified with Cbp3His7. Native purification of the Cbp3 complex from cbp3his7 mitochondria was performed as described in A. (C) Cbp3 and Cbp6 can be cross-linked to each other. Mitochondria harboring Cbp3His7 were incubated with the cross-linker MBS or a control (DMSO). Cbp3 and cross-linked proteins were purified as described in Fig. 1 B. The elution fractions were analyzed by Western blotting using antibodies against Cbp3 (left) and Cbp6 (right). The asterisk indicates the cross-link of Mrpl4 to Cbp3His7. (D) Cbp6 co-migrates with mitochondrial ribosomes in a salt-sensitive manner. Triton X-100 lysates of wild-type mitochondria were fractionated as described in Fig. 1 C. MM, molecular mass; P, pellet; S, supernatant; T, 100% total before ribosome fractionation.
Because we could efficiently copurify both Cbp3 and Cbp6 in one complex, we asked whether it was possible to confirm this interaction with an independent approach. We therefore repeated the cross-linking analysis with Cbp3His7 (Fig. 1 B) and decorated the elution fractions with Cbp6 antibodies. This analysis revealed that the band migrating at 52 kD is a cross-linking product of Cbp6 and Cbp3His7 (Fig. 4 C), substantiating that both factors interact with each other in mitochondria. Next, we tested whether Cbp6 co-migrates with Cbp3 and mitochondrial ribosomes upon centrifugation of mitochondrial lysates through a high density sucrose cushion. Similar to Cbp3, Cbp6 cosedimented in a salt-sensitive manner with ribosomes (Fig. 4 D). We therefore concluded that Cbp3 interacts with Cbp6 in a complex that binds to mitochondrial ribosomes.
Synthesis and assembly of cytochrome b requires an intact Cbp3–Cbp6 complex
We next deleted CBP6 in the strain containing the intronless mitochondrial genome and analyzed synthesis and stability of mitochondrially encoded proteins. The absence of Cbp6 provoked defects in both the production and stability of cytochrome b (Fig. 5 A) that were indistinguishable from the defects observed in the absence of Cbp3 (Fig. 2, C and D). Next, we deleted CBP6 in the strain containing the cob::ARG8m mitochondrial genome and found that the absence of CBP6 reduces growth on media requiring translation of the COB-ARG8m-COB mRNA (Fig. 5 B). Concordantly, synthesis of Arg8 in this strain was significantly reduced (Fig. 5 C), confirming the primary hypothesis that Cbp6 is a translational activator of the COB mRNA (Dieckmann and Tzagoloff, 1985).
Figure 5.
Deletion of CBP6 provokes a phenotype similar to that of Δcbp3 cells. (A) Cytochrome b is destabilized in the absence of Cbp6. Mitochondrial translation products from wild-type or Δcbp6 cells were analyzed as described in Fig. 2 (C and D). (B) Cbp6 is critical for efficient translation of a cob::ARG8m reporter. Cells of wild type, Δcbp3, or Δcbp6 harboring the modified mitochondrial genome described in Fig. 3 A were plated in serial 10-fold dilutions onto synthetic media (SD) containing or lacking arginine and incubated for 2 d. (C) The absence of Cbp6 impairs translation of COB-ARG8m-COB mRNA. Mitochondrial translation products of wild-type and Δcbp6 cob::ARG8m cells were labeled with [35S]methionine and analyzed by autoradiography. MM, molecular mass.
Because the phenotypes of disruption of either CBP3 or CBP6 are strikingly similar, we asked whether both proteins fulfill the same functions or whether they have distinct roles in the biogenesis of cytochrome b. Because both proteins interact in one complex, we first tested whether they are required to stabilize each other. When analyzing Cbp3 and Cbp6 in strains in which one of both components was missing, we found that the absence of Cbp3 destabilizes Cbp6 and vice versa (Fig. 6 A). In contrast, both proteins remained stable in the absence of Cbp4 (Fig. 6 A). Moreover, the absence of either Cbp3 or Cbp6 had no influence on the stability of the other COB mRNA–specific translational activators Cbs1 and Cbs2 (Fig. 6 B).
Figure 6.
Cbp3 and Cbp6 act in one complex. (A) Cbp3 is destabilized when Cbp6 is absent and vice versa. Cell lysates of the indicated yeast strains were analyzed by Western blotting using the indicated antibodies. The asterisk indicates an unspecific cross-reaction of the Cbp6 antibody. (B) The levels of the other translational activators of the COB mRNA, Cbs1 and Cbs2, are not affected when CBP3 or CBP6 is deleted. Cells expressing ProteinAhis7-tagged variants of Cbs1 or Cbs2 were lysed and analyzed by Western blotting. (C and D) Overexpression of either CBP3 or CBP6 cannot complement deletion of the other. The indicated yeast strains harboring the wild-type (C) or the cob::ARG8m mitochondrial DNA (D) were transformed with 2µ plasmids encoding CBP3 or CBP6 under control of the TPI promoter or an empty vector. Cell lysates were analyzed by Western blotting using the indicated antibodies. MM, molecular mass.
To investigate whether increased amounts of Cbp3 or Cbp6 could complement the deletion of CBP6 and CBP3, respectively, we transformed plasmids allowing the overexpression of CBP3 and CBP6 into Δcbp3 and Δcbp6 cells that contain either wild-type (intronless) or cob::ARG8m mitochondrial genomes. Next, cells were scored for expression of cytochrome b or the cob::ARG8m reporter (Fig. 6, C and D). Although the plasmids encoding CBP3 or CBP6 could restore accumulation of cytochrome b and Arg8 of the respective deletion mutant, overexpression of either CBP6 in Δcbp3 cells or CBP3 in Δcbp6 had no effect on steady-state levels of cytochrome b or Arg8 (Fig. 6, C and D). These observations point to an obligatory shared function of both proteins in synthesis and assembly of cytochrome b.
Interaction of Cbp3 and Cbp6 with the ribosome requires formation of a Cbp3–Cbp6 complex
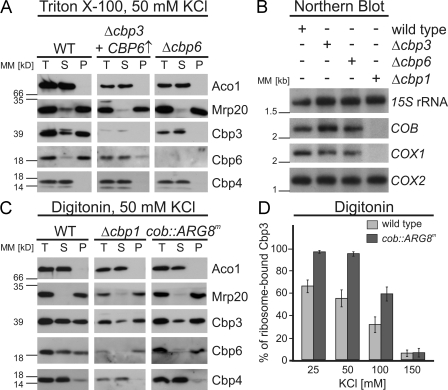
Next, we investigated whether Cbp3 or Cbp6 can interact independently of each other with mitochondrial ribosomes. We therefore lysed mitochondria of these strains with Triton X-100 and tested for co-migration of Cbp3 and Cbp6 with ribosomes. Although in wild-type mitochondria both factors interacted with ribosomes, Cbp3 and Cbp6 were unable to bind to ribosomes in Δcbp6 and Δcbp3 cells, respectively (Fig. 7 A). We therefore concluded that Cbp3 and Cbp6 can only interact with mitochondrial ribosomes when both proteins form a complex.
Figure 7.
Interaction of Cbp3 and Cbp6 with the ribosome requires formation of a Cbp3–Cbp6 complex and is influenced by cytochrome b. (A) Cbp3 and Cbp6 cannot bind to mitochondrial ribosomes in the absence of the other factor. Triton X-100 lysates of mitochondria from the indicated strains were fractionated as described in Fig. 1 C in the presence of 50 mM KCl. Fractions were analyzed by Western blotting. Because Cbp6 is not detectable in the Δcbp3 strain, mitochondria from Δcbp3 cells overexpressing CBP6 were used. (B) COB mRNA is absent in Δcbp1 but not in Δcbp3 and Δcbp6 cells. Total RNA was isolated from mitochondria of the indicated strains and analyzed by Northern blotting. 15S ribosomal RNA served as a loading control. (C) The Cbp3–Cbp6 complex can bind to mitochondrial ribosomes in the absence of cytochrome b but is mainly present in a non–ribosome-bound form in the wild type (WT). Mitochondria of the indicated strains were lysed with digitonin in the presence of 50 mM KCl and fractionated as described in Fig. 1 C. P, pellet; S, supernatant; T, 100% total before ribosome fractionation. (D) Cbp3 is more tightly bound to ribosomes of mitochondria harboring a cob::ARG8m mitochondrial genome and is released upon treatment with increasing salt concentrations. Mitochondria of wild-type and cob::ARG8m cells were lysed with digitonin and increasing KCl concentrations as indicated on the x axis and fractionated as in Fig. 1 C. The percentage of Cbp3 that was found in the pellet fraction was determined densitometrically from three independently performed experiments. The error bars represent the standard deviation of these experiments. MM, molecular mass.
We next asked whether the diminished synthesis of cytochrome b in Δcbp3 and Δcbp6 cells is caused by a destabilization of the COB mRNA. Cbp1 is a protein required for the stabilization and translation of the COB transcript (Dieckmann et al., 1984; Islas-Osuna et al., 2002) and thus for synthesis of cytochrome b. We deleted CBP1, CBP3, and CBP6 in cells with an intron-containing mitochondrial genome and analyzed the relative amounts of COB mRNA in these cells. The COB transcript failed to accumulate in the absence of Cbp1 (Fig. 7 B; Dieckmann et al., 1982), thereby also accounting for the decrease in mature COX1 mRNA that requires an intron of the COB mRNA precursor for maturation (Dhawale et al., 1981; De La Salle et al., 1982). In contrast, the COB mRNA is stable in cells lacking Cbp3 or Cbp6, excluding that the defect to synthesize cytochrome b in these strains is caused by lower amounts of the mRNA.
Interaction of the Cbp3–Cbp6 complex with the ribosome is influenced by the presence of cytochrome b
Although the Cbp3–Cbp6 complex was almost quantitatively recovered with ribosomes when mitochondria were lysed with Triton X-100 (Fig. 1 C), we observed that a substantial quantity of the complex did not cofractionate with ribosomes when mitochondria were lysed with the mild detergent digitonin (Fig. 7 C). This behavior could reflect a Cbp3–Cbp6 complex that contains additional proteins and is preserved in digitonin but disrupted by the more stringent detergent Triton X-100. To characterize this digitonin-stable complex in more detail, we asked whether the binding of Cbp3–Cbp6 to ribosomes is altered when cytochrome b cannot be produced. We therefore prepared mitochondria from strains lacking CBP1 (and as a consequence COB mRNA) and from a strain carrying the cob::ARG8m mitochondrial genome that cannot synthesize cytochrome b but contains an mRNA with the 5′ and 3′ UTRs of COB. In contrast to the wild-type situation, the Cbp3–Cbp6 complex was mainly ribosome bound in mitochondria from these two mutants (Fig. 7 C). On the contrary, Cbp4 was mainly found in the soluble fraction of mitochondria from both mutants and the wild type (Fig. 7 C). These results show that the presence of the COB mRNA or its translation product cytochrome b is not required for binding of the Cbp3–Cbp6 complex to mitochondrial ribosomes.
To further characterize the divergent binding behavior of the Cbp3–Cbp6 complex, we tested the salt sensitivity of Cbp3–Cbp6 co-migration with ribosomes of the wild type and the strain with the cob::ARG8m mitochondrial genome. Increasing the salt concentration from 25 to 150 mM KCl provoked a detachment of Cbp3 from ribosomes in the presence or absence of cytochrome b synthesis. Importantly, more Cbp3 was found at the ribosome when cytochrome b was absent. Collectively, these data show that the Cbp3–Cbp6 complex interacts with ribosomes in a cytochrome b–dependent fashion. Because Cbp3 can directly interact with cytochrome b (Fig. 2 B), it is likely that the non–ribosome-bound form represents an assembly intermediate of the bc1 complex.
The Cbp3–Cbp6 complex is released from ribosomes upon cytochrome b binding
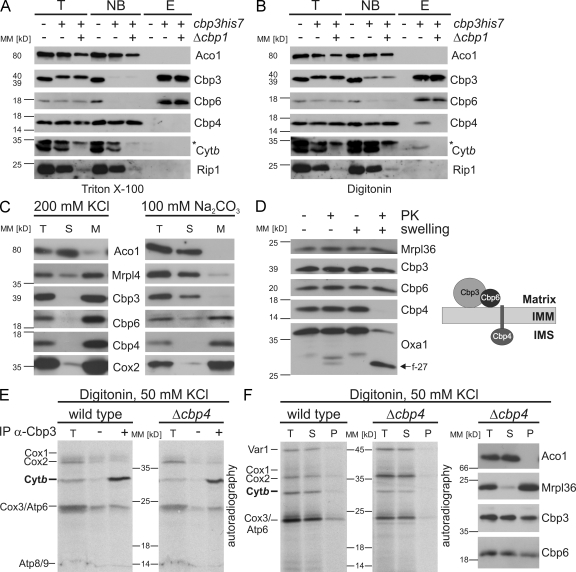
The finding that the Cbp3–Cbp6 complex also exists in a non–ribosome-bound state inspired us to characterize its composition in the presence or absence of cytochrome b. We therefore purified the complex from mitochondria of cbp3his7 strains containing or lacking CBP1. When the complex was purified from Triton X-100 lysates, Cbp6 was efficiently copurified, and the interaction was not changed by the absence of COB mRNA (Fig. 8 A). Cbp4 was again not purified with Cbp3 from Triton X-100 lysates (Figs. 4 A and 8 A). Next, we purified the complex using digitonin as detergent. Under these conditions, Cbp6, Cbp4, and cytochrome b were purified with Cbp3His7 from wild-type mitochondria (Fig. 8 B). Importantly, Rip1, a subunit of the bc1 complex, was not copurified, suggesting that the Cbp3–Cbp6–Cbp4–cytochrome b complex reflects an assembly intermediate of the bc1 complex. However, the absence of COB mRNA provoked a loss of interaction of Cbp4 with the Cbp3His7–Cbp6 complex (Fig. 8 B), indicating that Cbp4 binds to the Cbp3–Cbp6 complex only when cytochrome b can be synthesized.
Figure 8.
The Cbp3–Cbp6 complex is released from ribosomes upon cytochrome b binding. (A and B) Cbp4 interacts with the Cbp3–Cbp6 complex only in the presence of cytochrome b. Mitochondria of the indicated strains were lysed with Triton X-100 (A) or digitonin (B), and Cbp3His7 was purified under native conditions as described in Fig. 4 A. The asterisks indicate a nonspecific cross-reaction of the antibody against cytochrome b. E, elution after Ni-NTA purification; NB, 20% of unbound material after Ni-NTA purification; T, 20% total before Ni-NTA purification. (C) Cbp6 and Cbp4 are tightly associated with the inner mitochondrial membrane (IMM). Mitochondrial membranes were extracted with high salt (left) or carbonate (right) as described in the Materials and methods section. M, membrane fraction. (D) Cbp3 and Cbp6 are located in the mitochondrial matrix, whereas Cbp4 faces the intermembrane space (IMS). Wild-type mitochondria were incubated in isoosmotic or hypoosmotic buffers (swelling) and treated with proteinase K (PK) or left untreated. The arrow points to a fragment of Oxa1 (f-27) that confirms efficient swelling. (right) A model of the topology of Cbp3, Cbp6, and Cbp4. (E) The Cbp3–Cbp6 complex can bind newly synthesized cytochrome b in the absence of Cbp4. Translation products from wild-type and Δcbp4 mitochondria were labeled with [35S]methionine. The organelles were reisolated, lysed with digitonin, and subjected to coimmunoprecipitation with antibodies against Cbp3 or preimmune serum. The samples were analyzed by autoradiography. T, 5% total before immunoprecipitation (IP). (F) The Cbp3–Cbp6 complex is mainly present in a non–ribosome-bound state in the absence of Cbp4, and newly synthesized cytochrome b is liberated from ribosomes independent of Cbp4. Translation products from wild-type and Δcbp4 mitochondria were labeled with [35S]methionine. Mitochondria were reisolated, lysed with digitonin, and fractionated as described in Fig. 1 C in the presence of 50 mM KCl. Samples were analyzed by autoradiography (left) and Western blotting using the indicated antibodies (right). MM, molecular mass; P, pellet; S, supernatant.
Next, we aimed to identify the topology of the Cbp3–Cbp6 complex containing cytochrome b and Cbp4. We first extracted mitochondrial membranes with either high salt or sodium carbonate. The Cbp3–Cbp6 complex and Cbp4 could not be extracted from the membrane with high salt (Fig. 8 C, left). When membranes were extracted with alkaline solution, Cbp3 was recovered in the soluble fraction, whereas Cbp4 and Cbp6 remained in the pellet, indicating that both proteins interact more tightly with the membrane than Cbp3 (Fig. 8 C, right). Cbp6 does not contain a predictable transmembrane segment, whereas Cbp4 has such a domain close to the N terminus followed by a large hydrophilic region. We next asked how Cbp4 and Cbp6 might be oriented in the membrane. To answer this, we exposed mitochondria or mitoplasts (mitochondria whose outer membranes are ruptured by hypotonic swelling) to proteinase K treatment. Cbp3 and Cbp6 were protected from proteolytic degradation (Fig. 8 D), demonstrating that both proteins are located in the mitochondrial matrix. Cbp4 was degraded in mitoplasts, showing that large parts of the protein are present in the intermembrane space (Fig. 8 D, right).
The presence of Cbp4 in the cytochrome b–containing Cbp3–Cbp6 complex inspired us to test whether addition of Cbp4 might induce release of the Cbp3–Cbp6 complex from ribosomes. We therefore first checked whether an interaction of Cbp3 with cytochrome b occurs in the absence of Cbp4. Translation products of mitochondria from wild-type and Δcbp4 cells were labeled with [35S]methionine. Next, mitochondria were lysed with digitonin, and proteins associated with Cbp3 were precipitated with antibodies against Cbp3. This approach revealed that the Cbp3–Cbp6 complex interacts with newly synthesized cytochrome b even in the absence of Cbp4 (Fig. 8 E). This newly synthesized cytochrome b was similarly released from ribosomes in the presence or absence of Cbp4 (Fig. 8 F, left). Moreover, when we checked for ribosome co-migration of the Cbp3–Cbp6 complex in digitonin lysates, we found that a comparable fraction of the complex was present in the supernatant of mitochondria from Δcbp4 and wild-type cells (Figs. 7 C and 8 F, right). We therefore conclude that it is the binding of cytochrome b to the Cbp3–Cbp6 complex (but not the interaction of Cbp4 with the Cbp3–Cbp6–cytochrome b complex) that triggers release of the complex from the ribosome.
Discussion
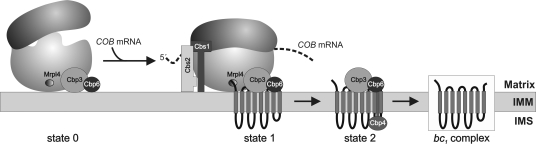
In this study, we have shown that the Cbp3–Cbp6 complex is a novel ligand of the tunnel exit of the yeast mitochondrial ribosome with a dual role in the biogenesis of cytochrome b. Our data support a working model (Fig. 9) in which dynamic interactions of this complex with the ribosome and newly synthesized cytochrome b allow a coordinated synthesis and assembly of this central component of the bc1 complex. The Cbp3–Cbp6 complex can bind to the mitochondrial ribosome in the absence of cytochrome b synthesis (Fig. 9, state 0), and ribosome binding is required for efficient translation of the COB mRNA (Fig. 9, state 1). Upon synthesis of cytochrome b, the Cbp3–Cbp6 complex interacts with the newly synthesized protein, and this provokes liberation of the complex from the ribosome (Fig. 9, state 2). By binding to the tunnel exit of mitochondrial ribosomes, the Cbp3–Cbp6 complex is well positioned to allow an efficient interaction with the newly synthesized cytochrome b. This interaction of Cbp3–Cbp6 with cytochrome b is important to protect the newly synthesized polypeptide from proteolytic degradation and to promote its assembly. Interestingly, Cbp4 is part of the Cbp3–Cbp6–cytochrome b complex. Cbp4 exposes a large soluble domain into the intermembrane space. It is therefore possible that Cbp4 is part of an assembly step that acts on cytochrome b from the intermembrane space side of the inner membrane.
Figure 9.
Hypothetical model for the role of the Cbp3–Cbp6 complex during biogenesis of cytochrome b. In the absence of an mRNA, the Cbp3–Cbp6 complex is bound to mitochondrial ribosomes (state 0). COB mRNA binds to the ribosome, and translation can only be initiated efficiently when the Cbp3–Cbp6 complex is ribosome bound (state 1). Cbp3 and Cbp6 then interact with the newly synthesized cytochrome b. Cbp4 is recruited (state 2), and this complex mediates the assembly of cytochrome b into a functional bc1 complex. IMM, inner mitochondrial membrane; IMS, intermembrane space.
In addition to an involvement in assembly of the bc1 complex, the Cbp3–Cbp6 complex plays an important role in promoting synthesis of cytochrome b. The Cbp3–Cbp6 complex shares the latter function with Cbs1 and Cbs2, two factors acting on the 5′ UTR of the COB mRNA (Rödel, 1986; Rödel and Fox, 1987). Although the binding sites of Cbs1 and Cbs2 on the ribosome (Krause-Buchholz et al., 2004, 2005) are still enigmatic, we show here that the Cbp3–Cbp6 complex binds in the vicinity of the tunnel exit. This interaction with the ribosome is important to allow efficient synthesis of cytochrome b. At present, we can only speculate about how binding of the complex to the tunnel exit influences cytochrome b synthesis. A possible scenario is that the long 5′ UTR of the COB mRNA contacts the Cbp3–Cbp6 complex. Consequently, such a contact might be necessary for efficient initiation of translation.
To date, not much is known about the molecular functions of translational activators in mitochondria. Genetic evidence suggests that at least some of them are implicated in initiation (Green-Willms et al., 1998; Nouet et al., 2007; Williams et al., 2007), in line with reported interactions of these factors with the small ribosomal subunit (McMullin et al., 1990; Haffter et al., 1991; Williams et al., 2005, 2007). In addition, recent studies showed that translational activators can regulate protein synthesis in the context of respiratory chain assembly, and feedback loops have been identified for both Cox1 (Perez-Martinez et al., 2003; Barrientos et al., 2004) and Atp6 (Rak and Tzagoloff, 2009). This is best understood for the case of Cox1 synthesis that specifically requires two translational activators, Pet309 and Mss51 (Mick et al., 2011). Apart from activating translation of the COX1 mRNA, Mss51 also mediates assembly of the newly synthesized Cox1. In case of a blocked cytochrome c oxidase assembly, Mss51 is sequestered in an assembly intermediate and is therefore not available to activate a new round of COX1 translation. By this mechanism, synthesis of Cox1 is adjusted to levels that can successfully be incorporated into the enzyme.
The Cbp3–Cbp6 complex might work in a similar way to Mss51 because it promotes both synthesis and assembly of cytochrome b. Its impact on cytochrome b synthesis is nevertheless different than the situation of COX1 translation because, here, deletion of Mss51 results in a complete block of Cox1 synthesis (Perez-Martinez et al., 2003), whereas translation of COB mRNA can still proceed in the absence of Cbp3 and Cbp6, albeit at reduced levels. Further work will reveal whether synthesis of cytochrome b is influenced by the efficiency of bc1 complex assembly.
The tunnel exit of cytoplasmic ribosomes is the site where a variety of general biogenesis factors accept the newly synthesized proteins for further maturation (Kramer et al., 2009). To do so, these factors interact with a conserved set of proteins located at the rim of the tunnel exit. Homologues of these bacterial ribosomal proteins are also found in mitochondrial ribosomes, and it was exciting to see that Mrpl4, the homologue of the bacterial L29 protein, can be cross-linked to Cbp3. In contrast to the interaction partners of the tunnel exit of cytoplasmic ribosomes, the Cbp3–Cbp6 complex is specifically required for the biogenesis of cytochrome b and therefore for only one translation product. The finding that a product-specific biogenesis factor interacts with the tunnel exit of mitochondrial ribosomes might suggest that synthesis of distinct translation products in mitochondria requires a specialized organization for each encoded protein. Such a specialization on specific translation products might be possible because of the extremely low number of different proteins that are synthesized in mitochondria. It will be exciting for future research to unravel the molecular mechanisms by which mitochondria synthesize these proteins.
Materials and methods
Yeast strains and growth media
All strains used in this study (Table S1) were isogenic to either the wild-type strain YPH499 (Sikorski and Hieter, 1989) or W303. The strains carrying the intronless or the cob::ARG8m mitochondrial genome were contributed by A. Tzagoloff (Columbia University, New York, NY), and the strain carrying the cox2::ARG8m mitochondrial genome was contributed by N. Bonnefoy (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). His7- or ProteinAHis7-tagged variants of Mrpl4, Cbp3, Cbp6, Cbs1, and Cbs2 were generated by replacing the stop codons of the endogenous ORFs by a sequence encoding a heptahistidine or ProteinA-heptahistidine tag using a HIS3 selection cassette (Lafontaine and Tollervey, 1996). CBP3, CBP6, CBP4, and CBP1 were disrupted with a Kanamycin resistance cassette. Yeast cultures were grown at 30°C in lactate medium and YP (1% yeast extract and 2% peptone) medium supplemented with 2% dextrose, 2% galactose, or 2% glycerol.
Isolation of mitochondria
Yeast cells were grown to midexponential phase (OD595nm = 1.3), harvested by centrifugation (3,000 g for 5 min), washed once with distilled water, resuspended (2 ml/g of cell wet weight) in MP1 buffer (0.1 M Tris base and 10 mM dithiothreitol), and incubated for 10 min at 30°C. Cells were then washed once with 1.2 M sorbitol, resuspendend (6.7 ml/g of cell wet weight) in MP2 buffer (20 mM KPi, pH 7.4, 0.6 M sorbitol, and 3 mg/g of cell wet weight zymolyase 20T [Seikagaku Biobusiness]), and incubated, shaking for 1 h at 30°C to digest the cell wall (spheroplastation). Spheroplasts were harvested (3,000 g for 5 min at 4°C) and resuspended (13.4 ml/g of cell wet weight) in homogenization buffer (10 mM Tris, pH 7.4, 0.6 M sorbitol, 1 mM EDTA, 1 mM PMSF, and 0.2% Albumin bovine Fraction V, fatty acid free [Serva]). All subsequent steps were performed on ice; centrifugations were performed at 4°C. The spheroplast suspension was homogenized in two portions by 10–15 strokes of a Teflon plunger in a tight-fitting homogenizer (Sartorius Stedim Biotech S.A.). The homogenate was centrifuged at 3,000 g for 5 min, and the supernatants were combined. Centrifugation was repeated until no residual cell debris was visible. Then mitochondria were harvested by centrifugation at 17,000 g for 12 min. The pellet was resuspended in isotonic buffer (0.6 M sorbitol and 20 mM Hepes, pH 7.4) to give a final concentration of 10 mg/ml.
Construction of a mitochondrial genome with the COB coding sequence replaced by ARG8m
The cob::ARG8m mitochondrial genome was constructed by A. Tzagoloff using PCR amplification of the 5′ and 3′ UTR of COB using the primer pairs 5′-GGCGAATTCGATATCATTAATATTAATATAATCGTC-3′/5′-GGCGGATCCTGATTTTCTAAATGCCATATTATTATT-3′ and 5′-GGCGGATCCCGGTAGAGTTAATAAATAATATAT-3′/5′-GGGTCTAGAGATTCTATAATAATTATGCTTTATG-3′. The two fragments were amplified from mitochondrial DNA of the respiratory-competent haploid strain MR6 (Rak et al., 2007) as the template. The PCR products were digested with EcoRI–BamHI and XbaI–BamHI, respectively, and ligated to the EcoRI and XbaI sites of pJM2 (Steele et al., 1996). The resultant plasmid was termed pCOB/ST3. ARG8m was amplified using the primer pair 5′-GGCGGATCCTTCAAAAGATATTTATCATCAAC-3′/5′-GGCGGATCCTTAAGCATATACAGC-3′ and cloned into the BamHI site of pCOB/ST3. The resultant plasmid pCOB/ST5 was introduced into the kar1-1 strain αDFS160ρ0 by biolistic transformation (Bonnefoy and Fox, 2007) with the PDS-1000/He particle delivery system (Bio-Rad Laboratories). Transformants were selected for their ability to rescue the cox2 mutation of M9-94/A3 (Tzagoloff et al., 1975). A transformant verified to have acquired the modified cob::ARG8m gene (αDFS160/COB/ST5) was crossed to the ARG8-deficient mutant MRSI0, a derivative of MR6 and SDC22 (Duvezin-Caubet et al., 2003; Rak et al., 2007) that had previously been cytoduced with an intronless mitochondrial genome from strain MCC109 (a gift from P. Perlman, University of Texas Southwestern Medical Center, Dallas, TX). Cytoductants in which the coding sequence of COB had been replaced by that of ARG8m were identified by their ability to grow on media lacking arginine and by lack of respiratory growth.
Cross-linking and denaturing purification
Mitochondria from yeast strains expressing His7-tagged proteins were incubated in isotonic buffer (0.6 M sorbitol and 20 mM Hepes, pH 7.4). The membrane-permeable, noncleavable cross-linker MBS was dissolved in DMSO and used at a final concentration of 200 µM. DMSO without cross-linker served as a vehicle control. Cross-linking was performed at 25°C for 45 min. For analysis of cross-linking to newly synthesized proteins encoded in the mitochondrial genome, in organello translation was performed at 30°C as described in the Labeling of mitochondrial translation products in organello section. The membrane-permeable, cleavable cross-linker DSP was added 10 min after addition of [35S]methionine, and labeling was allowed to go on for another 30 min. Cross-linking was stopped by addition of 100 mM Tris/HCl, pH 8.0, and 100 mM β-mercaptoethanol and incubating the samples for 10 min at 25°C. Mitochondria were then reisolated by centrifugation at 25,000 g for 10 min at 4°C and for purification of His7-tagged proteins on nickel–nitrilotriacetic acid (Ni-NTA) beads lysed in 50 µl of 1% SDS and briefly heated at 98°C. Next, the sample was adjusted to 1 ml with binding buffer (1% Triton X-100, 300 mM NaCl, 20 mM imidazole, pH 7.4, and 20 mM KPi, pH 7.4), and the His-tagged proteins and their cross-linking partners were purified on Ni-NTA beads.
Ni-NTA purification was performed by incubating the respective samples with 10 µl Ni-NTA beads for 2 h at 4°C. The beads were then washed three times with binding buffer and eluted with binding buffer containing 500 mM imidazole, pH 7.4. In the case of DSP, the cross-linker was cleaved by 100 mM DTT for 30 min at 37°C. The samples were then separated via SDS-PAGE and analyzed by Western blotting and immunodetection or autoradiography.
Native purification of Cbs1, Cbs2, Cbp6, or Cbp3
Mitochondria from the wild type or mutants expressing ProteinAHis7-tagged Cbs1 or Cbs2 or His7-tagged Cbp6 or Cbp3 (1 mg each) were lysed for 30 min in 1 ml of buffer containing 1% Triton X-100 or 1% digitonin, 150 mM KCl, 20 mM Hepes/KOH, pH 7.4, 1 mM PMSF, 1× Complete Protease Inhibitor mix, 20 mM imidazole, pH 7.4, and 20% glycerol. Tagged Cbs1, Cbs2, Cbp6, or Cbp3 and complexed proteins were purified by incubating the respective samples with 10 µl Ni-NTA beads tumbling for 3 h at 4°C. The beads were then washed three times with lysis buffer containing 0.1% detergent and were eluted with lysis buffer containing 0.1% detergent and 500 mM imidazole, pH 7.4. Alternatively, Cbp3 was purified from mitochondrial digitonin lysates by immunoprecipitation using serum against Cbp3 or preimmune serum (as a negative control) and Protein A–Sepharose beads. When interactions with newly synthesized mitochondrially encoded proteins were analyzed, in organello translation was performed at 30°C as described in the Labeling of mitochondrial translation products in organello section, radioactive labeling was stopped by adding 8 mM of unlabeled methionine, mitochondria were reisolated, and immunoprecipitation of Cbp3 was performed. Beads were washed as previously described and an additional time with 20 mM Hepes/KOH, pH 7.4. Bound proteins were eluted with sample buffer. Samples were separated via SDS-PAGE and analyzed by Western blotting and/or autoradiography.
Labeling of mitochondrial translation products in vivo
Cells were grown on minimal medium containing 2% galactose, 20 µg/ml arginine, methionine, threonine, and tyrosine, 30 µg/ml isoleucine, 50 µg/ml phenylalanine, 100 µg/ml valine, and 0.1% glucose (Prestele et al., 2009). A cell amount corresponding to an optical density (OD595nm) of 0.5 was collected, washed twice with minimal medium containing 2% galactose, and incubated at 30°C for 10 min in the same buffer supplemented with 0.12 mg/ml of all amino acids except methionine. After inhibiting cytosolic protein synthesis with 0.15 mg/ml cycloheximide, labeling of mitochondrial proteins was started by addition of 6 µCi [35S]methionine. Pulse labeling was stopped after the indicated time points by addition of lysis buffer (1.85 M NaOH and 1.1 M β-mercaptoethanol) containing 8 mM of unlabeled methionine. After incubation for 10 min on ice, proteins were precipitated with 14% TCA. The stability of mitochondrial translation products was chased by stopping a 15-min labeling reaction by the addition of 8 mM of unlabeled methionine and by taking samples after the indicated time points, which were lysed and TCA precipitated as previously described. Pellets were resolved in sample buffer, subjected to SDS-PAGE on 16/0.2% acrylamide/bisacrylamide gels, and analyzed by autoradiography and Western blotting.
Labeling of mitochondrial translation products in organello
Mitochondria were resuspended at a final concentration of 1 mg/ml in translation buffer (0.6 M sorbitol, 150 mM KCl, 15 mM KPi, pH 7.4, 20 mM Hepes, pH 7.4, 12.67 mM MgCl2, 4 mM ATP, 0.5 mM GTP, 5 mM phosphoenolpyruvate, 5 mM α-ketoglutarate, 12.13 µg/ml alanine, arginine, aspartic acid, asparagine, glutamic acid, glutamine, glycine, histidine, isoleucine, leucine, lysine, phenylalanine, proline, serine, threonine, tryptophane, tyrosine, and valine, 66.67 µM cysteine, and 10 µg/ml pyruvate kinase). After 5 min of incubation at 30°C, 4 µCi [35S]methionine/100 µg mitochondria was added, and incubation was pursued for 20 min. Labeling of mitochondrial translation products was stopped by the addition of unlabeled methionine to a final concentration of 8 mM, and mitochondria were reisolated by centrifugation (10,000 g at 4°C for 10 min).
Fractionation of mitochondrial lysates
300 µg of isolated mitochondria was lysed for 30 min on ice in lysis buffer containing 1% Triton X-100 or 1% digitonin, 50 or 150 mM KCl, 1× Complete Protease Inhibitor mix, 0.5 mM MgCl2, and 20 mM Hepes/KOH, pH 7.4. After a clarifying spin for 10 min at 25,000 g at 4°C, one half of the extract was precipitated with 12% TCA (100% total), and the rest was underlayered with 50 µl sucrose solution (1.2 M sucrose and 20 mM Hepes/KOH, pH 7.4) and centrifuged for 105 min at 200,000 g in a TLA100 rotor (Beckman Coulter) at 4°C. The supernatant was then collected, and the ribosome-containing pellet was resuspended in lysis buffer. Proteins of both fractions were precipitated with 12% TCA. The resulting pellets were dissolved in sample buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Analysis of mitochondrial transcripts by Northern blotting
For RNA isolation, mitochondria were resuspended in AE buffer (50 mM sodium acetate, pH 5.3, and 10 mM EDTA, pH 8) and lysed by addition of 1% SDS. After addition of an equal volume of phenol, pH 4.5–5, the mixture was briefly heated (65°C), rapidly chilled on ice, and stored at −80°C until the phenol was frozen. Next, the aqueous and phenolic phase was separated by centrifugation (13,000 g for 5 min), and the aqueous phase was collected, mixed with an equal amount of phenol/chloroform/isoamylalcohol solution (25:24:1), and centrifuged again. The RNA was precipitated from the aqueous phase in the presence of 0.3 M sodium acetate, pH 5.3, and isopropanol (2 vol) by incubation at −20°C for at least 20 min and centrifugation at 16,100 g for 5 min. The RNA pellet was washed once with 80% ethanol, dried, and resuspended in diethylpyrocarbonate-H2O. For Northern blotting analysis, 2 µg mitochondrial RNA of each strain was separated on a denaturing 1.2% formaldehyde-agarose gel, washed with diethylpyrocarbonate-H2O and TBE (45 mM Tris, 45 mM borate, and 1 mM EDTA), and subsequently transferred to a nylon membrane. After blotting, the membrane was briefly washed in 2× SSC (0.3 M NaCl and 30 mM Na3-citrate, pH 7.0) and dried overnight. Before hybridization, RNA was UV cross-linked and prehybridized with Church buffer (1 mM EDTA, 7% SDS, 1% BSA, and 0.25 M NaPi, pH 7.2). RNA was hybridized with gene-specific 32P-labeled probes overnight and analyzed by autoradiography.
Analysis of the topology of Cbp3, Cbp6, and Cbp4
Alkaline extraction of proteins was performed from mitochondria resuspended in 0.1 M Na2CO3 and incubated for 30 min, shaking at 4°C. Membranes and associated proteins were separated by centrifugation at 30,000 g for 45 min. Proteins from the resulting supernatant were precipitated with 12% TCA. Membrane pellets were directly resuspended in sample buffer. Fractions were analyzed by SDS-PAGE followed by Western blotting.
Alternatively, membrane-spanning and -associated proteins were fractionated from soluble proteins by salt extraction. Mitochondria were resuspended in buffer containing 200 mM KCl and 20 mM Hepes/KOH, pH 7.4, and subjected to three cycles of freeze thawing in liquid nitrogen and 37°C. Membranes were collected by centrifugation, and the fractions were further processed as described for the alkaline extraction procedure.
To analyze the submitochondrial localization of proteins, mitochondria were either resuspended in isoosmotic (600 mM sorbitol and 20 mM Hepes/KOH, pH 7.4) or hypoosmotic (20 mM Hepes/KOH, pH 7.4) buffer to generate mitoplasts. Mitochondria or mitoplasts were then incubated with 100 µg/ml proteinase K for 30 min on ice or left untreated. The protease was inhibited by the addition of 1 mM PMSF, and mitochondria (or mitoplasts) were reisolated, washed three times with a buffer containing 600 mM sorbitol, 80 mM KCl, 20 mM Hepes/KOH, pH 7.4, and 1 mM PMSF, resuspended in sample buffer, and subjected to SDS-PAGE and Western blotting.
Miscellaneous
The antibody against the His7 tag was purchased from QIAGEN, and the antibody against the ProteinA tag was purchased from Sigma-Aldrich. Antibodies against Cbp3, Cbp4, and Cbp6 were obtained by immunizing rabbits with purified mature Cbp3His6, Cbp4(61–171), and MBP-Cbp6, respectively. The Arg8 antibody was a gift from T. Fox (Cornell University, Ithaca, NY).
Online supplemental material
Table S1 contains a summary of the yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201103132/DC1.
Acknowledgments
We especially thank Alex Tzagoloff for sharing reagents, strains (W303 containing either an intronless [MRSI0] or the cob::ARG8m [MRSI0ΔCOB] mitochondrial genome), unpublished results, and much helpful advice. We thank Inge Kühl (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France) and Nathalie Bonnefoy for the cox2::ARG8m strain, their help with the construction of the cox2::ARG8m Δcbp3 strain, and comments on the manuscript. We thank Tom Fox for contributing the Arg8 antibody and Martin Jung for help with antibody production. We are grateful for Andrea Trinkaus and Sabine Knaus for expert technical assistance, the members of our group for stimulating discussions, and Markus Hildenbeutel for critically reading the manuscript.
This research was supported by the Stiftung Rheinland-Pfalz für Innovation. Moreover, work in the authors’ laboratory was supported by grants from the Deutscher Akademischer Austausch Dienst, Deutsche Forschungsgemeinschaft, Swedish Research Council, Center for Biomembrane Research at Stockholm University, and Rheinland-Pfalz Initiative für Membrantransport. K. Kehrein is a recipient of a predoctoral stipend from the Carl Zeiss Foundation.
Footnotes
Abbreviations used in this paper:
- DSP
- dithiobis(succinimidyl propionate)
- MBS
- m-maleimidobenzoyl-N-hydroxysuccinimide ester
- Ni-NTA
- nickel–nitrilotriacetic acid
- UTR
- untranslated region
References
- Barrientos A., Zambrano A., Tzagoloff A. 2004. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23:3472–3482 10.1038/sj.emboj.7600358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingel-Erlenmeyer R., Kohler R., Kramer G., Sandikci A., Antolic´ S., Maier T., Schaffitzel C., Wiedmann B., Bukau B., Ban N. 2008. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature. 452:108–111 10.1038/nature06683 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Fox T.D. 2000. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet. 262:1036–1046 10.1007/PL00008646 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Fox T.D. 2007. Directed alteration of Saccharomyces cerevisiae mitochondrial DNA by biolistic transformation and homologous recombination. Methods Mol. Biol. 372:153–166 10.1007/978-1-59745-365-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Grivell L.A. 1978. The mitochondrial genome of yeast. Cell. 15:705–723 10.1016/0092-8674(78)90257-X [DOI] [PubMed] [Google Scholar]
- Chacinska A., Koehler C.M., Milenkovic D., Lithgow T., Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell. 138:628–644 10.1016/j.cell.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M.C., Fox T.D. 1988. Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc. Natl. Acad. Sci. USA. 85:2677–2681 10.1073/pnas.85.8.2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivellone M.D. 1994. Characterization of CBP4, a new gene essential for the expression of ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J. Biol. Chem. 269:21284–21292 [PubMed] [Google Scholar]
- De La Salle H., Jacq C., Slonimski P.P. 1982. Critical sequences within mitochondrial introns: pleiotropic mRNA maturase and cis-dominant signals of the box intron controlling reductase and oxidase. Cell. 28:721–732 10.1016/0092-8674(82)90051-4 [DOI] [PubMed] [Google Scholar]
- Dhawale S., Hanson D.K., Alexander N.J., Perlman P.S., Mahler H.R. 1981. Regulatory interactions between mitochondrial genes: interactions between two mosaic genes. Proc. Natl. Acad. Sci. USA. 78:1778–1782 10.1073/pnas.78.3.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C.L., Tzagoloff A. 1985. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J. Biol. Chem. 260:1513–1520 [PubMed] [Google Scholar]
- Dieckmann C.L., Pape L.K., Tzagoloff A. 1982. Identification and cloning of a yeast nuclear gene (CBP1) involved in expression of mitochondrial cytochrome b. Proc. Natl. Acad. Sci. USA. 79:1805–1809 10.1073/pnas.79.6.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C.L., Koerner T.J., Tzagoloff A. 1984. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5′ end processing of cytochrome b pre-mRNA. J. Biol. Chem. 259:4722–4731 [PubMed] [Google Scholar]
- Duvezin-Caubet S., Caron M., Giraud M.F., Velours J., di Rago J.P. 2003. The two rotor components of yeast mitochondrial ATP synthase are mechanically coupled by subunit delta. Proc. Natl. Acad. Sci. USA. 100:13235–13240 10.1073/pnas.2135169100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green-Willms N.S., Fox T.D., Costanzo M.C. 1998. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol. Cell. Biol. 18:1826–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschke S., Gröne K., Heublein M., Hölz S., Israel L., Imhof A., Herrmann J.M., Ott M. 2010. Proteins at the polypeptide tunnel exit of the yeast mitochondrial ribosome. J. Biol. Chem. 285:19022–19028 10.1074/jbc.M110.113837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S.Q., Peske F., Wieden H.J., Rodnina M.V., Wintermeyer W. 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 9:566–573 10.1261/rna.2196403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., McMullin T.W., Fox T.D. 1991. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 127:319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas-Osuna M.A., Ellis T.P., Marnell L.L., Mittelmeier T.M., Dieckmann C.L. 2002. Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 277:37987–37990 10.1074/jbc.M206132200 [DOI] [PubMed] [Google Scholar]
- Kramer G., Rauch T., Rist W., Vorderwülbecke S., Patzelt H., Schulze-Specking A., Ban N., Deuerling E., Bukau B. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature. 419:171–174 10.1038/nature01047 [DOI] [PubMed] [Google Scholar]
- Kramer G., Boehringer D., Ban N., Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16:589–597 10.1038/nsmb.1614 [DOI] [PubMed] [Google Scholar]
- Krause-Buchholz U., Barth K., Dombrowski C., Rödel G. 2004. Saccharomyces cerevisiae translational activator Cbs2p is associated with mitochondrial ribosomes. Curr. Genet. 46:20–28 10.1007/s00294-004-0503-y [DOI] [PubMed] [Google Scholar]
- Krause-Buchholz U., Schöbel K., Lauffer S., Rödel G. 2005. Saccharomyces cerevisiae translational activator Cbs1p is associated with translationally active mitochondrial ribosomes. Biol. Chem. 386:407–415 10.1515/BC.2005.049 [DOI] [PubMed] [Google Scholar]
- Kronekova Z., Rödel G. 2005. Organization of assembly factors Cbp3p and Cbp4p and their effect on bc(1) complex assembly in Saccharomyces cerevisiae. Curr. Genet. 47:203–212 10.1007/s00294-005-0561-9 [DOI] [PubMed] [Google Scholar]
- Lafontaine D., Tollervey D. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469–3471 10.1093/nar/24.17.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu L., Marsy S., Saint-Georges Y., Jacq C., Dujardin G. 2011. A transcriptome screen in yeast identifies a novel assembly factor for the mitochondrial complex III. Mitochondrion. 11:391–396 10.1016/j.mito.2010.12.002 [DOI] [PubMed] [Google Scholar]
- McMullin T.W., Haffter P., Fox T.D. 1990. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol. Cell. Biol. 10:4590–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret J.F., Neuhof A., Morgan D.G., Plath K., Radermacher M., Rapoport T.A., Akey C.W. 2000. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 6:1219–1232 10.1016/S1097-2765(00)00118-0 [DOI] [PubMed] [Google Scholar]
- Mick D.U., Fox T.D., Rehling P. 2011. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12:14–20 10.1038/nrm3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 10.1146/annurev.biochem.76.052705.163409 [DOI] [PubMed] [Google Scholar]
- Nobrega F.G., Nobrega M.P., Tzagoloff A. 1992. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 11:3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouet C., Bourens M., Hlavacek O., Marsy S., Lemaire C., Dujardin G. 2007. Rmd9p controls the processing/stability of mitochondrial mRNAs and its overexpression compensates for a partial deficiency of oxa1p in Saccharomyces cerevisiae. Genetics. 175:1105–1115 10.1534/genetics.106.063883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X., Broadley S.A., Fox T.D. 2003. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22:5951–5961 10.1093/emboj/cdg566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestele M., Vogel F., Reichert A.S., Herrmann J.M., Ott M. 2009. Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol. Biol. Cell. 20:2615–2625 10.1091/mbc.E08-12-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., Tzagoloff A. 2009. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl. Acad. Sci. USA. 106:18509–18514 10.1073/pnas.0910351106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M., Tetaud E., Godard F., Sagot I., Salin B., Duvezin-Caubet S., Slonimski P.P., Rytka J., di Rago J.P. 2007. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 282:10853–10864 10.1074/jbc.M608692200 [DOI] [PubMed] [Google Scholar]
- Rödel G. 1986. Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5′-untranslated COB leader. Curr. Genet. 11:41–45 10.1007/BF00389424 [DOI] [PubMed] [Google Scholar]
- Rödel G. 1997. Translational activator proteins required for cytochrome b synthesis in Saccharomyces cerevisiae. Curr. Genet. 31:375–379 10.1007/s002940050219 [DOI] [PubMed] [Google Scholar]
- Rödel G., Fox T.D. 1987. The yeast nuclear gene CBS1 is required for translation of mitochondrial mRNAs bearing the cob 5′ untranslated leader. Mol. Gen. Genet. 206:45–50 10.1007/BF00326534 [DOI] [PubMed] [Google Scholar]
- Sharma M.R., Koc E.C., Datta P.P., Booth T.M., Spremulli L.L., Agrawal R.K. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 115:97–108 10.1016/S0092-8674(03)00762-1 [DOI] [PubMed] [Google Scholar]
- Sharma M.R., Booth T.M., Simpson L., Maslov D.A., Agrawal R.K. 2009. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl. Acad. Sci. USA. 106:9637–9642 10.1073/pnas.0901631106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D.F., Butler C.A., Fox T.D. 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA. 93:5253–5257 10.1073/pnas.93.11.5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R.B. 1975. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 250:8228–8235 [PubMed] [Google Scholar]
- Williams E.H., Bsat N., Bonnefoy N., Butler C.A., Fox T.D. 2005. Alteration of a novel dispensable mitochondrial ribosomal small-subunit protein, Rsm28p, allows translation of defective COX2 mRNAs. Eukaryot. Cell. 4:337–345 10.1128/EC.4.2.337-345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.H., Butler C.A., Bonnefoy N., Fox T.D. 2007. Translation initiation in Saccharomyces cerevisiae mitochondria: functional interactions among mitochondrial ribosomal protein Rsm28p, initiation factor 2, methionyl-tRNA-formyltransferase and novel protein Rmd9p. Genetics. 175:1117–1126 10.1534/genetics.106.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. 1989. Identification and characterization of a new gene (CBP3) required for the expression of yeast coenzyme QH2-cytochrome c reductase. J. Biol. Chem. 264:11122–11130 [PubMed] [Google Scholar]
- Zara V., Conte L., Trumpower B.L. 2009. Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta. 1793:89–96 10.1016/j.bbamcr.2008.04.011 [DOI] [PubMed] [Google Scholar]