Direct interaction between the catalytic domain of Epac1 and the nuclear pore component RanBP2 blocks Epac1 catalytic activity and downstream cAMP signaling.
Abstract
Cyclic adenosine monophosphate (cAMP) is a second messenger that relays a wide range of hormone responses. In this paper, we demonstrate that the nuclear pore component RanBP2 acts as a negative regulator of cAMP signaling through Epac1, a cAMP-regulated guanine nucleotide exchange factor for Rap. We show that Epac1 directly interacts with the zinc fingers (ZNFs) of RanBP2, tethering Epac1 to the nuclear pore complex (NPC). RanBP2 inhibits the catalytic activity of Epac1 in vitro by binding to its catalytic CDC25 homology domain. Accordingly, cellular depletion of RanBP2 releases Epac1 from the NPC and enhances cAMP-induced Rap activation and cell adhesion. Epac1 also is released upon phosphorylation of the ZNFs of RanBP2, demonstrating that the interaction can be regulated by posttranslational modification. These results reveal a novel mechanism of Epac1 regulation and elucidate an unexpected link between the NPC and cAMP signaling.
Introduction
Small G proteins act as molecular switches that couple extracellular signals to diverse cellular responses by cycling between an inactive GDP-bound and active GTP-bound conformation. This activation cycle is regulated by guanine nucleotide exchange factors (GEFs), which induce dissociation of the bound nucleotide and its replacement by the more abundant GTP. The resulting conformational change allows the binding of effector proteins and induction of downstream signaling. Conversely, GTPase-activating proteins (GAPs) stimulate GTP hydrolysis and, thereby, inactivation of the G protein. Together, GEFs and GAPs establish both temporal and spatial control of G protein signaling. They are typically multidomain proteins, and signaling pathways that impinge on G proteins do so mainly by regulating the activity and localization of the associated GEFs and GAPs. This regulation includes the binding of second messengers, posttranslational modifications, and their interaction with proteins and lipids (Bos et al., 2007).
Exchange protein directly activated by cAMP1 (Epac1) and Epac2 are GEFs for small G proteins of the Rap family (de Rooij et al., 1998; Kawasaki et al., 1998) and, thereby, function in cellular processes ranging from exocytosis to cell–cell junction formation and cell–extracellular matrix adhesion (Gloerich and Bos, 2010). The activity of Epac is directly regulated by the second messenger cAMP. cAMP is produced by hormone receptor–activated adenylate cyclases and becomes compartmentalized because of its local degradation by phosphodiesterases (Baillie, 2009). Similar to other GEFs for Ras-like small G proteins, activity of Epac is mediated by a CDC25 homology domain (CDC25-HD), which is stabilized by a Ras exchange motif (REM) domain. In the autoinhibited state, the catalytic site within the CDC25-HD is sterically covered by the N-terminal regulatory region, which harbors a DEP (Disheveled, Egl-10, and Pleckstrin) domain and one or two cyclic nucleotide-binding domains in Epac1 and Epac2, respectively. As demonstrated by the crystal structures of both active and inactive Epac2, autoinhibition is released by a conformational change induced by binding of cAMP (Rehmann et al., 2006, 2008). In addition, cAMP establishes spatial control of Epac1, as the cAMP-induced conformational change induces the translocation of Epac1 to the plasma membrane (Ponsioen et al., 2009). This membrane recruitment is mediated by the DEP domain and is required for efficient Rap activation at the plasma membrane and, consequently, Rap-mediated cell adhesion (Ponsioen et al., 2009). Alternative anchoring mechanisms further control the cellular distribution of Epac1, which may reflect the diverse functions assigned to this GEF. For instance, plasma membrane recruitment by activated ERM (ezrin, radixin, and moesin) proteins also couples Epac1 activity to cell adhesion (Gloerich et al., 2010), whereas nuclear Epac1 regulates the DNA damage-responsive protein kinase (DNA-PK; Huston et al., 2008).
RanBP2 (Nup358) is a cytosolic component of the nuclear pore complex (NPC; Wu et al., 1995; Yokoyama et al., 1995). RanBP2 was originally described as a regulator of nucleocytoplasmic transport based on its link with the small G protein Ran and the presence of docking motifs for nuclear transport receptors (Melchior et al., 1995; Singh et al., 1999; Yaseen and Blobel, 1999; Bernad et al., 2004; Forler et al., 2004; Hutten et al., 2008, 2009). The multidomain structure of RanBP2 (Fig. 1 A) suggested a more pleiotropic function for this nucleoporin, and RanBP2 is now recognized as a regulator of various proteins that each associate with a selective domain of RanBP2. For instance, the cyclophilin homology domain is implicated in the interconversion of retinal opsins (Ferreira et al., 1996, 1997), the leucine-rich domain suppresses activity of mitochondrial Cox11 (Aslanukov et al., 2006), and the internal repeat domain binds Ubc9 and functions as an E3 SUMO ligase (Pichler et al., 2002). By regulating the localization and function of these proteins, RanBP2 serves in a multitude of cellular processes that range from nuclear envelope assembly (Salina et al., 2003; Prunuske et al., 2006) to mitosis (Salina et al., 2003; Dawlaty et al., 2008; Klein et al., 2009; Splinter et al., 2010) and glucose metabolism (Aslanukov et al., 2006). Here, we demonstrate a novel function for RanBP2 as a negative regulator of cAMP signaling through the tethering of Epac1 to the NPC and direct inhibition of its catalytic activity.
Figure 1.
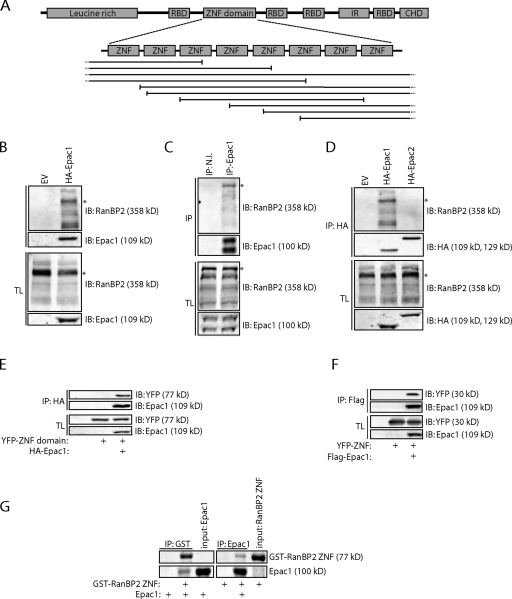
Epac1 directly interacts with the ZNF domain of RanBP2. (A) Domain architecture of RanBP2 with the fragments of RanBP2 isolated in a yeast two-hybrid screen using full-length Epac1 as bait shown below. RBD, Ran-binding domain; ZNF, zinc finger; IR, internal repeat domain; CHD, cyclophilin homology domain. (B) Coimmunoprecipitation of endogenous RanBP2 with HA-tagged Epac1 in HEK293T cells. Note that besides a major band of 358 kD (asterisks in B–D), multiple additional bands of RanBP2 are present on a 6% SDS-PAGE gel, which all disappear upon siRNA-mediated depletion of RanBP2 (Fig. S1 B). (C) Coimmunoprecipitation of endogenous RanBP2 with Epac1, which was immunoprecipitated with a rabbit polyclonal Epac1 antibody but not with preimmune serum (N.I., nonimmune) in Ovcar3 cells. (D) Coimmunoprecipitation of endogenous RanBP2 with HA-tagged Epac1 but not with HA-tagged Epac2 in HEK293T cells. (E) Coimmunoprecipitation of the YFP-tagged ZNF domain of RanBP2 with Flag-tagged Epac1 in HEK293T cells. (F) Coimmunoprecipitation of a YFP-tagged version of one on the individual ZNFs (ZNF #2) of RanBP2 with HA-tagged Epac1 in HEK293T cells. (G) Pull-down of bacterially purified Epac1 with the GST-tagged bacterially purified ZNF domain of RanBP2 and, conversely, the ZNF domain with Epac1. Proteins were visualized by simply blue staining. EV, empty vector; IB, immunoblot; IP, immunoprecipitation; TL, total lysate.
Results
Epac1 directly interacts with the zinc finger (ZNF) domain of RanBP2
To identify novel Epac1-interacting proteins, a yeast two-hybrid screen was performed using a human placenta cDNA library and full-length Epac1 as bait. Multiple positive clones were isolated containing partial cDNAs that encode for the nuclear pore component RanBP2. All isolated cDNAs of RanBP2 contained a fragment of its ZNF domain that consists of eight individual ZNFs, implying that binding to Epac1 is mediated by the ZNFs of RanBP2 (Fig. 1 A). To confirm the interaction of Epac1 with RanBP2 in mammalian cells, HA-tagged Epac1 was immunoprecipitated from HEK293T cells, which showed the coimmunoprecipitation of endogenous RanBP2 (Fig. 1 B and Fig. S1, A and B). Also, in other mammalian cell lines, including U2OS cells, we could confirm the interaction between exogenous Epac1 and RanBP2 (Fig. S1 C). Similarly, immunoprecipitation of endogenous Epac1 from Ovcar3 cells revealed the binding of RanBP2, demonstrating that RanBP2 and Epac1 interact in cells endogenously (Fig. 1 C). In contrast, HA-tagged Epac2 was unable to coimmunoprecipitate RanBP2 from HEK293T cells, indicating that the binding of RanBP2 is selective for Epac1 (Fig. 1 D). As the yeast two-hybrid results implied the binding of Epac1 to the ZNFs of RanBP2, HEK293T cells were transfected with Flag-tagged Epac1 together with the YFP-tagged ZNF domain or with an individual ZNF of RanBP2 (YFP-ZNF). Indeed, immunoprecipitation of Flag-Epac1 demonstrated the binding of both of these fragments (Fig. 1, E and F). To exclude that the interaction requires additional proteins that are conserved in Saccharomyces cerevisiae, we tested the binding between both proteins in vitro using bacterially purified Epac1 and the ZNF domain of RanBP2. This demonstrated the ability of both purified proteins to interact (Fig. 1 G), confirming the direct interaction between the ZNFs of RanBP2 and Epac1.
RanBP2 recruits Epac1 to the NPC
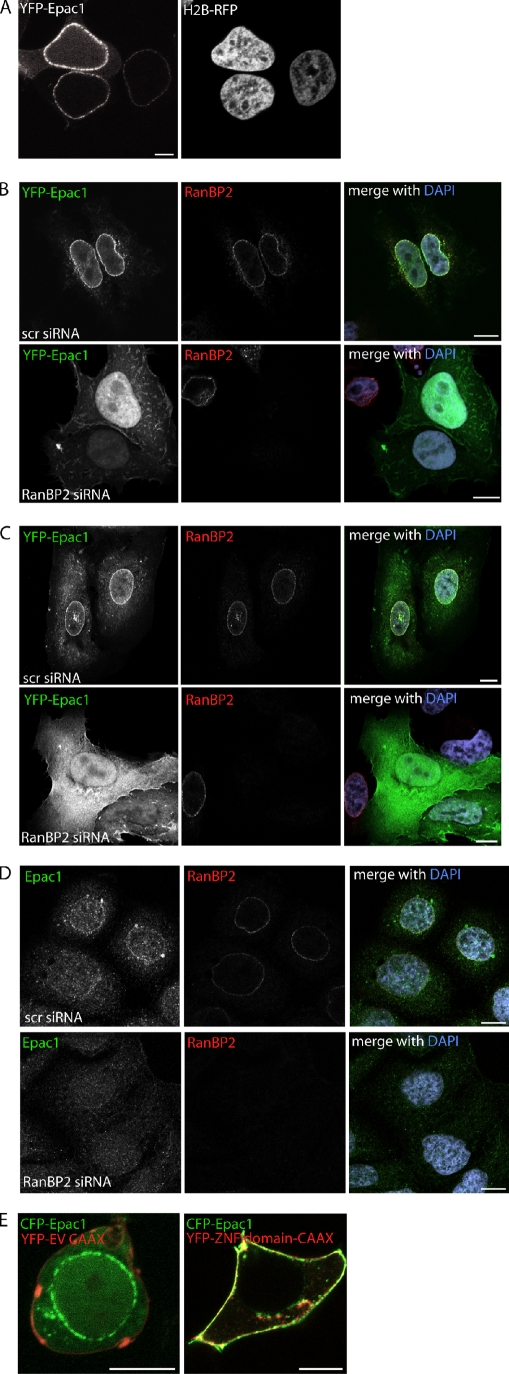
RanBP2 constitutes the cytoplasmic filaments of the NPC (Delphin et al., 1997; Walther et al., 2002). Also, Epac1 has been reported to localize to the NPC (Wang et al., 2006; Brock et al., 2008), and indeed, in addition to the cytosol and the nucleus, a large fraction of YFP-Epac1 is present at the nuclear envelope in HEK293T cells (Fig. 2 A). Strikingly, at low expression levels, YFP-Epac1 is detectable solely at this compartment, implying a high affinity of Epac1 for its anchor at the nuclear envelope (Fig. 2 A). We examined whether RanBP2 functions as an anchoring protein for Epac1 at this cellular compartment. For this, HEK293T cells were transfected with YFP-tagged Epac1, and upon fixation, cells were immunolabeled for endogenous RanBP2. This revealed the presence of exogenous Epac1 within the cytosol and nucleus and a large fraction of Epac1 that colocalized with RanBP2 at the nuclear envelope (Fig. 2 B). To test whether RanBP2 is required for the targeting of Epac1 to the NPC, HEK293T cells were depleted of RanBP2 by siRNA-mediated knockdown of the protein. Importantly, other nuclear pore components are not affected by cellular depletion of RanBP2 (Walther et al., 2002). RanBP2 down-regulation resulted in a complete loss of YFP-Epac1 from the nuclear envelope and the enrichment of Epac1 in the cytosol and nucleus (Fig. 2 B). Similarly, YFP-Epac1 localized in a RanBP2-dependent manner at the nuclear envelope in a variety of other cell lines tested, including U2OS cells (Fig. 2 C). As expected, Epac2, which does not interact with RanBP2 (Fig. 1 D), was not present at the nuclear envelope (Fig. S2). To confirm that RanBP2-mediated targeting of Epac1 to the NPC also occurs endogenously, fixed Ovcar3 cells were immunolabeled for both Epac1 and RanBP2. This revealed the presence of endogenous Epac1 at the nuclear envelope, which disappeared upon siRNA-mediated depletion of RanBP2 from these cells (Fig. 2 D). To validate this role of RanBP2 in the targeting of Epac1, we introduced the ZNFs of RanBP2 linked to the CAAX motif of K-Ras into HEK293Ts. This chimera localizes to the plasma membrane and, indeed, recruits Epac1 to this compartment as well (Fig. 2 E). In conclusion, these data show that RanBP2 functions as an anchoring protein for Epac1 at the NPC.
Figure 2.
RanBP2 recruits Epac1 to the NPC. (A) Live imaging of YFP-Epac1 together with the nuclear marker H2B-RFP in HEK293T cells showing the localization of Epac1 in the cytosol, the nucleus, and at the nuclear envelope. (B–D) Localization of YFP-Epac1 in HEK293T cells (B) and U2OS cells (C) and of endogenous Epac1 in Ovcar3 cells (D). Cells were transfected with either control (scr, scrambled) or RanBP2 siRNAs, and the HEK293T and U2OS cells were transfected the next day with YFP-Epac1. 60 h after siRNA transfection, cells were fixed and stained for endogenous RanBP2 with the goat polyclonal RanBP2 antibody and also for endogenous Epac1 in the Ovcar3 cells. Epac1 colocalizes with RanBP2 at the nuclear envelope, which is dependent on the presence of the RanBP2. (E) Live imaging of CFP-Epac1 together with YFP–empty vector–CAAX or YFP-ZNF-CAAX in HEK293T cells. Targeting of the ZNFs of RanBP2 to the plasma membrane by addition of the CAAX motif of K-Ras recruits Epac1 to the plasma membrane as well. Bars, 10 µm.
Epac1 remains bound to RanBP2 at the NPC upon cAMP binding
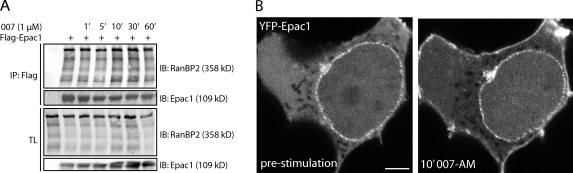
The cAMP-induced conformational change of Epac1 not only results in its activation but also in its translocation to the plasma membrane (Ponsioen et al., 2009). Potentially, RanBP2-bound Epac1 also may be released upon cAMP binding and become redistributed to the plasma membrane. To test whether activated Epac1 remains associated with RanBP2, HEK293T cells were transfected with Flag-tagged Epac1, and its binding to RanBP2 was determined upon stimulation with the Epac-selective cAMP analogue 8-pCPT-2′-O-Me-cAMP (007). After prolonged stimulation (up to 1 h) with 007, Flag-Epac1 retained its ability to coimmunoprecipitate RanBP2 to a similar extent as in unstimulated cells (Fig. 3 A). In addition, the presence of Epac1 at the NPC during its activation by cAMP was visualized by live imaging of HEK293T cells transfected with YFP-tagged Epac1. Although 007 stimulation resulted in the relocalization of the cytosolic fraction of Epac1 to the plasma membrane, the localization of Epac1 at the NPC remained unaffected (Fig. 3 B). These data indicate that Epac1 remains targeted to NPC by RanBP2 independent of cAMP signaling.
Figure 3.
Epac1 remains bound to RanBP2 at the NPC upon cAMP binding. (A) Coimmunoprecipitation of endogenous RanBP2 with Flag-tagged Epac1 in HEK293T cells after stimulation with 100 µM 8-pCPT-2′-O-Me-cAMP (007) for the indicated time points. (B) Confocal live imaging of HEK293T cells transfected with YFP-Epac1 before and 10 min after stimulation with 1 µM 8-pCPT-2′-O-Me-cAMP-AM (007-AM). Although the cytosolic fraction of Epac1 relocalizes to the plasma membrane upon 007-AM stimulation, its localization at the nuclear envelope remains unaltered. Bar, 5 µm. IB, immunoblot; IP, immunoprecipitation; TL, total lysate.
Phosphorylation of the ZNFs of RanBP2 releases Epac1 from the NPC
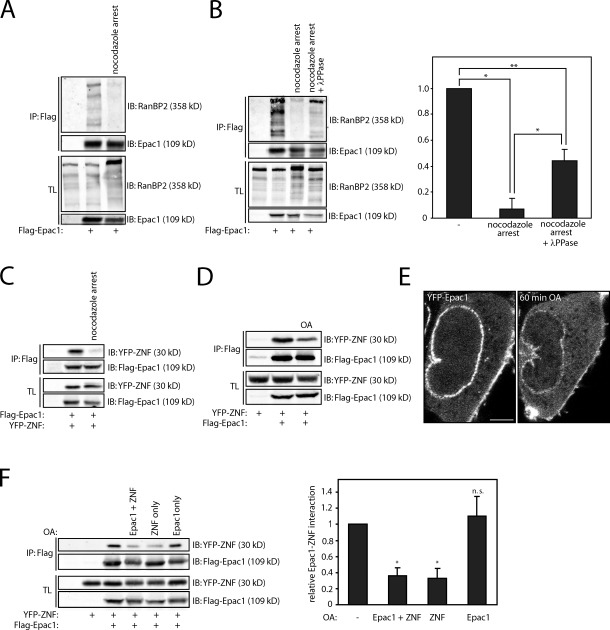
Epac1 remains associated with RanBP2 upon cAMP binding, but alternative signaling pathways may impinge on RanBP2 or Epac1 to affect their interaction. During mitosis, the nuclear envelope is broken down, and NPCs are disintegrated, which involves the posttranslational modification of diverse nucleoporins by mitotic kinases (Tran and Wente, 2006; Antonin et al., 2008). RanBP2 and Epac1 display a distinct localization pattern in mitosis, with RanBP2 being enriched at the kinetochores (Salina et al., 2003; Joseph et al., 2004) and Epac1 localizing at the mitotic spindle and centrosomes (Qiao et al., 2002). This suggests that Epac1 is released from RanBP2 during this phase of the cell cycle, which might reveal a potential regulatory mechanism for their interaction. Therefore, we examined the binding of Epac1 to RanBP2 in mitotic cells. U2OS cells were transfected with Flag-tagged Epac1 and arrested in mitosis by treatment with the microtubule-destabilizing agent nocodazole after a thymidine release. Subsequently, mitotic cells were collected by mitotic shake off, and the binding of RanBP2 to Flag-Epac1 was tested. As hypothesized, mitotic arrested cells displayed a large decrease in the Epac1–RanBP2 interaction compared with unsynchronized cells (Fig. 4 A). Likewise, when U2OS cells were arrested in mitosis by taxol or noscapine, the interaction between Epac1 and endogenous RanBP2 was diminished (Fig. S3). Interestingly, RanBP2 displayed a decreased electrophoretic mobility in mitotic cells, suggesting its increased phosphorylation in mitosis similar to other nucleoporins (Fig. 4 A and Fig. S3). The contribution of this potential phosphorylation to the loss of Epac1 binding was tested by treatment of cell lysates from mitotic cells with λ-phosphatase before coimmunoprecipitation. λ-Phosphatase reverted the band shift of endogenous RanBP2 and, moreover, restored the interaction between Epac1 and RanBP2 (Fig. 4 B). Importantly, YFP-ZNF also displayed a decreased electrophoretic mobility and reduced binding to Epac1 in nocodazole-arrested cells (Fig. 4 C). This suggests that Epac1 is released from RanBP2 by phosphorylation of the ZNFs. To verify this, binding of Flag-Epac1 to YFP-ZNF was tested upon stimulation of cells with the phosphatase inhibitor okadaic acid (OA). By inhibiting protein phosphatase 1 and 2A (Cohen et al., 1990), OA stimulation decreases protein dephosphorylation and results in the elevated phosphorylation state of proteins. Indeed, YFP-ZNF as well as Flag-tagged Epac1 displayed a band shift upon treatment of HEK293T cells with OA (Fig. 4 D). Immunoprecipitation of Flag-Epac1 after OA treatment demonstrated a significant decrease in the binding of YFP-ZNF (Fig. 4 D). Similar results were obtained with an alternative protein phosphatase inhibitor Calyculin A (unpublished data). In line with these binding experiments, live imaging of YFP-Epac1 in HEK293T cells revealed the release of Epac1 from the nuclear envelope upon OA stimulation (Fig. 4 E). To confirm that Epac1 is indeed released by phosphorylation of the ZNFs and exclude a contribution of Epac1 modification, HEK293T cells were transfected with either Flag-tagged Epac1 or the YFP-tagged ZNF. Next, either the Epac1- or ZNF-transfected cells, or both, were subjected to OA treatment, resulting in selective phosphorylation of Flag-Epac1 or YFP-ZNF. Upon mixture of the lysates, the subsequent coimmunoprecipitation revealed that OA stimulation of YFP-ZNF–expressing cells, but not Flag-Epac1–expressing cells, decreases the Epac1–ZNF interaction (Fig. 4 F). Thus, we demonstrate that the pool of Epac1 bound to RanBP2 at the NPC can be released by phosphorylation of the ZNFs. It should be noted that although we have identified this regulation to occur downstream of mitotic signaling pathways, the involved kinases are currently unclear, and other kinase-mediated pathways may impinge on the interaction as well.
Figure 4.
Phosphorylation of the ZNFs of RanBP2 releases Epac1 from the NPC. (A) Coimmunoprecipitation of endogenous RanBP2 with Flag-tagged Epac1 in U2OS cells that were arrested in mitosis by 24-h incubation with 2.5 µM thymidine, washed three times with PBS, and incubated for 16 h with 250 ng/ml nocodazole. Mitotic cells were subsequently collected by mitotic shake off and subjected to coimmunoprecipitation. (B) Coimmunoprecipitation of endogenous RanBP2 with Flag-tagged Epac1 in U2OS cells that were arrested in mitosis similar to panel A but with treatment of the lysate with 1 µM λ-phosphatase for 30 min before immunoprecipitation of Flag-Epac1. The quantification shows the mean with standard deviation of the relative binding of RanBP2 to Flag-Epac1 from three independent experiments. Statistical analysis was performed using a one-tailed Student’s t test. Asterisks indicate the p-value of the respective sample with the associated control sample. *, P < 0.007; **, P < 0.0005. (C) Coimmunoprecipitation of the YFP-tagged individual ZNF (ZNF #2) of RanBP2 (YFP-ZNF) with Epac1 in U2OS cells that were arrested in mitosis similar to panel A. (D) Coimmunoprecipitation of the YFP-tagged individual ZNF (ZNF #2) of RanBP2 (YFP-ZNF) with Flag-tagged Epac1 in HEK293T cells after stimulation with the phosphatase inhibitor okadaic acid (OA; 1 µM) for 1 h. OA results in a decrease in electrophoretic mobility of both YFP-ZNF and Flag-Epac1 and a decrease in association of the two proteins. (E) Confocal live imaging of HEK293T cells transfected with YFP-Epac1 before and 1 h after stimulation with 1 µM OA. This shows the decreased presence of Epac1 at the nuclear envelope upon OA stimulation. Note that also the plasma membrane localization of Epac1 is affected by OA stimulation, whereas an increase in cytosolic YFP-Epac1 is observed. Bar, 2 µm. (F) Coimmunoprecipitation of YFP-ZNF with Flag-tagged Epac1 after selective induction of phosphorylation of either YFP-ZNF or Flag-Epac1 by OA. Two separate dishes of HEK293T cells were transfected with either YFP-ZNF or Flag-Epac1 where indicated, and either one of the two or both were stimulated with 1 µM OA. Subsequently, the cell lysates of YFP-ZNF– and Flag-Epac1–expressing cells were mixed and subjected to coimmunoprecipitation. This demonstrates that OA stimulation of YFP-ZNF–transfected cells, but not Flag-Epac1–transfected cells, inhibits the binding between the two proteins. The quantification shows the mean with standard deviation of the relative binding of YFP-ZNF to Flag-Epac1 from three independent experiments. Statistical analysis was performed using a one-tailed Student’s t test. *, P < 0.0001. The minus signs indicate unstimulated cells. IB, immunoblot; IP, immunoprecipitation; TL, total lysate.
RanBP2 binds to the CDC25-HD of Epac1
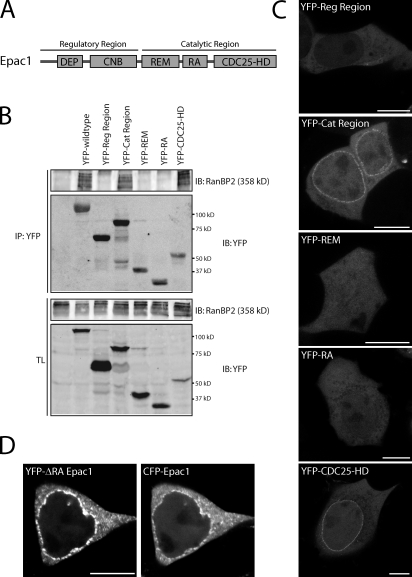
To examine which region of Epac1 is responsible for NPC anchoring, several truncated versions of Epac1 were tested for RanBP2 binding. In contrast to the regulatory region, the catalytic region was able to coimmunoprecipitate RanBP2 from HEK293T cells similar to full-length Epac1 (Fig. 5, A and B). Further truncation of this region into the individual domains revealed that the CDC25-HD is mediating the interaction with RanBP2 (Fig. 5 B). Accordingly, both the YFP-tagged catalytic region as well as the CDC25-HD localized to the nuclear membrane in these cells (Fig. 5 C). Although the Ras association (RA) domain of Epac1 has been implicated previously in NPC targeting through binding to the small G protein Ran (Liu et al., 2010), the individual RA domain does not localize to the nuclear envelope (Fig. 5 C). Furthermore, a mutant of Epac1 in which the RA region is replaced by the region linking the REM domain and the CDC25-HD of the Ras GEF Sos (son of sevenless) still localizes to the nuclear envelope similar to wild-type Epac1 (Fig. 5 D). In line with this, siRNA-mediated depletion of Ran did not affect the presence of Epac1 at the nuclear envelope (Fig. S4). This indicates that RanBP2 is sufficient for the localization of Epac1 at the NPC and functions independently of Ran.
Figure 5.
RanBP2 binds to the CDC25-HD of Epac1. (A) Domain architecture of Epac1 showing its catalytic region with the CDC25-homology domain (CDC25-HD) responsible for the catalysis of Rap, which is stabilized by the Ras exchange motif (REM) domain. In addition, the catalytic region contains a Ras association (RA) motif. The regulatory region of Epac1 contains the cAMP-binding (CNB) domain and a Disheveled, Egl-10, and Pleckstrin (DEP) domain. (B) Coimmunoprecipitation of endogenous RanBP2 with YFP-tagged full-length Epac1, the individual regulatory region (Reg Region), catalytic region (Cat Region), and the individual domains from the catalytic region of Epac1 in HEK293T cells. The CDC25-HD of Epac1 mediates the binding to RanBP2. (C) Confocal live imaging of HEK293T cells transfected with the YFP-tagged individual regulatory region, catalytic region, and the individual domains from the catalytic region of Epac1. Both the catalytic region and the CDC25-HD localize to the nuclear envelope. (D) Confocal live imaging of HEK293T cells transfected with YFP-tagged Epac1 lacking its RA domain (YFP-Epac1-ΔRA) together with CFP-tagged wild-type Epac1 showing that Epac1 lacking its RA domain is recruited to the nuclear envelope similar to wild-type Epac1. To maintain structural integrity, the RA domain of Epac1 is replaced by the region linking the REM domain and the CDC25-HD of the RasGEF Sos, which lacks an RA domain. Bars, 10 µm. IB, immunoblot; IP, immunoprecipitation; TL, total lysate.
RanBP2 binding inhibits GEF activity of Epac1
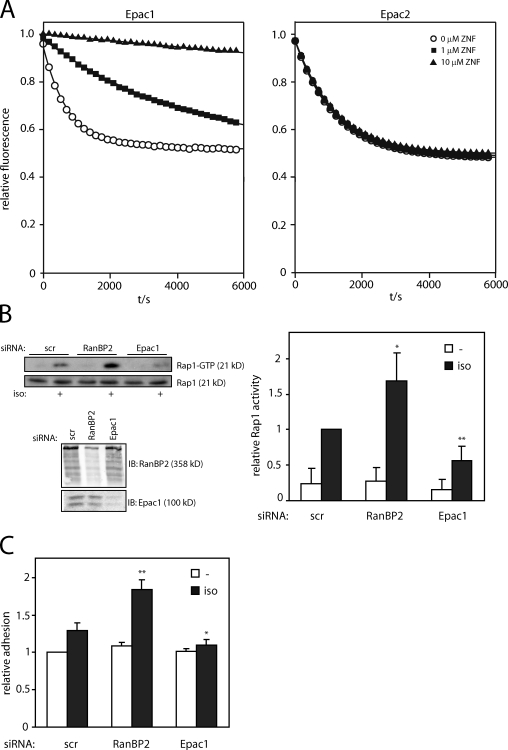
Because the CDC25-HD is catalyzing the nucleotide exchange of Rap, the interaction of RanBP2 with this domain might affect the ability of Epac1 to activate Rap. Therefore, the effect of RanBP2 binding on the activity of Epac1 was tested in vitro. For this, bacterially purified Epac1 was incubated with Rap1B loaded with fluorescently labeled mantGDP in the presence of excess unlabeled GDP. The exchange of Rap-bound mantGDP by cAMP-activated Epac1 was measured as a decrease in fluorescence, both in the absence or presence of increasing amounts of the ZNF domain of RanBP2. This revealed that the ZNF domain inhibited the activity of Epac1 toward Rap (Fig. 6 A). Importantly, the activity of Epac2 was not altered by the presence of the RanBP2 ZNF domain (Fig. 6 A), which is compatible with the notion that Epac2 does not interact with RanBP2 (Fig. 1 D). To confirm this inhibitory function for RanBP2 in vivo, we tested the activity of Epac1 after its activation by physiological levels of cAMP in Ovcar3 cells upon depletion of RanBP2. Activation of Epac1 by stimulation of the β adrenergic receptor with isoproterenol increased Rap1-GTP levels in these cells, and this was diminished upon siRNA-mediated depletion of Epac1 (Fig. 6 B). Instead, Epac1-induced Rap1 activation was significantly enhanced after knockdown of RanBP2, indicating the increased pool of cellular Epac1 that is capable of activating Rap1 upon RanBP2 depletion (Fig. 6 B). To verify that RanBP2 binding also functionally impedes Epac1 signaling, Epac1-induced adhesion of Ovcar3 cells to the extracellular matrix protein fibronectin was measured. In accordance with the Rap-GTP pull-down experiment, depletion of Epac1 abolished isoproterenol-induced adhesion of Ovcar3 cells, whereas this was significantly enhanced by knockdown of RanBP2 (Fig. 6 C). Thus, by interacting with the CDC25-HD of Epac1, RanBP2 functions as a negative regulator of Epac1 and establishes an inactive pool of this GEF at the NPC.
Figure 6.
RanBP2-binding inhibits GEF activity of Epac1. (A) Measurement of Epac1 activity in vitro. Rap1B was loaded with fluorescent mantGDP and the release of mantGDP by Epac1Δ1–148 (left) or Epac2 (right) in the presence of 10 µM cAMP was measured in real time in the absence or presence of 1 or 10 µM of the zinc finger (ZNF) domain of RanBP2 (which, as shown in Fig. 2, targets Epac1 to the NPC in vivo). The presented data are representatives of three independent experiments. (B) Pull-down of Rap1-GTP from Ovcar3 cells upon stimulation for 5 min with 5 µM isoproterenol (iso) 60 h after transfection with control (scr, scrambled), RanBP2, or Epac1 siRNAs. The bottom blot shows the efficiency of the knockdown of RanBP2 and Epac1. The quantification shows the mean with standard deviation of Rap1 activity from four independent experiments. Statistical analysis was performed using a one-tailed Student’s t test. Asterisks indicate the p-value of the respective sample with the associated control sample. *, P < 0.006; **, P < 0.0003. (C) Adhesion of Ovcar3 cells transfected with control (scrambled), RanBP2, or Epac1 siRNAs. 60 h after siRNA transfection, cells were allowed to adhere to a fibronectin-coated surface for 45 min in the absence or presence of 5 µM isoproterenol, and adhesion was subsequently detected by the measure of endogenous phosphatase activity. Shown are mean data with standard deviation from three individual experiments, which were normalized to the adhesion of control siRNA-transfected cells. Statistical analysis was performed using a Student’s t test. Asterisks indicate the p-value of the respective sample with the associated control sample. *, P < 0.02; **, P < 0.0003. The minus signs indicate unstimulated cells. IB, immunoblot.
Discussion
cAMP is a second messenger that is essential for relaying hormonal responses in many biological processes. Spatial regulation of cAMP signaling is established by local degradation of cAMP by spatially restricted phosphodiesterases and, furthermore, by the confined targeting of the cAMP effector proteins. We have previously described several aspects of the spatiotemporal control of the cAMP effector Epac1. Epac1 is released from autoinhibition by the binding of cAMP (de Rooij et al., 1998; Rehmann, 2006, 2008), and in addition, diverse anchoring mechanisms, including plasma membrane targeting (Ponsioen et al., 2009; Gloerich et al., 2010), recruit Epac1 to specific subcellular localizations. This compartmentalization of Epac1 results in local activation of Rap, which couples Epac1 signaling to specific cellular processes. We now show that Epac1, in addition, is negatively regulated by its interaction with the nucleoporin RanBP2. We found that Epac1 directly interacts with the ZNFs of RanBP2, which recruits Epac1 to the NPC. RanBP2 interacts with the catalytic CDC25-HD of Epac1, resulting in the inhibition of the catalytic activity of Epac1 toward Rap1 in vitro. Finally, we show that depletion of RanBP2 liberates Epac1 from the NPC and enhances Epac1-dependent cAMP responses, i.e., induction of Rap1 activity and, consequently, cell–extracellular matrix adhesion. Thus, RanBP2 establishes an inactive pool of Epac1 at the NPC, which upon release, may enhance cAMP-Epac1-Rap signaling. Collectively, these results show, for the first time, that components of the NPC function to maintain an inactive state of GEFs in the cell.
Our results show that the ZNFs of RanBP2 directly inhibit Epac1 activity by binding to the catalytic CDC25-HD of Epac1. Elucidation of the mechanism awaits structural analysis of the complex, but RanBP2 may reorient the Rap-binding surfaces within the CDC25-HD or, alternatively, prevent Rap binding by steric hindrance. Clearly, this interaction is regulated, and our data indicate that this regulation includes the phosphorylation of the ZNFs of RanBP2 (Fig. 4). First, Epac1 is released from RanBP2 during mitosis, which correlates with a mobility shift of RanBP2 as well as of the individual ZNF of RanBP2. Second, both this decrease in electrophoretic mobility and the release of Epac1 can be reverted by incubation with λ-phosphatase. Finally, the phosphatase inhibitor OA induces the release of Epac1 from the NPC by elevating the phosphorylation state of the ZNFs. Thus, phosphorylation of the ZNFs is the most likely trigger to induce the release of Epac1 from RanBP2. Currently, the involved residues within RanBP2 are elusive, but because RanBP2 contains multiple ZNFs, it is likely that multiple phosphorylation sites are involved. Several phosphorylation sites have been reported within the ZNFs of RanBP2 (Beausoleil et al., 2004; Dephoure et al., 2008; Gauci et al., 2009; Mayya et al., 2009), but a more detailed analysis is required to identify the critical sites for Epac1 release and the involved upstream kinases. In addition to the phosphorylation of RanBP2, alternative mechanisms may also induce the release of Epac1 from the NPC. For instance, the DNA-binding capacity of ZNFs within transcription factors can be regulated by the oxidation of the zinc-binding cysteines (Webster et al., 2001), and a similar mechanism may exist for RanBP2.
Our results show that depletion of RanBP2 results in an increase in cAMP-induced Epac1-dependent Rap1 activation and, consequently, cell adhesion, which is in accordance with a function for RanBP2 as a negative regulator of Epac1. One intriguing question is why this inactive pool of Epac1 is maintained at the NPC. Potentially, RanBP2 might contribute further to the regulation of Epac1 than merely inhibiting its GEF activity. RanBP2 functions as an E3 SUMO ligase (Pichler et al., 2002); however, thus far, we were unable to detect sumoylation of Epac1 by RanBP2 in vivo or in vitro (unpublished data). RanBP2 also plays a role in nuclear import (Yaseen and Blobel, 1999; Hutten et al., 2008, 2009); however, RanBP2 likely does not control nuclear import of Epac1, as Epac1 remains nuclearly localized in cells depleted of RanBP2 (Fig. 2, A and B). Alternatively, Epac1 itself may function at the NPC independent of its Rap nucleotide exchange activity: for instance, by directly affecting the function of RanBP2 in nuclear import or sumoylation or as an adaptor protein to recruit additional proteins to the NPC. Indeed, several Rap-independent effects of Epac1 have previously been described (López De Jesús et al., 2006; Métrich et al., 2008; Sehrawat et al., 2008). Finally, Epac1 may exert a local function at the NPC upon its release from RanBP2.
Recently, it was suggested that the localization of Epac1 at the NPC is mediated by binding of its putative RA domain to the NPC-associated small G protein Ran (Liu et al., 2010). Our results, however, show that RanBP2 is the anchor for Epac1 at the NPC and that RanBP2 functions independently of Ran. First, siRNA-mediated depletion of RanBP2 completely disrupts the presence of Epac1 at the nuclear envelope (Fig. 2), whereas siRNA-mediated depletion of Ran does not show any effect on the localization of Epac1 at the nuclear envelope (Fig. S4). Second, the individual CDC25-HD of Epac1 localizes to the nuclear envelope, whereas the individual RA domain is not able to do so (Fig. 2 C). In addition, Epac1 lacking its RA domain is targeted to the nuclear envelope similar to wild-type Epac1 (Fig. 2 D). Using a distinct RA domain mutant, Liu et al. (2010) did observe effects on the localization of Epac1 at the nuclear envelope. This effect might be indirect, as based on our structural understanding of Epac, this mutant also comprises parts of the REM domain, which stabilizes the CDC25-HD. Third, the ZNF region of RanBP2 that is targeted to the plasma membrane by addition of the CAAX motif of K-Ras is able to recruit Epac1 to the plasma membrane (Fig. 2 E), whereas RanV19-CAAX is not able to do so (Fig. S5). Because RanBP2 binding inhibits Epac1 activity (Fig. 6), this indicates Epac1 is kept at the NPC in an inactive state. In line with this, we have previously shown that cAMP-Epac1 signaling activates Rap predominantly at the plasma membrane and not at the nuclear envelope (Ponsioen et al., 2009). Indeed, siRNA-mediated depletion of RanBP2 releases Epac1 from the NPC and enhances cAMP-induced Rap activation and cell adhesion (Fig. 6). Thus, RanBP2 maintains an inactive pool of Epac1 at the NPC.
Our data indicate that phosphorylation of the ZNFs and, consequently, their decrease in affinity for Epac1 occurs in mitosis when the NPC is broken down (Fig. 5). RanBP2 is pivotal for various aspects of mitosis, which involves both its E3 SUMO ligase as well as adaptor protein function, and ablation of RanBP2 function results in various mitotic defects (Salina et al., 2003; Joseph et al., 2004; Prunuske et al., 2006; Dawlaty et al., 2008; Klein et al., 2009; Splinter et al., 2010). Potentially, the liberation of Epac1 from RanBP2 may be a prerequisite for proper mitosis to take place, as release of Epac1 may allow the binding of other proteins to the ZNFs. For instance, binding of the coatomer complex COPI to the ZNFs of RanBP2 has been implicated in nuclear envelope breakdown (Prunuske et al., 2006). Alternatively, RanBP2-released Epac1, itself, may contribute to aspects of this phase of the cell cycle. cAMP levels rapidly drop during the onset of mitosis and rise again during the transition into interphase (Grieco et al., 1996), which would restrict a role for RanBP2-released Epac1 and, consequently, Rap1 activation to the end of mitosis. In line with this, it was recently demonstrated that Rap1 activity is required for respreading of rounded cells at the end of mitosis (Dao et al., 2009), which may involve its activation by Epac1.
In summary, we have demonstrated the regulated recruitment of Epac1 to the NPC by RanBP2, thereby maintaining an inactive cellular pool of Epac1. RanBP2 might function as a generic node that transduces different cellular inputs to regulate the availability of Epac1 and, consequently, to modulate the cAMP-Epac1-Rap1–signaling network. Very recently, it was shown for the Dbl family of Rho GEFs that binding of myosin II to its catalytic site suppresses GEF activity (Lee et al., 2010). This implies that the formation of inhibitory protein complexes mediated by the catalytic domain might represent a common mechanism for the regulation of GEFs.
Materials and methods
Reagents and antibodies
007 was obtained from Biolog Life Sciences; isoproterenol, nocodazole, taxol, thymidine, and noscapine were obtained from Sigma-Aldrich; OA was obtained from Enzo Life Sciences; and λ-phosphatase was obtained from New England Biolabs, Inc. The following antibodies were used: mouse monoclonal GFP (Roche), Flag M2 (Sigma-Aldrich), rabbit polyclonal RanBP2 (Abcam), Rap1 (Santa Cruz Biotechnology, Inc.), and Ran (BD). The monoclonal Epac1 antibody (Cell Signaling Technology) has been described previously (Ponsioen et al., 2009). The goat polyclonal RanBP2 antibody was a gift from F. Melchior (Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany) and has previously been described (Pichler et al., 2002). The rabbit polyclonal Epac1 antibody (2293) was generated in house by injection of purified Δ1–148-Epac1. ON-TARGETplus SMARTpool siRNAs targeting RanBP2 (L-004746), Epac1 (L-007676), and Ran (L-010353) and control siRNAs were obtained from Thermo Fisher Scientific.
DNA constructs
Full-length Epac1 (RapGEF3, Homo sapiens; available from GenBank/EMBL/DDBJ under accession no. 3978530) and Epac2 (RapGEF4, Mus musculus; available from GenBank/EMBL/DDBJ under accession no. 9790086), the separate regulatory (amino acids 1–328) and catalytic (amino acids 330–881) region of Epac1, the individual REM (amino acids 330–486), RA (amino acids 487–593), and CDC25-HD (amino acids 598–867) of Epac1, and the ZNF domain (amino acids 1,341–1,819) and individual ZNF (ZNF #2; amino acids 1,407–1,456) of RanBP2 were C-terminally cloned to either a Citrine YFP or Flag-His tag in a pcDNA3 vector or an HA tag in a pMT2 vector using the Gateway system (Invitrogen). cDNA for RanBP2 was a gift from T. Nishimoto (Kanazawa University Cancer Research Institute, Kanazawa, Japan), and RFP-H2B was a gift from R. Medema (University Medical Center Utrecht, Utrecht, Netherlands). The YFP-ΔRA-Epac1 mutant has been described previously (Ponsioen et al., 2009). In this mutant, the RA domain between the REM and CDC25-HD of Epac1 has been replaced by the homologous region of the RasGEF Sos (which lacks an RA domain) to maintain the structural integrity of Epac1. The C-terminal CAAX motif of K-Ras (van der Wal et al., 2001) was used to generate the ZNF domain–CAAX and RanV19-CAAX chimeras.
Yeast two-hybrid screening
Human full-length Epac1 cloned in a pB27 vector was screened with a randomly primed human placenta library by Hybrigenics S.A. as previously described (Rain et al., 2001).
Protein purification
Rap1B (amino acids 1–167), Epac1 (amino acids 149–882), and Epac2 (amino acids 280–993) proteins were purified as previously described (Rehmann, 2006; Rehmann et al., 2006). The ZNF domain was cloned into a pGEX6P3 vector (GE Healthcare) and expressed in the bacterial strain CK600K as previously described for Epac (Rehmann, 2006) but in medium supplemented with 100 µM ZnSO4. The protein was essentially purified as previously described for Epac (Rehmann, 2006), but all buffers were supplemented with 10 µM ZnCl2, and instead of on column cleavage, the protein was eluted with 20 mM gluthatione, cleaved with PreScission Protease, dialyzed, and reloaded to a GST column to remove the cleaved GST.
Cell culture and transfection
HEK293T (human embryonic kidney) and U2OS (human osteosarcoma) cells were cultured in DME, and Ovcar3 (ovarian carcinoma) cells were cultured in RPMI 1640 medium. All media were supplemented with 10% FBS and antibiotics. Cells were transfected with expression plasmids using a transfection reagent (FuGENE; Roche) and with ON-TARGETplus SMARTpool siRNA using HiPerFect (QIAGEN) according to manufacturer’s protocol.
Coimmunoprecipitations
Immunoprecipitations were performed in lysis buffer containing 1% Triton X-100, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and protease and phosphatase inhibitors. Cell lysates were cleared by centrifugation and incubated with protein A agarose beads (GE Healthcare) coupled to the indicated antibody. After extensive washing with lysis buffer, bound proteins were eluted in Laemmli buffer and analyzed by SDS-PAGE and Western blotting with the indicated antibodies. Where indicated (Fig. 4 D), cells were treated for 1 h with 1 µM OA before lysis. For coimmunoprecipitations in mitotic cells, cells were treated for 24 h with 2.5 mM thymidine followed by extensive washing with PBS and 16-h incubation with 250 ng/ml nocodazole, 2 µM taxol, or 25 µM noscapine. Mitotic cells were subsequently collected by mitotic shake off. The in vitro interaction between bacterially purified Epac1 and GST-tagged RanBP2 ZNF domain was performed in a buffer containing 100 mM NaCl, 5 mM dithiothreitol, 2.5% glycerol, and 0.005% Tween using either protein A agarose beads coupled to the Epac1 antibody or glutathione agarose beads.
Confocal microscopy
For visualization of YFP-Epac1 and RanBP2 in fixed cells, cells were grown on 12-mm glass coverslips (for experiments with HEK293T, cells were precoated with poly-l-lysine [Sigma-Aldrich]), fixed with 3.8% formaldehyde, permeabilized using 0.1% Triton X-100, and blocked in 2% BSA. For visualization of endogenous Epac1, cells were fixed in ice-cold methanol followed by 3.8% formaldehyde. Cells were incubated with the indicated primary antibodies and, subsequently, with Alexa Fluor–conjugated secondary antibodies (Invitrogen). Mounted slides were examined using a confocal laser-scanning microscope (Axioskop2; Carl Zeiss; 63× magnification lenses, NA 1.4). For live imaging, transfected cells were grown in Willco wells (Willco Wells B.V.) and examined at 37°C in L-15 medium (Leibovitz; Sigma-Aldrich) using an confocal laser-scanning microscope (Axioskop2; 63× magnification lenses, NA 1.4). Postacquisition image adjustments were performed using ImageJ software (National Institutes of Health).
In vitro Rap activation assay
Loading of Rap with the fluorescent GDP analogue mantGDP (2′-/3′-O-[N′-methylanthraniloyl]guanosine-5′-O-diphosphate) and the general assay set up were performed as previously described (Rehmann, 2006) but with 10 µM ZnCl2 in the reaction buffer. RanBP2 ZNF was added to the reaction at concentrations as indicated in the figures.
In vivo Rap activation assay
Rap activity was assayed as described previously (van Triest and Bos, 2004). In brief, 60 h after siRNA transfection of Ovcar3 cells grown in 6-well plates, cells were stimulated for 5 min with 5 µM isoproterenol and, subsequently, lysed in buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 10% glycerol, 2 mM MgCl2, and protease and phosphatase inhibitors. Lysates were cleared by centrifugation, and active Rap was precipitated with a GST fusion protein of the Ras-binding domain of Ral guanine nucleotide dissociation stimulator precoupled to glutathione–Sepharose beads. Bound proteins were eluted in Laemmli buffer and analyzed by SDS-PAGE and Western blotting with the indicated antibodies.
Adhesion assay
Adhesion of Ovcar3 cells was measured as described previously (Lyle et al., 2008). In brief, 48-well polystyrene cell culture dishes were precoated with 5 µg/ml fibronectin and, subsequently, blocked with bovine serum albumin (Sigma-Aldrich). 60 h after siRNA transfection, cells were trypsinized, washed once in RPMI 1640 containing 10% FBS, and allowed to recover surface proteins for 1.5 h in suspension in RPMI 1640 containing 0.5% FBS, glutamine, and 10 mM Hepes, pH 7.4, at 37°C with gentle rolling. Subsequently, 6.0 × 105 cells were plated per well in the absense or presence of 5 µM isoproterenol. Adhesion was allowed to proceed for 45 min at 37°C, and unbound cells were discarded by washing with PBS. Adhered cells were lysed in buffer containing 0.4% Triton X-100, 50 mM sodium citrate, and 10 mg/ml phosphatase substrate (Sigma-Aldrich). The reaction was incubated at 37°C, and the total amount of cellular protein was determined by measuring the absorption at 405 nm.
Online supplemental material
Fig. S1 shows coimmunoprecipitation of RanBP2 with Epac1. Fig. S2 shows localization of YFP-Epac2 in HEK293T cells. Fig. 3 shows the coimmunoprecipitation of RanBP2 with Epac1 in cells arrested in mitosis by incubation with nocodazole, taxol, or noscapine. Fig. S4 shows the localization of YFP-Epac1 in cells depleted of Ran. Fig. S5 shows the localization of CFP-Epac1 in cells expressing YFP-RanV19-CAAX. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011126/DC1.
Acknowledgments
We thank John de Koning, Hybrigenics S.A., and University Medical Center Utrecht for the yeast two-hybrid screen, Takeharu Nishimoto for RanBP2 cDNA, Frauke Melchior for the RanBP2 antibody and help with in vitro sumoylation experiments, Kees Jalink and Bas Ponsioen for helpful discussions, and the members of our laboratory for stimulating discussions and critical reading of the manuscript.
This study is supported by Chemical Sciences (to M. Gloerich), Earth and Life Sciences (to H. Rehmann), Netherlands Proteomics Centre (to L.A.T. Meijer), and the Netherlands Genomics Initiative (to J.L. Bos) of the Netherlands Organization for Scientific Research.
Footnotes
Abbreviations used in this paper:
- Epac
- exchange protein directly activated by cAMP
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- NPC
- nuclear pore complex
- OA
- okadaic acid
- RA
- Ras association
- REM
- Ras exchange motif
- ZNF
- zinc finger
References
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582:2004–2016 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Aslanukov A., Bhowmick R., Guruju M., Oswald J., Raz D., Bush R.A., Sieving P.A., Lu X., Bock C.B., Ferreira P.A. 2006. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2:e177 10.1371/journal.pgen.0020177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie G.S. 2009. Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 276:1790–1799 10.1111/j.1742-4658.2009.06926.x [DOI] [PubMed] [Google Scholar]
- Beausoleil S.A., Jedrychowski M., Schwartz D., Elias J.E., Villén J., Li J., Cohn M.A., Cantley L.C., Gygi S.P. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 101:12130–12135 10.1073/pnas.0404720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad R., van der Velde H., Fornerod M., Pickersgill H. 2004. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 24:2373–2384 10.1128/MCB.24.6.2373-2384.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., Wittinghofer A. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 129:865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Brock T.G., Serezani C.H., Carstens J.K., Peters-Golden M., Aronoff D.M. 2008. Effects of prostaglandin E2 on the subcellular localization of Epac-1 and Rap1 proteins during Fcgamma-receptor-mediated phagocytosis in alveolar macrophages. Exp. Cell Res. 314:255–263 10.1016/j.yexcr.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., Holmes C.F., Tsukitani Y. 1990. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem. Sci. 15:98–102 10.1016/0968-0004(90)90192-E [DOI] [PubMed] [Google Scholar]
- Dao V.T., Dupuy A.G., Gavet O., Caron E., de Gunzburg J. 2009. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J. Cell Sci. 122:2996–3004 10.1242/jcs.041301 [DOI] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C., Guan T., Melchior F., Gerace L. 1997. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell. 8:2379–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. 2008. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 105:10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J., Zwartkruis F.J., Verheijen M.H., Cool R.H., Nijman S.M., Wittinghofer A., Bos J.L. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 396:474–477 10.1038/24884 [DOI] [PubMed] [Google Scholar]
- Ferreira P.A., Nakayama T.A., Pak W.L., Travis G.H. 1996. Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature. 383:637–640 10.1038/383637a0 [DOI] [PubMed] [Google Scholar]
- Ferreira P.A., Nakayama T.A., Travis G.H. 1997. Interconversion of red opsin isoforms by the cyclophilin-related chaperone protein Ran-binding protein 2. Proc. Natl. Acad. Sci. USA. 94:1556–1561 10.1073/pnas.94.4.1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D., Rabut G., Ciccarelli F.D., Herold A., Köcher T., Niggeweg R., Bork P., Ellenberg J., Izaurralde E. 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol. Cell. Biol. 24:1155–1167 10.1128/MCB.24.3.1155-1167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauci S., Helbig A.O., Slijper M., Krijgsveld J., Heck A.J., Mohammed S. 2009. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81:4493–4501 10.1021/ac9004309 [DOI] [PubMed] [Google Scholar]
- Gloerich M., Bos J.L. 2010. Epac: defining a new mechanism for cAMP action. Annu. Rev. Pharmacol. Toxicol. 50:355–375 10.1146/annurev.pharmtox.010909.105714 [DOI] [PubMed] [Google Scholar]
- Gloerich M., Ponsioen B., Vliem M.J., Zhang Z., Zhao J., Kooistra M.R., Price L.S., Ritsma L., Zwartkruis F.J., Rehmann H., et al. 2010. Spatial regulation of cyclic AMP-Epac1 signaling in cell adhesion by ERM proteins. Mol. Cell. Biol. 30:5421–5431 10.1128/MCB.00463-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieco D., Porcellini A., Avvedimento E.V., Gottesman M.E. 1996. Requirement for cAMP-PKA pathway activation by M phase-promoting factor in the transition from mitosis to interphase. Science. 271:1718–1723 10.1126/science.271.5256.1718 [DOI] [PubMed] [Google Scholar]
- Huston E., Lynch M.J., Mohamed A., Collins D.M., Hill E.V., MacLeod R., Krause E., Baillie G.S., Houslay M.D. 2008. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc. Natl. Acad. Sci. USA. 105:12791–12796 10.1073/pnas.0805167105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Flotho A., Melchior F., Kehlenbach R.H. 2008. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import. Mol. Biol. Cell. 19:2300–2310 10.1091/mbc.E07-12-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Wälde S., Spillner C., Hauber J., Kehlenbach R.H. 2009. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell Sci. 122:1100–1110 10.1242/jcs.040154 [DOI] [PubMed] [Google Scholar]
- Joseph J., Liu S.T., Jablonski S.A., Yen T.J., Dasso M. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617 10.1016/j.cub.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Springett G.M., Mochizuki N., Toki S., Nakaya M., Matsuda M., Housman D.E., Graybiel A.M. 1998. A family of cAMP-binding proteins that directly activate Rap1. Science. 282:2275–2279 10.1126/science.282.5397.2275 [DOI] [PubMed] [Google Scholar]
- Klein U.R., Haindl M., Nigg E.A., Muller S. 2009. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol. Biol. Cell. 20:410–418 10.1091/mbc.E08-05-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Choi C.K., Shin E.Y., Schwartz M.A., Kim E.G. 2010. Myosin II directly binds and inhibits Dbl family guanine nucleotide exchange factors: a possible link to Rho family GTPases. J. Cell Biol. 190:663–674 10.1083/jcb.201003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Takahashi M., Li Y., Dillon T.J., Kaech S., Stork P.J. 2010. The interaction of Epac1 and Ran promotes Rap1 activation at the nuclear envelope. Mol. Cell. Biol. 30:3956–3969 10.1128/MCB.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López De Jesús M., Stope M.B., Oude Weernink P.A., Mahlke Y., Börgermann C., Ananaba V.N., Rimmbach C., Rosskopf D., Michel M.C., Jakobs K.H., Schmidt M. 2006. Cyclic AMP-dependent and Epac-mediated activation of R-Ras by G protein-coupled receptors leads to phospholipase D stimulation. J. Biol. Chem. 281:21837–21847 10.1074/jbc.M604156200 [DOI] [PubMed] [Google Scholar]
- Lyle K.S., Raaijmakers J.H., Bruinsma W., Bos J.L., de Rooij J. 2008. cAMP-induced Epac-Rap activation inhibits epithelial cell migration by modulating focal adhesion and leading edge dynamics. Cell. Signal. 20:1104–1116 10.1016/j.cellsig.2008.01.018 [DOI] [PubMed] [Google Scholar]
- Mayya V., Lundgren D.H., Hwang S.I., Rezaul K., Wu L., Eng J.K., Rodionov V., Han D.K. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2:ra46 10.1126/scisignal.2000007 [DOI] [PubMed] [Google Scholar]
- Melchior F., Guan T., Yokoyama N., Nishimoto T., Gerace L. 1995. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J. Cell Biol. 131:571–581 10.1083/jcb.131.3.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métrich M., Lucas A., Gastineau M., Samuel J.L., Heymes C., Morel E., Lezoualc’h F. 2008. Epac mediates beta-adrenergic receptor-induced cardiomyocyte hypertrophy. Circ. Res. 102:959–965 10.1161/CIRCRESAHA.107.164947 [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Ponsioen B., Gloerich M., Ritsma L., Rehmann H., Bos J.L., Jalink K. 2009. Direct spatial control of Epac1 by cyclic AMP. Mol. Cell. Biol. 29:2521–2531 10.1128/MCB.01630-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunuske A.J., Liu J., Elgort S., Joseph J., Dasso M., Ullman K.S. 2006. Nuclear envelope breakdown is coordinated by both Nup358/RanBP2 and Nup153, two nucleoporins with zinc finger modules. Mol. Biol. Cell. 17:760–769 10.1091/mbc.E05-06-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Mei F.C., Popov V.L., Vergara L.A., Cheng X. 2002. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 277:26581–26586 10.1074/jbc.M203571200 [DOI] [PubMed] [Google Scholar]
- Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., et al. 2001. The protein-protein interaction map of Helicobacter pylori. Nature. 409:211–215 10.1038/35051615 [DOI] [PubMed] [Google Scholar]
- Rehmann H. 2006. Characterization of the activation of the Rap-specific exchange factor Epac by cyclic nucleotides. Methods Enzymol. 407:159–173 10.1016/S0076-6879(05)07014-X [DOI] [PubMed] [Google Scholar]
- Rehmann H., Das J., Knipscheer P., Wittinghofer A., Bos J.L. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature. 439:625–628 10.1038/nature04468 [DOI] [PubMed] [Google Scholar]
- Rehmann H., Arias-Palomo E., Hadders M.A., Schwede F., Llorca O., Bos J.L. 2008. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature. 455:124–127 10.1038/nature07187 [DOI] [PubMed] [Google Scholar]
- Salina D., Enarson P., Rattner J.B., Burke B. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001 10.1083/jcb.200304080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrawat S., Cullere X., Patel S., Italiano J., Jr, Mayadas T.N. 2008. Role of Epac1, an exchange factor for Rap GTPases, in endothelial microtubule dynamics and barrier function. Mol. Biol. Cell. 19:1261–1270 10.1091/mbc.E06-10-0972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.B., Patel H.H., Roepman R., Schick D., Ferreira P.A. 1999. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J. Biol. Chem. 274:37370–37378 10.1074/jbc.274.52.37370 [DOI] [PubMed] [Google Scholar]
- Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350 10.1371/journal.pbio.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053 10.1016/j.cell.2006.05.027 [DOI] [PubMed] [Google Scholar]
- van der Wal J., Habets R., Várnai P., Balla T., Jalink K. 2001. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J. Biol. Chem. 276:15337–15344 10.1074/jbc.M007194200 [DOI] [PubMed] [Google Scholar]
- van Triest M., Bos J.L. 2004. Pull-down assays for guanoside 5′-triphosphate-bound Ras-like guanosine 5′-triphosphatases. Methods Mol. Biol. 250:97–102 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Pickersgill H.S., Cordes V.C., Goldberg M.W., Allen T.D., Mattaj I.W., Fornerod M. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63–77 10.1083/jcb.200202088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dillon T.J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P.J. 2006. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol. Cell. Biol. 26:2130–2145 10.1128/MCB.26.6.2130-2145.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster K.A., Prentice H., Bishopric N.H. 2001. Oxidation of zinc finger transcription factors: physiological consequences. Antioxid. Redox Signal. 3:535–548 10.1089/15230860152542916 [DOI] [PubMed] [Google Scholar]
- Wu J., Matunis M.J., Kraemer D., Blobel G., Coutavas E. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209–14213 10.1074/jbc.270.23.14209 [DOI] [PubMed] [Google Scholar]
- Yaseen N.R., Blobel G. 1999. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin nup358. J. Biol. Chem. 274:26493–26502 10.1074/jbc.274.37.26493 [DOI] [PubMed] [Google Scholar]
- Yokoyama N., Hayashi N., Seki T., Panté N., Ohba T., Nishii K., Kuma K., Hayashida T., Miyata T., Aebi U., et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature. 376:184–188 10.1038/376184a0 [DOI] [PubMed] [Google Scholar]