Polarized fluorescence microscopy reveals that septins across diverse species assemble into similar higher-order structures consisting of dynamic, paired filaments.
Abstract
The septins are conserved, GTP-binding proteins important for cytokinesis, membrane compartmentalization, and exocytosis. However, it is unknown how septins are arranged within higher-order structures in cells. To determine the organization of septins in live cells, we developed a polarized fluorescence microscopy system to monitor the orientation of GFP dipole moments with high spatial and temporal resolution. When GFP was fused to septins, the arrangement of GFP dipoles reflected the underlying septin organization. We demonstrated in a filamentous fungus, a budding yeast, and a mammalian epithelial cell line that septin proteins were organized in an identical highly ordered fashion. Fluorescence anisotropy measurements indicated that septin filaments organized into pairs within live cells, just as has been observed in vitro. Additional support for the formation of pairs came from the observation of paired filaments at the cortex of cells using electron microscopy. Furthermore, we found that highly ordered septin structures exchanged subunits and rapidly rearranged. We conclude that septins assemble into dynamic, paired filaments in vivo and that this organization is conserved from yeast to mammals.
Introduction
The septins are conserved from fungi through humans, form heteromeric filaments in vitro, and are assembled into a variety of higher order structures in cells (Pan et al., 2007). Septins serve as membrane diffusion barriers and as scaffolds for the localization of proteins, and contribute to cytokinesis and cell migration (Hartwell, 1971; Barral et al., 2000; Longtine et al., 2000; Takizawa et al., 2000; Gladfelter et al., 2001; McMurray and Thorner, 2009; Tooley et al., 2009). Aberrant septin function has been implicated in neurodegenerative disorders, such as Alzheimer’s and Parkinson’s diseases, and also in forms of cancer (Peterson and Petty, 2010). Recently, septins have been found to function as diffusion barriers at the base of primary cilia, implicating their participation in ciliopathies (Hu et al., 2010; Kim et al., 2010).
The ability of septins to assemble into hetero-oligomeric complexes and higher-order structures is critical for their function in many cells. Septins can be made to form nonpolar polymers in vitro from recombinant or purified protein (Frazier et al., 1998; Sirajuddin et al., 2007; Bertin et al., 2008). However, it is not yet known if these linear filaments reflect how septins are organized within larger superstructures observed in vivo. Electron micrographs of putative septin complexes at the yeast cell cortex have yielded contrasting impressions depicting septin rings arranged as an ordered array of filaments of unknown orientation (Byers and Goetsch, 1976), a meshwork, or as an apparently random “gauze” of septin subunits (Rodal et al., 2005). The differences in appearance could represent coexisting, differentially organized populations of septins or arise as artifacts because of the processing needed for such techniques. Many questions about septin organization in cells remain: Do all higher-order septin-based structures share a common organization at the molecular level? Are septins assembled into paired polymers in cells, similar to those formed in vitro? Are mobile and immobile populations of septins organized differently in higher-order structures? How does septin organization change when rings reorganize? Discerning the organization of higher-order septin structures is crucial to understanding their assembly and function in normal cells and their malfunction in disease.
In this paper, we implement a quantitative, rapid imaging and analysis method using polarized fluorescence microscopy. We use this to assess the organization of septin proteins within morphologically distinct septin structures in living cells. We have built our study upon the work of Vrabioiu and Mitchison (2006, 2007), who used a polarized fluorescence technique to analyze septins in the budding yeast neck. These experiments led to a model of ordered, nonpolar organization of septin filaments. However, this study was limited to analysis of budding yeast and lacked automation, which restricted spatial and temporal resolution and the ability to measure large numbers of cells (Vrabioiu and Mitchison, 2006, 2007). Polarized fluorescence microscopy takes advantage of the fact that GFP has a dipole moment and is preferentially excited by and emits linearly polarized light that is parallel to its dipole moment (Inoué et al., 2002). By restricting the movement of GFP relative to its fusion protein, the angle of preferential absorption or emission can be correlated to the orientation of the tagged protein.
We fused a constrained GFP (with either 3 or 4 aa deleted from the N terminus) to the C terminus of a septin to limit the rotation of the GFP relative to the septin. This allows the GFP orientation to report the orientation of the septins because of their relatively inflexible connection. We expressed these constructs in the budding yeast, Saccharomyces cerevisiae, in the filamentous fungus, Ashbya gossypii, and in a mammalian tissue culture cell. A. gossypii is related on the genome level to S. cerevisiae, and it expresses a similar number of septin proteins that are 50–80% identical (Dietrich et al., 2004). A. gossypii, in contrast to S. cerevisiae, has a different cell shape (tube vs. sphere), differently appearing septin rings (discrete bars vs. contiguous hourglass), different size structures (4–5 µm in diameter compared with ∼1.0 µm), and septin rings that persist and are dynamic for many hours after assembly (DeMay et al., 2009). Additionally, we analyzed a mammalian septin, SEPT2, in MDCK cells where septins can colocalize with microtubules and form filaments at the cell cortex and about the nucleus (Spiliotis and Nelson, 2006; Hu et al., 2008; Spiliotis et al., 2008). We applied polarized fluorescence analysis, transmission EM (TEM), time-lapse, and photobleaching approaches in these different cell types. With this combination of approaches, we arrive at a molecular model of paired, dynamic septin filaments conserved across diverse species of different cell morphologies and septin structures.
Results
Polarized fluorescence measurement and analysis
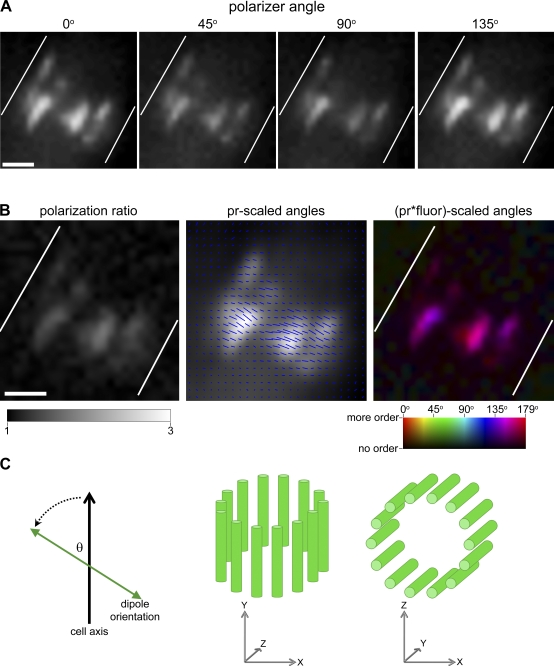
We have developed a polarized fluorescence imaging system that incorporates a liquid crystal (LC) universal polarizer into the excitation light path of a standard wide-field microscope. The LC polarizer generates linearly polarized light oscillating at an angle that is determined by applied voltages. The voltages are controlled from a computer, enabling rapid, sequential, and sensitive switching between different polarization angles. To detect anisotropy in a fluorescence signal, a series of five fluorescence images were recorded, each excited by polarized light of a different polarization angle (0°, 45°, 90°, 135°, 0°; counter-clockwise off the horizon of the image plane). Using this microscope setup, we acquired image series of live A. gossypii cells expressing a constrained GFP (-conGFP) fused to the septin Cdc12p (Fig. 1 A). To distinguish between order and disorder in GFP-labeled protein assemblies, each image series was analyzed for systematic changes in fluorescence intensity. The fluorescence of ordered GFP populations reaches a maximum when the polarization of the excitation light is aligned with a majority of the GFP dipole moments. We describe the image acquisition and processing algorithms, including corrections for photobleaching, cytoplasmic background, and differential transmission of polarized light by microscope components in the Materials and methods text and in detail in a second manuscript in preparation (unpublished data).
Figure 1.
Analysis of constrained septin–GFP fusions using polarized fluorescence microscopy shows that septin rings in A. gossypii are ordered. (A) Raw fluorescence acquired by exciting GFPs in an A. gossypii septin ring with four angles of polarized light in a cell expressing Cdc12-conGFP4. Cell outline is shown in white. Bar, 1 µm. (B) Maps of polarization ratio (pr) and GFP dipole angle (azimuth) were calculated for every camera pixel based on the data in A. The pr across the ring is expressed with the angle as blue lines of a length proportional to the pr, oriented according to the calculated azimuth and overlayed on the sum fluorescence (“pr-scaled” image). Alternatively, the amount and orientation of ordered protein is represented using a color scheme (“spectrum” look-up table in ImageJ) in which each angle is represented as a color whose intensity is the product of the fluorescence and the pr (“pr*fluor-scaled angles” image). Bar, 1 µm. (C) Representation of the “offset” angle (θ), which is the distance counterclockwise from the cell growth axis to the mean azimuth. Two perspectives of an A. gossypii septin ring viewed in the xy or top view (left ring) and the xz or cross section view (right ring).
The image acquisition and processing algorithms generate two maps of the fluorescence anisotropy measured in every pixel (Fig. S1). The first map represents the angle of polarization at which a maximum of fluorescence signal is detected. The second map represents the ratio of maximum to minimum fluorescence intensity, which we call the polarization ratio. We call the map of angles the azimuth image and the second map the ratio image. Both maps can be merged either as a color-coded image or a grid of short lines, in which the line orientation represents the angle and the line length represents the polarization ratio measured at each location (Figs. 1 B, middle; and Fig. S2). In color-coded images, hue represents the angle and intensity represents the measured ratio and quantity of protein (Fig. 1 B, right; and Fig. S2). We calculated angle and ratio values for a septin structure by averaging the angles for the population of pixels and assessing the variability within a ring (ring var) and also in a population of rings (pop var; Fig. S2 and Table I; see Materials and methods for description of analysis). A complicating factor in comparing anisotropy measurements between septin rings was the variable orientation of cells and rings in the image plane. To compare the measured azimuth of fluorescence anisotropy of rings that differ in their orientation within the field, the mean azimuth angle of each ring was normalized to its cell growth/ring axis in degrees of counter-clockwise travel. We call this the “offset angle” of the ring azimuth and use it to identify and evaluate septin rings with similar orientations of the septin–GFP constructs (Fig. 1 C).
Table I.
Order properties of septin structures analyzed by polarized fluorescence
| Strain | Type | Offset | Pop var | Ring var | polRatio | n |
| Ag Cdc12-conGFP4 | ir | 87° | 0.04 | 0.14 | 1.95 ± 0.47 | 13 |
| Ag Cdc12-conGFP4 | split | 180° | 0.17 | 0.34 | 1.43 ± 0.18 | 10 |
| Ag Cdc12-conGFP0 | ir | 86° | 0.07 | 0.3 | 1.52 ± 0.18 | 11 |
| Ag Cdc12-conGFP0 | split | 4° | 0.07 | 0.41 | 1.27 ± 0.14 | 10 |
| Ag Cdc12-conGFP3 | ir | 0° | 0.15 | 0.27 | 1.41 ± 0.16 | 12 |
| Ag Cdc12-conGFP3 | split | 88° | 0.18 | 0.48 | 1.24 ± 0.10 | 10 |
| Ag Cdc3-conGFP | ir | 86° | 0.06 | 0.1 | 1.90 ± 0.71 | 11 |
| Ag Cdc3-conGFP | split | 180° | 0.11 | 0.24 | 1.33 ± 0.18 | 10 |
| Sc Cdc12-conGFP4 | hg | 92° | 0.02 | 0.03 | 1.97 ± 0.51 | 13 |
| Sc Cdc12-conGFP4 | split | 3° | 0.01 | 0.15 | 1.74 ± 0.23 | 10 |
| Sc Cdc12-conGFP0 | hg | 96° | 0.07 | 0.1 | 1.39 ± 0.16 | 11 |
| Sc Cdc12-conGFP0 | split | 1° | 0.02 | 0.17 | 1.49 ± 0.29 | 10 |
| Sc Cdc12-conGFP3 | hg | 175° | 0.06 | 0.07 | 1.53 ± 0.18 | 10 |
| Sc Cdc12-conGFP3 | split | 96° | 0.05 | 0.22 | 1.32 ± 0.10 | 10 |
| Sc Cdc3-conGFP | hg | 93° | 0.03 | 0.06 | 1.51 ± 0.10 | 10 |
| Sc Cdc3-conGFP | split | 2° | 0.02 | 0.11 | 1.82 ± 0.35 | 10 |
| MDCK SEPT2 1-344 | fibers | 89° | 0.05 | 0.12 | 1.21 ± 0.08 | 10 |
| MDCK SEPT2 1-340 | fibers | 90° | 0.02 | 0.06 | 1.27 ± 0.14 | 10 |
| MDCK SEPT2 | fibers | 92° | 0.04 | 0.18 | 1.17 ± 0.06 | 10 |
| Ag Cdc12-conGFP4 | FCF | 91° | 0.01 | 0.01 | 3.91 ± 1.39 | 10 |
| Ag Cdc3-conGFP | fibers | 60° | 0.01 | 0.04 | 1.44 ± 0.32 | 11 |
| Ag Cdc3-conGFP | fibers | 122° | 0.03 | 0.04 | 1.69 ± 0.33 | 14 |
Ir, “inter-region” septin rings formed along the hyphae of A. gossypii, made of discrete bars (Fig. 1); hg, “hourglass” septin rings in S. cerevisiae, appear contiguous by light microscopy; split, hg or ir rings which have undergone septation; FCF/fibers, septins organized into fibrous structures, “FCF” fibers induced by treatment with FCF; offset, angle between polarized fluorescence and ring axis, averaged over the population of rings analyzed; pop var, “population variance” in the average dipole orientation of the population of rings analyzed. For pop var and ring var, scale = 0–1; 0 = no variance; 1 = complete variance/randomness. ring var, “ring variance” denotes the average variability of dipole coherence across a single structure; each individual ring var was found using weighted averaging, “vars” were arithmetically averaged for final “ring var”. For pop var and ring var, scale = 0–1; 0 = no variance; 1 = complete variance/randomness. polRatio, “polarization ratio” denotes the average strength of order for septin structures; 1 = not ordered.
Septin rings contain ordered proteins
We first applied this technique to septins expressed in the filamentous fungus A. gossypii, in which they assemble into rings made of discrete bars at regular intervals at the cortex of tubular hyphae (Fig. 1 C; DeMay et al., 2009). To evaluate septin order, we expressed a panel of constrained septin–GFP fusions in which 3 aa have been deleted from the N terminus of GFP and between 0–4 aa have been deleted from the C terminus of Cdc12. These truncations are predicted to lead to an α-helical structure that extends from the septin into the GFP without a flexible linker region between the septin and GFP. Constructs were expressed from the endogenous promoter on a replicating plasmid. All constrained septin–GFPs incorporated into normal looking septin-based structures, and cells grew well.
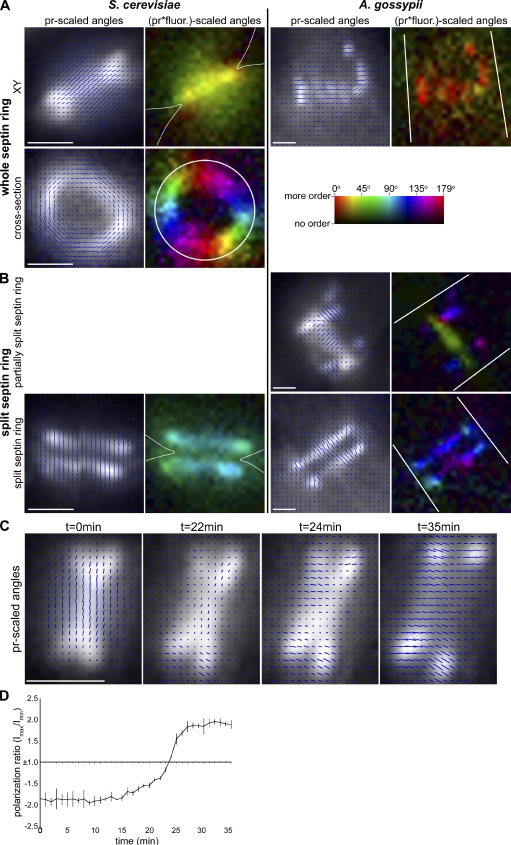
All septin rings in cells expressing Cdc12-conGFP4 contain ordered proteins, based on the consistent offset angles among the many cells imaged. This is apparent as a coherence of angles in pixels that represent the septin ring and is easily seen quantitatively as vectors or colors with the same orientation (Fig. 1, A and B; and Fig. 2), or qualitatively in the flicker of fluorescence intensity as the image series of different polarization angles is viewed (Video 1). All rings observed in all cells expressing this Cdc12 construct (Cdc12-conGFP4) have GFP dipoles with a net orientation ∼90° to the cell growth/septin ring axis (Fig. 2 and Table I). An ∼90° offset angle was observed regardless of whether the focal plane was near the top or bottom of the cell (Fig. 2 A). Notably, anisotropic signal could be seen even within the diffuse septin signal that accumulates at growing tips of A. gossypii, which suggests that the septins may be polymerizing before construction of higher order bar assemblies (Fig. 2 B). Order was not lost with time after assembly such that newly assembled (closer to hyphal tips) and mature rings (closer to center of mycelia) were similarly and comparably ordered (Table I; a mixture of new, intermediate, and old rings was analyzed).
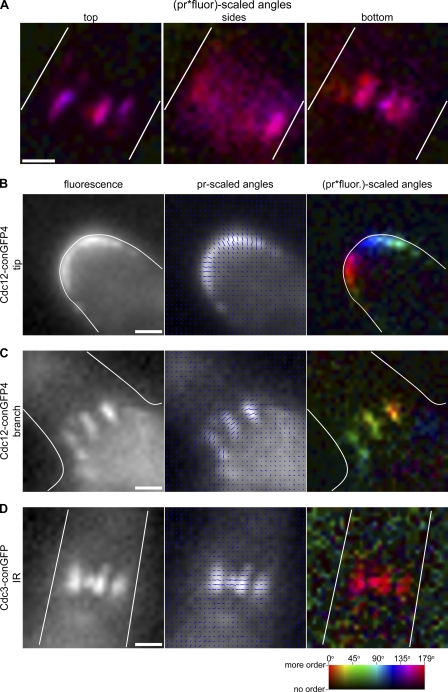
Figure 2.
Septin rings and multiple septin subunits are ordered throughout A. gossypii development. (A) A. gossypii septin rings analyzed at their top, middle, and bottom planes were measured to have GFP dipoles oriented perpendicular (or parallel, depending on the strain imaged) to the cell growth axis. The same septin ring shown in Fig. 1 (A and B), from a cell expressing Cdc12-conGFP4, is pictured here at different focal planes. The amount and azimuth of ordered septin protein is shown using a color scale (“pr*fluor-scaled angles” image). Cells expressing Cdc12-conGFP4 or Cdc3-conGFP from a replicating plasmid were imaged using polarized excitation and analyzed. Fluorescence (left), fluorescence overlayed with blue lines scaled in length by pr and oriented at the calculated GFP dipole angle (azimuth) for each camera pixel (middle), and the amount and azimuth of ordered septin protein using a color scale (right) are displayed. (B) Septins before ring assembly are ordered at growing hyphal tips. The typical azimuth orientation is perpendicular to the cell cortex. (C) Septin rings formed at branch points are highly ordered. The typical azimuth orientation is perpendicular to the cell growth axis. (D) An inter-region (IR) septin ring, assembled in the wake of a growing tip in a cell expressing Cdc3-conGFP. Septin rings in cells expressing this Cdc3-conGFP construct exhibit similar order, with the typical azimuth orientation perpendicular to the cell growth axis. Cell outlines are shown in white. Bars, 1 µm.
To ensure that the observed order was not an artifact of the particular Cdc12-conGFP4 construct, additional Cdc12 constructs were created. These are predicted to change the orientation of the GFP dipole relative to the septin axis because each amino acid of an α-helix represents an ∼100° rotation with the twist of the helix. Remarkably, the septin rings of Cdc12-conGFP0 also exhibit an offset angle of ∼90° despite the changed linker length (Fig. S3). Additionally, we produced a third construct of Cdc12 (Cdc12-conGFP3) in which we observed a GFP dipole angle that was 0° offset from the cell growth axis (Fig. S3). It is possible that the order we observe is unique to Cdc12p or, alternatively, is a property of all the septins. Therefore, we tagged a different septin (Cdc3p) with constrained GFP. Septin rings incorporating Cdc3-conGFP also had an offset angle of ∼90° (Figs. 2 D and S4). The measured anisotropies depended on the GFP linker being shortened because cells expressing full-length Cdc12-GFP or Cdc3-GFP did not display anisotropy (Fig. S3 and unpublished data). Thus, using four different constructs with two different septin proteins, we see highly stereotypical order in all septin rings within and between cells (Table I).
Septin filaments align parallel to growth axis
The order we detected could arise in septin bars or fibers from filaments aligned in parallel or perpendicular to the septin structure. It is important to note that the orientation of incorporated GFP dipoles does not necessarily reflect the orientation of a septin filament because the septin C terminus linked to the GFP could be positioned in any orientation relative to the septin. To determine how septin filaments may be oriented in the bars in A. gossypii, A. gossypii cells were treated with the small molecule forchlorfenuron (FCF), which leads to the formation of numerous septin fibers throughout the cell (Fig. 3 A; DeMay et al., 2010). These fibers possess each of the five septin proteins and are considered to be elongated, stabilized septin polymers or bundles of polymers (Hu et al., 2008; DeMay et al., 2010). We hypothesized that we could predict the orientation of ordered filaments in normal septin rings if we measured the anisotropy of septin–GFP constructs in these elongated fibers. If the same offset angle is observed in both septin rings and in these septin fibers, this would support the finding that septin filaments are aligned parallel to the growth axis within the short bars of a normal ring. After treating with FCF, septin fibers in cells expressing the Cdc12-conGFP4 construct were found to have GFP dipole offset angles of ∼90° (Fig. 3 A and Table I). This measurement matches what is observed in septin rings, indicating that cortical bars in A. gossypii rings are likely composed of septin filaments aligned parallel to the cell axis.
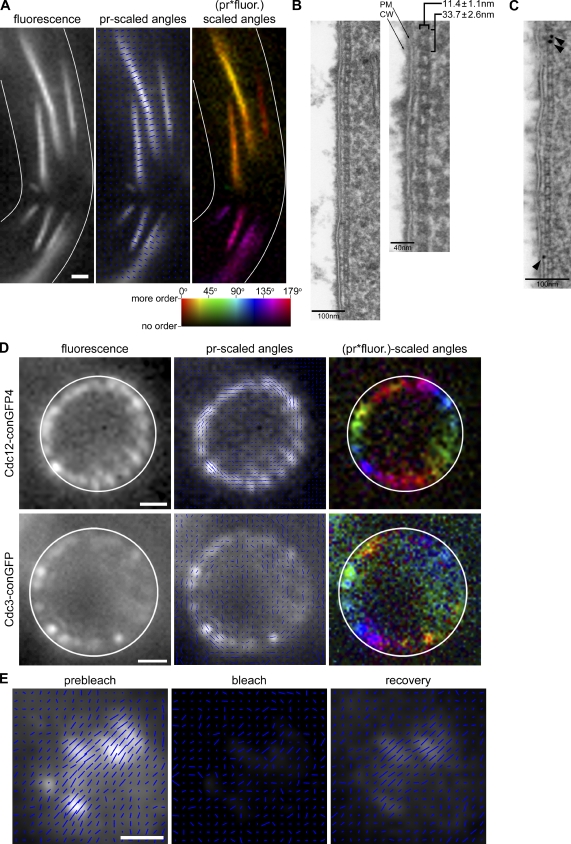
Figure 3.
Ordered septin filaments are paired, align parallel to the cell growth axis, and are dynamic. (A) A. gossypii cells expressing Cdc12-conGFP4 were treated with the septin filament stabilizing drug FCF. The mean GFP dipole orientation in septin fibers was found to be perpendicular to the fiber axis. Cell outlines are shown in white. Bar, 1 µm. (B) A. gossypii cells (AG127) were treated for 3 h with FCF and processed for TEM. A typical filament and a magnified section are displayed. Brackets point to the measured width and periodicity of a population of septin filaments gathered from many different sections accompanied by standard deviations (statistics are given in Table S1). PM, plasma membrane; CW, cell wall. (C) An immunolabeled section of SHS1-6HA–expressing cells (AG296) in which septins have been localized using an anti-HA primary and a 10-nm gold conjugated secondary antibody. Arrowheads point to gold particles. Bar, 100 nm. (D) A. gossypii cells expressing Cdc12-conGFP4 or Cdc3-conGFP exhibit GFP dipoles oriented parallel to the cell cortex when imaged in cross section (xz plane). Cell outlines are shown in white. Bars, 1 µm. (E) A septin ring in A. gossypii expressing Cdc12-conGFP4 was analyzed, photobleached and analyzed, and allowed to recover before repeat of analysis. The recovered signal is ordered and reports the same GFP dipole orientation (perpendicular to the cell growth axis) as the starting ring. Bar, 1 µm.
Paired septin filaments observed in cells
It is striking that different conGFP constructs and varied septin structures all exhibit azimuth angles either in parallel or perpendicular to the cell growth or fiber axes. Given that this technique is measuring the fluorescence of a population of molecules projected on a single camera pixel, we hypothesize that the common offset orientations of 90° or 0° we observe might actually be caused by combining light emitted from distinct populations of septins. In this scenario, the individual GFP dipole angles vary from construct to construct but the recurring azimuthal measurements of 90° or 0° arise from superimposing the light from several populations of dipoles related by rotational symmetries. To find further evidence for symmetries in the ultrastructure of filaments in cells, we visualized septins in A. gossypii by TEM. As the bars in normal septin rings are only 1–2 µm in length, it is very difficult to get the appropriate thin section to see septins in native rings because hyphae are oriented at all angles in the sections. However we were able to detect filaments at the cortex in cells treated with FCF (Figs. 3 B and S5 C). These structures are symmetric across their width, which is 11.4 ± 1.1 nm, and possess regular electron-dense components down their length with a periodicity of 33.7 ± 2.6 nm (n = 176). These dimensions and periodicity match measurements of paired septin filaments formed in vitro (Table S1; Byers and Goetsch, 1976; Frazier et al., 1998; Bertin et al., 2008, 2010). Finally, immuno-gold labeling, directed against an HA-tagged Shs1, demonstrates that these filaments contain septin proteins (Figs. 3 C and S5 D). Interestingly, in a subset of images, filaments appear “stacked” or “bundled” together laterally (Fig. S5 C). These additional, associated filaments possess the same width and periodicity as the filament directly associated with the membrane. Thus, paired septin filaments can simultaneously form lateral associations with the plasma membrane and other septin filaments in cells.
Septins are rotationally constrained at the cortex and are unlikely to be twisted
The GFP dipole measurements and TEM data support the existence of paired septin filaments but do not distinguish between twisted or straight polymers, although our TEM images do not possess a twisted appearance. Rodal et al. (2005), using rapid freeze-deep etch methods, visualized gauzes of septins in unroofed cortices of yeast, and in some cases the filaments in these appeared to be slightly twisted. Additionally, a recent paper from the Nogales and Thorner groups has shown septin filaments in vitro on membranes in what may be a twisted conformation when low concentrations of protein are present (Bertin et al., 2010).
We used measurements of fluorescence anisotropy in cross section (the xz plane) to distinguish between twisted and straight polymers in cells. If polymers twist, then, when septin-conGFP signals are viewed in cross section (on end), the signal will appear isotropic. This is because as the filament twists, the GFP dipoles would be at many different positions depending on the pitch of the twisting polymer and, when viewed in aggregate, would appear as disordered signal (Fig. 6 D). However, if the polymers are straight then we should detect that the septin rings are ordered in cross section relative to the membrane. Septins in rings viewed in cross section are anisotropic and report GFP dipole angles parallel or perpendicular to the local cell cortex (Figs. 3 D and S4 and Video 1). Anisotropy in cross section holds for all of the constrained GFP strains of A. gossypii. We conclude that septin filaments do not twist in cells and are straight relative to the cell cortex.
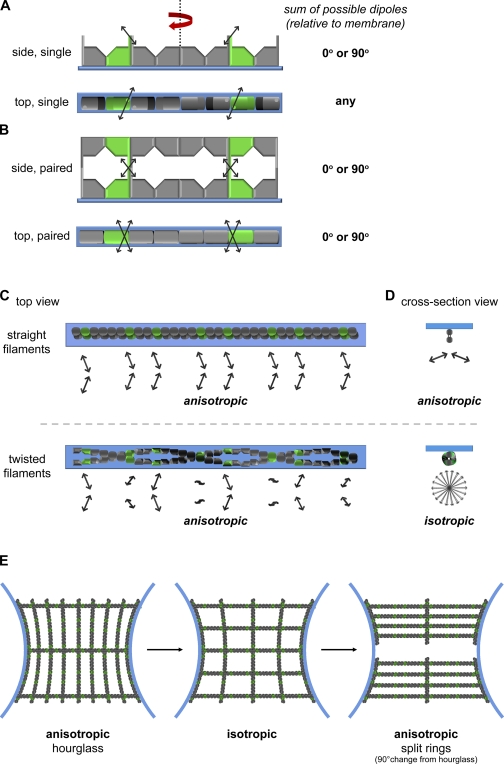
Figure 6.
Septin filament organization models and corresponding changes in dipole orientation. (A) Representations of a single septin protofilament shown associated with the cortex (blue) with the conGFP-tagged septin (green) shown with a possible GFP dipole orientation (black). C-terminal coiled-coil domains are shown as rectangles extending from the three septins that are predicted to have these domains. The left and right half of a filament are related by a 180° rotation around a vertical axis shown in the side view as a dotted line and red arrow. The summed angle for all possible dipole orientations is noted on the right. (B) Representations of paired septin filaments. Top and bottom filaments are related by a 180° rotation around a horizontal axis. The summed angle for all possible dipole orientations is noted on the right. (C) Paired filament models show polymerization of filament if it processes straight or twisted relative to the cortex. Corresponding dipole angles of tagged subunits are represented by the arrows. (D) The same filament procession and relative dipole angles that could be captured in the axial resolution of a 1.4 NA objective (∼500 nm) when viewed in cross section. The dipole orientations for each organizational scenario is shown below the filament and noted for anisotropy. (E) Model for filament organization in the S. cerevisiae hourglass (left), transition (middle), and split ring (right) states that is consistent with anisotropy measurements through time.
Ordered septins are dynamic
This polarized microscopy technique enables the detection and analysis of order but cannot rule out a coexisting disordered population. Septins have been shown to exchange between the higher-order structures and the free, cytosolic pools in different cell types and structures (Caviston et al., 2003; Dobbelaere et al., 2003; Hu et al., 2008; DeMay et al., 2010). In A. gossypii, there are distinct mobile and immobile populations of septins such that only a portion of a ring structure is turning over (DeMay et al., 2010). We hypothesized that the mobile fraction is arranged in a disordered, random fashion, whereas the ordered population is static. To test this hypothesis, A. gossypii septin rings incorporating constrained septin–GFPs were photobleached, and the anisotropy of the fluorescence signal was measured after recovery. Surprisingly, the fluorescence that returned to the septin rings was highly ordered and possessed the same offset azimuth as before the photobleach (Fig. 3 E). This indicates that even dynamic populations of septins are ordered.
Septins are similarly organized in A. gossypii and S. cerevisiae
There is still controversy as to the organization of septin filaments within the bud neck in yeast when in the hourglass state. The center of the debate is fixed upon the orientation of the filaments relative to the mother–bud axis, with data supporting filaments running parallel to the axis (Vrabioiu and Mitchison, 2006) and some speculation they run perpendicular to the axis (Byers and Goetsch, 1976; McMurray and Thorner, 2009). The unique bar ring composition of A. gossypii along with FCF and TEM measurements suggest that paired filaments run along the growth axis. We predicted that we could make use of comparisons between measurements in A. gossypii and S. cerevisiae to add insight into the arrangement in yeast. Therefore, we introduced the same panel of septin–GFP fusions analyzed in A. gossypii into yeast cells. Strikingly, we see the same 90° or 0°, construct-specific dipole orientation relative to the growth axis in the yeast septin hourglass as was seen in A. gossypii (Figs. 4 A, S3, and S4; Table I; and Video 2). Similarly, S. cerevisiae rings in cross section were also highly anisotropic, which indicates that in this system, the polymers are also not likely to twist (Figs. 4 A and S4 and Video 2). These combined data from A. gossypii and S. cerevisiae suggest that the higher order structures may be similarly organized in these divergent species with morphologically distinct septin rings.
Figure 4.
Septin rings in S. cerevisiae and A. gossypii are similarly ordered and accomplish an ∼90° change in orientation without concerted rotation. (A) Septin rings assembled in S. cerevisiae and A. gossypii expressing Cdc12-conGFP4 exhibit septins organized such that, on average, GFP dipoles are oriented perpendicular to the cell growth axis. As in A. gossypii (Fig. 3C), the S. cerevisiae septin hourglass, when viewed in cross section, also has the average GFP dipole position oriented parallel to the cell cortex, and perpendicular to the growth axis. Cell outlines are shown in white. Bars, 1 µm. (B) Split septin rings show an ∼90° change in the orientation of the GFP dipoles. Some septin rings in A. gossypii expressing Cdc12-conGFP4 were captured in a partially split state, exhibiting the original (perpendicular) and reorganized (parallel) orientation of dipoles. (C) Septin rings in S. cerevisiae expressing Cdc12-conGFP4 were imaged using every minute, through the transition from the hourglass to a split ring. (D) Polarization ratio of the central region (12 pixels) of S. cerevisiae septin rings as they progressed through the septin reorganization were analyzed and plotted. Average orientations perpendicular to the ring axis are plotted as negative values and average orientations parallel to the mother–bud axis are plotted as positive values to represent the difference in orientation seen through time. n = 3; error bars are standard deviation.
Sharp switch between angles at septin transitions
Septin dynamics, as measured by the appearance of an apparently fluid state based on FRAP, are limited in S. cerevisiae to ring assembly at start and hourglass splitting before cytokinesis (Caviston et al., 2003; Dobbelaere et al., 2003). An intriguing and still not well understood observation from previous work using polarized fluorescence was that at cytokinesis, the angle of maximum anisotropy of a yeast septin ring is observed to undergo an ∼90° change (Vrabioiu and Mitchison, 2006). To assess if this change in orientation is an evolutionarily conserved property of septins, we evaluated if a similar transition was seen in A. gossypii cells, which septate infrequently and as a senescence and stress response rather than in coordination with the cell cycle. In fact, split rings in A. gossypii did exhibit a 90° change in the orientation of GFP dipoles as compared with unsplit rings, precisely as was observed in S. cerevisiae (Figs. 4 B, S3, and S4; and Table I). Notably, as A. gossypii bars are in the midst of these transitions, the orientation of GFP dipoles in the center of the ring is changed by 90° and now parallel with the ring axis, whereas the dipole orientation at the bars more distal to the septal plane are still in the initial state, i.e., oriented perpendicular to the ring axis (Fig. 4 B). Thus, two different dipole orientations can be detected simultaneously in the same transitioning ring. Importantly, however, we do not see intermediate angles between 0° and 90° in anisotropic regions in splitting rings. This suggests that these reorganizations do not involve a gradual rotation but instead are two states of orientation. The switch-like transition between these states may be caused by a conformational change in the proteins or a disassembly and reassembly of septins in different arrangements relative to the growth axis.
To examine this transition through time, we tracked several S. cerevisiae septin hourglass structures with polarized fluorescence time lapse. A. gossypii rings are not suitable for this kind of analysis because their splitting is unpredictable because of a lack of clear cell cycle control. Strikingly, we see a similar switch-like transition between the orientations in budding yeast (Fig. 4 C and Video 3). As the change from an hourglass to a split ring occurs, there is a loss of anisotropy followed by a reestablishment of anisotropy, with the measured GFP dipole orientation having undergone a 90° change (Fig. 4 D). Because intermediate angles were not observed, it seems unlikely that septin filaments rotate as a unit to achieve this reorientation, as was previously proposed (Weirich et al., 2008).
Paired filament organization conserved through mammals
It is possible that the similar order and organization detected by polarized fluorescence in yeast and hyphae is a consequence of septins associating with membranes that are bound by a rigid cell wall. To evaluate the orientation of septins in the absence of a cell wall and determine if the measurements we observe reflect septin organization conserved through evolution, we generated a series of truncated SEPT2 constructs and expressed these in MDCK tissue culture cells. The septin fibers incorporating the SEPT2-GFP constructs are heteromeric, containing SEPT6 and SEPT7 (Fig. S5 B). Remarkably, three different SEPT2 constructs reported a GFP dipole angle of 90° relative to the long axis of septin fibers (Figs. 5 and S5, B and E; Video 4; and Table I). These dipole orientations were consistent in all fibers measured regardless of the abundance of fibers in the cell, such that even cells with very low expression of SEPT2 also show the same dipole orientation perpendicular to the fiber axis (Fig. S5, B and E). The remarkable consistency between measurements in all three systems separated by vast evolutionary time supports the finding that the basic organization of septins within the higher order structures of cells is as paired, straight filaments.
Figure 5.
Mammalian epithelial cells exhibit conserved septin order. (A) MDCK cells expressing SEPT2–conGFP-1-344, which incorporates into septin fibers. The GFP dipoles are measured to be oriented perpendicular to the fiber axis. The white box identifies the magnified region shown in B. Bar, 10 µm. (B) Magnified region of A including panels with scaled azimuth lines and color-intensity schemes. Bar, 1 µm.
Discussion
Septin rings are similarly ordered across species
In this study, we have implemented a polarized fluorescence microscopy system and developed analysis methods to investigate the organization of septins in higher-order structures in fungal and animal cells. Surprisingly, we consistently observe fluorescence anisotropy oriented parallel or perpendicular to the higher order structures formed in budding yeast, A. gossypii, and MDCK cells, regardless of how many amino acids are truncated to constrain the linkage between GFP and the septin C-terminal tails. The septin constructs in our study were not intentionally designed to orient the GFP dipole perpendicular or parallel to the ring structure, and the probability that these constructs would yield these same angles by chance is extremely small. In agreement with our data, consistent parallel or perpendicular anisotropy was observed in yeast in a previous study with different constructs that was undertaken before knowledge of the symmetries suggested by the atomic structure (Vrabioiu and Mitchison, 2006, 2007; Sirajuddin et al., 2007). Given the predicted C-terminal α-helical structure linking a septin to GFP, each amino acid addition/subtraction to the helix in a construct could change the angle of the dipole in the image plane by 100°. Yet, the measurements are consistent even between constructs that differ by a single amino acid and thus should change the orientation of the GFP substantially. The binary nature of these results across all cell types indicates that septins are likely organizing similarly on the molecular level in cells that differ in shape (spheres vs. tubes vs. flat epithelial cells), septin appearance (hourglasses vs. bars vs. fibers), and that have experienced substantial time since sharing a common ancestor (100 million to more than a billion years).
Septins likely form paired filaments within higher order structures
The consistent observation of fluorescence anisotropy angles parallel or perpendicular to the septin structure axis gives clues as to how septins are organized within larger structures. One mechanism by which we may observe these common angles is through the presence of symmetries in a septin protofilament and its larger assemblies. Indeed, models of protofilament organization based on x-ray crystal structures of recombinant proteins suggest that protofilaments (which in yeast and A. gossypii are likely eight proteins with two copies of each septin) are nonpolar and contain a symmetry axis relating the two identical halves by a 180° rotation (Fig. 6 A; Sirajuddin et al., 2007; Bertin et al., 2008). Because of this symmetry, two GFP-tagged septin molecules of the same kind and located on opposite sides of the protofilament axis will be oriented so as to be either mirror images or 180° rotations of each other depending on the viewpoint (Fig. 6 A). When such a protofilament is viewed from the side, the two GFP dipoles are oriented at supplementary angles. These supplementary angles are detected in sum because of the resolution of the light microscope. When combined, supplementary angles give a measurement of 90° or 0° relative to the filament axis (Fig. 6 A). However, these supplementary angles are only created when viewed from this perspective. When viewed from the top (Fig. 6 A), i.e., parallel to the axis of rotation that relates the two halves, the symmetry of a protofilament instead serves to orient the two GFP dipoles at the same angle in the image plane. In that view, the observed GFP dipole angle should be unique to a given construct and would likely vary between them; however, our data do not support this model (Figs. 2 A and 6 A).
Instead, our observations of the 0° or 90° offset angles regardless of viewpoint reveal an additional symmetry axis. We propose, and the TEM data support, that this additional symmetry arises through a higher-order association in which two filaments pair (Figs. 3 B and 6 B). Paired filaments possess a second rotational symmetry axis that is perpendicular to the first one, and therefore would report consistent parallel or perpendicular offset angles regardless of viewpoint, which is what we observe. Cortical filaments captured by TEM have the dimensions and periodicity of paired septin filaments formed in vitro (Fig. 3 B and C; Fig. S5 C and D; and Table S1). In some cases, we were able to capture septin filaments that associated with each other laterally, which indicates an ability of septins to associate both with the membrane and other filaments in bundles.
Further support for the existence of two populations of proteins averaging to yield the consistent parallel and perpendicular measurements comes from fibers formed by Cdc3-conGFP in both A. gossypii and budding yeast. This fusion protein incorporates into normal structures but also gave rise to additional septin filaments along the cortex of hyphae, similar in appearance to fibers induced by FCF. Unlike normal rings or FCF-induced fibers, Cdc3-conGFP ectopic fibers were found to have GFP dipoles oriented at two distinct offset angles, ∼60° and ∼120° rather than the usual 90° (Fig. S5 A). It seems probable that these Cdc3-conGFP fibers are actually unpaired, single septin filaments. This is because the two populations have offset angles of either 60° or 120°, which are supplementary angles, and when combined would result in the 90° offset angle observed at septin rings.
What distinguishes constructs that report a 90° as opposed to 0° measurement? We expect that the differentiation between a parallel or perpendicular measurement is based on the actual orientation of the GFP dipole relative to the septin subunit. As the tagged septin molecule becomes part of the protofilament, the GFP dipole will subtend a specific angle with respect to the filament axis. If the dipole is oriented nearly parallel to the filament axis, the offset angle of the polarized fluorescence from the population of dipoles will be 0°, whereas dipoles oriented nearly perpendicular to the filament axis will result in an offset angle of 90°. Dipoles oriented around 45° to the filament axis result in fluorescence that will show little or no anisotropy. In a second paper in preparation (unpublished data), we explore the range of angles predicted for the dipole orientation based on the anisotropy maps measured in the yeast bud neck.
In yeast, it remains unclear how septin filaments align relative to the growth axis. Models for filaments oriented perpendicular to the mother–bud axis have been proposed previously, based on the Byers and Goetsch (1976) electron micrographs, in which electron-dense elements appear to encircle the neck. These densities are ∼32 nm apart in the direction parallel to the mother–bud axis. It is intriguing that septin filaments in S. cerevisiae are likely made up of protofilaments that are 32 nm in length, and that filaments formed in vitro and visualized by negative stain can show a 32-nm periodicity along their length. These data support the finding that the original neck filaments observed decades ago may represent an array of filaments running parallel to the mother–bud axis that are in lateral register, thereby generating the bands of electron densities every 32 nm along the mother–bud axis. Based on the TEM presented here, combined with polarized fluorescence, we envision that the order we detect in cortical bars of A. gossypii septin rings, and in the hourglass of S. cerevisiae, most likely arises from one or more paired septin filaments aligned parallel to the cell axis (Fig. 6 E).
The majority of paired filaments are likely straight relative to the membrane
Septin polymers could conceivably associate with the plasma membrane as straight or twisted filaments, and both organizations would possess rotational symmetry when viewed in the xy plane (Figs. 1 C and 6 C). However, we favor a straight filament model because when rings are viewed in cross section, they are anisotropic (Figs. 3 D, 4 A, and S4). If the filaments (extending into or out of the page in the xz view) were twisted, the GFP dipole would be positioned at many different angles as the filament processed. This effect, at the resolution of these images, would result in the appearance of isotropy because of the observation of a population of molecules in aggregate (Fig. 6 D). The observation of anisotropy in the septin ring xz plane suggests that there is a majority population of straight, untwisted filaments that associate in a stereotypic manner with the cell cortex along a single face of the polymer. The only scenario in which a twisted filament would match the data are if there was a periodicity of the twist that was repeated every protofilament, which is predicted to be composed of eight septin proteins. This would mean an ∼45° rotation to each septin–septin interaction surface, which is possible but improbable. We predict that these straight, paired filaments must not be able to rotate about their long axis because this situation would fail to yield a consistent angle relative to the membrane curvature when viewed in cross section.
Another uncertainty about septin organization at the cell cortex is what face of the polymer associates with membranes. Two studies using recombinant septins and synthetic membranes have lead to a model in which both individual filaments that comprise a paired septin “railroad” track bind the membrane (Tanaka-Takiguchi et al., 2009; Bertin et al., 2010). In this model, the C-terminal coiled coils have been depicted as lying parallel to the membrane. However, in vivo it is not clear what faces of the paired filaments actually contact the plasma membrane. Given the multiple symmetry axes present in the filaments and the resolution of our imaging, we cannot distinguish between paired filaments in which both sides of the pair contact the membrane or a single side of the pair contacting the membrane. However, in the case of the Cdc3 ectopic fibers, which report offset angles of 60° and 120°, it is likely the C termini extend perpendicular rather than parallel to the membrane, in contrast to what has been proposed in vitro for paired filaments. Only in this arrangement would the Cdc3 fibers display this non-90° offset angle when viewed at the top and bottom of the cell. The questions of how paired septin filaments contact the membrane, what lateral associations may exist between paired filaments, and whether/how pairs can go on to form bundles in cells are fuel for future study.
Dynamic septins maintain order properties at the septin ring
We find that even the mobile population of septin proteins, which can exchange at the ring with free cytosolic septins, are highly ordered in A. gossypii. Proteins that exchange retain the same orientational properties as septins that are resident in the structure. Furthermore, based on the lack of polarity in the signal during recovery relative to the septin bars, we predict that the exchange is occurring within the polymers rather than solely at their ends. It is not yet clear what the unit of exchange is because order persists despite the individual molecules changing within the higher-order structure. One possibility is that there are layers of filaments extending from the membrane, and that those most proximal to the membrane are resident and act as templates for filaments polymerizing and depolymerizing at the cytosolic face. Another possibility is that individual monomers, dimers, protofilaments, or even paired protofilaments are coming and going within polymers analogous to individual bricks being lost and replaced within an intact wall. The overall integrity of the structure is maintained even when pieces of it are unstable. The function of such septin dynamics is not yet clear but potentially may relate to a septin ring in A. gossypii always being poised to form a septum, a transition that may require enhanced septin dynamics.
No concerted rotation of septins observed during hourglass–to–split rings transition
Our observations of the septin hourglass transition in S. cerevisiae have enabled analysis at a much higher temporal and spatial resolution than previous studies because we have been able to follow each pixel in a ring and calculate an offset angle based on fast rotation of the light polarization, instead of a slow rotation of the specimen itself (Vrabioiu and Mitchison, 2006, 2007). In light of these new data, it is highly unlikely that a rotation of intact septin filaments at the mother bud neck is the mechanism underlying the splitting transition in yeast as has been proposed previously (Weirich et al., 2008). The time-lapse data presented here accommodates the results of previous polarized fluorescence time-lapse work, with the critical difference being that the decrease and subsequent increase in anisotropy shown in both studies is not caused by a concerted rotation of anisotropic elements (filaments) but by an actual loss of observable anisotropy within the structure. The filament rotation was one possible interpretation of the data acquired by Vrabioiu and Mitchison (2006) because that experiment was performed using polarizers/analyzers parallel and perpendicular to the cell growth axis, and the loss of anisotropy they observe could be attributed to a rotation. The fixed positions of the polarizer/analyzers restricted the ability to detect whether or not the peak anisotropy was rotating to another angle or was actually decreasing to an isotropic state.
We favor a model in which the transition between different orientational states involves disassembly of filaments running parallel to the growth axis and reassembly of septin filaments perpendicular to the growth axis (Fig. 6 E). In this case, no intermediate dipole angle would be detected and there would likely be a period of isotropy at the peak of exchange (Fig. 6 E, middle). This is also consistent with the increase in septin dynamics reported at cytokinesis based on recovery from photobleaching (Caviston et al., 2003; Dobbelaere et al., 2003). Intriguingly, the disassembly and reassembly of the septin ring during septation is clearly separated in time in the dimorphic fungus Ustilago maydis (Böhmer et al., 2009). It may be that this same activity is taking place in S. cerevisiae, albeit simultaneously so that the gross fluorescence in the bud neck region does not noticeably change. Although this assembly mechanism appears to be the most probable way to achieve the observed transition results, similar results could be achieved by reorienting the septin C termini upon septation. However, this scenario is not favored because this conformational change need not always result in a 90° reorientation of the offset angles, which is what we observed. Alternatively, an isotropic intermediate might be seen if there were two populations of crossing polymers in the plane of the membrane; however, there is no evidence for that scenario.
What directs the septins to align perpendicular to the growth axis after the transition? Work recently published by the Nogales and Thorner groups shows recombinant septins on membranes polymerizing into a crossed pattern, whereby filaments are present perpendicular to and running between longer septin filaments (Bertin et al., 2010). The ability of septin filaments to be bridged by orthogonally oriented filaments may provide the base mechanism for the hourglass to split ring transition and the accompanying anisotropy change. It may be that at the transition, septin filaments that run parallel to the growth axis through most of the cell cycle are dismantled and reassembled perpendicular to the growth axis using a subpopulation of filaments, analogous to the crossing filaments seen in vitro, as a template or guide to orient (Fig. 6 E). These cross filaments oriented perpendicular to the mother bud axis may be present through the entire cell cycle; however, they would need to be fewer in number than the filaments running parallel to the growth axis or there would be no anisotropy detected in any stage of the cell cycle. The reorganized filaments after ring splitting, now likely running perpendicular to the cell growth axis (Fig. 6 E, right), can then function to promote cytokinesis (Dobbelaere and Barral, 2004).
Use of polarized fluorescence microscopy has enabled us to demonstrate the conserved molecular organization of septin proteins within higher order structures from fungi to mammals. Importantly, these data demonstrate that paired septin “railroad” tracks seen in vitro are the state of the polymers in vivo as well. Future work will determine mechanisms underlying rearrangements and dynamics so as to understand how cells remodel the cell cortex through changing septin organization.
Materials and methods
Plasmid and strain construction
A. gossypii culturing and transformation protocols were performed as detailed by Wendland et al. (2000) and Ayad-Durieux et al. (2000). Polymerase chain-reactions and DNA manipulation protocols were performed in accordance with Sambrook (2001). All restriction enzymes are from New England BioLabs, Inc., and all plasmids were sequenced at the Dartmouth MB CORE.
To create the CDC3-conGFP construct, GFP was amplified from plasmid AGB005 with oligos AG504 and AG505, which was truncated by four codons at the N terminus and contained homology to the C-terminal CDC3 such that 17 codons of CDC3 would be truncated upon fusion. A yeast cotransformation with amplification product and plasmid AGB127 yielded plasmid AGB204, which was verified by digestion with BamHI and EcoRI. This plasmid was transformed into wild-type A. gossypii or DHD5 via electroporation to yield strains 360 and Y010, respectively.
To create the CDC12-conGFP constructs, GFP was amplified from plasmid AGB005 using oligo pairs AGO525/197, AGO608/197, and AGO609/197, creating products containing GFP, truncated by four codons at the N terminus, which have homology to CDC12 such that fusion results in the truncation of zero, three, or four CDC12 C-terminal codons, respectively. Each product was yeast cotransformed with the plasmid AGB123, yielding AGB206 (CDC12-conGFP0), AGB228 (CDC12-conGFP3), and AGB229 (CDC12-conGFP4). These plasmids were verified by digestion with NdeI and XmaI. Plasmids AGB206, AGB228, and AGB229 were transformed into wt A. gossypii or DHD5 via electroporation to yield strains 363, 408, and 409; and Y012, Y015, and Y016, respectively.
To create strain AG296 (SHS1-6HA-Gen3), the 6HA-Gen tag was amplified off of plasmid AGB35 (pAGT125 6HA-Gen3) using primers with homology to the SHS1 locus (AGO328 [SHS1-6HA F] and AGO313 [6HA G3 R]) and transformed into ΔlΔt. Integration was verified by PCR using primers AGO365 (5′ SHS1 tag) and AGO37 (VG3), AGO36 (VG5) and AGO403 (Tef2T R), and AGO324 (SHS1 6HA F seq) and AGO325 (SHS1 6HA R seq).
Rigid SEPT2-GFP fusions were made by fusing amino acids 5–240 of EGFP (GFP-4) to amino acids 1–344 (SEPT2-17) and 1–340 (SEPT2-21) of the mouse SEPT2 (GI:228480250). The recombinant sequences were made by initially subcloning SEPT2 cDNA into the XhoI and BamHI sites of pEGFP-N1 using primers S2-F and S2-R. The pEGFP-SEPT2 plasmid served as a template for the construction of SEPT2-21-EGFP-4 using primers S2-21-F and S2-21-R, and of SEPT2-17-EGFP-4 using primers S2-17-F and S2-17-R. Note that the primers in each set are complementary to each other, 5′ halves of primers “-F” are complementary to the sequence encoding the C terminus of truncated SEPT2, and 3′ halves are complementary to the sequence encoding the N terminus of truncated EGFP; 5′ halves of primers “-R” are complementary to the sequence encoding the N terminus of truncated EGFP, and 3′ halves are complementary to the sequence encoding the C terminus of truncated SEPT2. To generate those recombinant plasmids, PCR was performed using these primers, and pEGFP-SEPT2 was used as the template. PCR-amplified fragments self-anneal into the final plasmids with several amino acids at the C terminus of SEPT2 and the N terminus of EGFP deleted. PCR products were treated with DpnI at 37°C for 1 h to digest the template plasmids and were transformed into XL-1 Blue competent cells (Agilent Technologies) for amplification. The pSEPT2-21-EGFP-4 and pSEPT2-17-EGFP-4 plasmids were sequenced with primer S2-C-seq to confirm C-terminal fusions of SEPT2 with EGFP.
Cell culture and preparation
For polarized fluorescence imaging, A. gossypii cells were grown in 15 ml Ashbya full media (AFM) with glass beads in a 50-ml conical tube (Sarstedt), shaking at 30°C for ∼16 h. A. gossypii culture was then transferred to another tube (free of beads) where cells were collected via gravity, resuspended in 25% AFM (to reduce the autofluorescence of the medium), placed on a slide, covered with a coverslip, sealed with VALAP, and imaged. S. cerevisiae cells were grown in 15 ml YPD in a 50-ml conical tube, shaking at 30° for ∼14 h. Cells were collected via gravity, resuspended in 25% YPD, transferred to a slide, covered with a coverslip, sealed with VALAP, and imaged. MDCK cells were grown on collagen-coated coverslips and transfected for transient plasmid expression using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After transfection, coverslips were placed face down on a standard slide, sealed with VALAP, and imaged. Imaging was performed at room temperature.
Polarized fluorescence microscopy setup and imaging parameters
The fluorescence of cells was excited with polarized light using a Microphot SA upright microscope stand (Nikon) equipped with oil immersion optics (60× Plan-Apochromat objective lens and Apochromat condenser, both 1.4 numerical aperture; Nikon) and a xenon arc lamp (Nikon). We used a trans-illumination instead of epi-illumination path to avoid the use of a dichroic mirror, which introduces polarization distortions and reduces efficiency. To minimize background light, we used highly discriminating and efficient interference filters for the illumination (482/18 nm) and imaging path (525/45 nm; both BrightLine bandpass filters were from Semrock). In the illumination path, after the excitation filter, we placed a LC universal compensator (LC-PolScope; Cambridge Research and Instrumentation Inc.), which was operated as a variable linear polarizer, called an LC polarizer. The LC polarizer and digital camera (Retiga EXi CCD camera; QImaging) were controlled and synchronized for image acquisition using custom software (CamAcqJ) developed in-house for the open source imaging platform ImageJ (National Institutes of Health).
Photobleaching of septin rings was performed by closing the field diaphragm of the microscope to restrict the excitation light to the ring region. Photobleaching of septin rings to <20% initial intensity typically took between 3 and 8 min. After this step, cells were allowed to recover for 15 min and were then imaged via polarized excitation.
Calculation of fluorescence anisotropy
Our acquisition and analysis routine is based on the observation that fluorescence anisotropy is manifested in a sinusoidal variation of the fluorescence intensity as a function of the angle of the transmission axis of the linear polarizer (Fig. S1). Because in our experiments we recorded the fluorescence emitted by the specimen without analyzing its polarization, the recorded intensity I versus the angle ϕ of the excitation polarizer is described by (Inoué et al., 2002): I = {Imax + Imin + [Imax − Imin] cos[2(ϕ − azimuth)]}/2.
The polarization ratio = Imax/Imin is 1 for isotropic fluorescence and increases with increasing anisotropy. Its value is affected by several circumstances, including the anisotropy of the individual fluorophores, their mutual alignment, and their inclination with respect to the microscope axis. In addition, the ratio is reduced with increasing numerical aperture of the excitation and emission optics. The azimuth, however, represents the axis of net alignment of the excitation transition moments as projected into the plane perpendicular to the microscope axis. With respect to a laboratory frame of reference, the azimuth can be any angle and will depend on the orientational constraint imposed on the fluorophores by the linker to their fusion proteins and the orientation distribution of the protein molecules as they are incorporated into a larger molecular complex.
We determined the fluorescence anisotropy based on intensities measured in images recorded at four specified polarizer angles (I0 through I135 in Fig. S1). Subsequently, image arithmetic was used to calculate the fluorescence anisotropy for each camera pixel using the following expressions:
The measured anisotropy, polarization ratio, and azimuth values were subject to systematic errors introduced by fluorescence bleaching, superposition of fluorescence from different cell structures or compartments, and differential transmission of optical components in the microscope. To minimize the influence of these systematic errors on our measurements, we have developed the following calibration and correction procedures.
Systematic error correction
Raw intensity values were corrected for systematic errors, including fluorescence bleaching and differential transmission of microscope optical components. A camera offset recorded at zero light intensity was also subtracted from every image. To be able to correct for fluorescence bleaching, we started the series of image acquisitions with the LC analyzer at setting 0°, followed by 135°, 90°, and 45°, and ended the series with an additional image , again at setting 0°. The time interval between each image was the same and was mainly determined by the exposure time (typ. ∼1 s). By comparing corresponding intensity values in images and , the bleach exponent was measured using the following relationship:
where and denote intensities averaged over a region that is expected to have the same bleach exponent. Subsequently, images for settings 135°, 90°, and 45° were corrected for bleaching by multiplying appropriate regions with:
where the superscript BleachCorr indicates the applied bleach correction.
In our experiments, we observed bleach exponents that differed between GFP-labeled septins suspended in the cytosol and those assembled into complexes such as the hourglass structure. Furthermore, the fluorescence of cortical septin complexes was superimposed on the fluorescence of soluble proteins in the cytosol. Therefore, we first analyzed the cytosol fluorescence located near a septin assembly and determined the cytosol’s fluorescence magnitude and bleach exponent. We then subtracted the cytosol fluorescence from the fluorescence observed in septin assemblies and determined the magnitude, bleach exponent, and anisotropy of the excess fluorescence of septin assemblies.
A further step in identifying instrument bias makes use of the fluorescence of randomly oriented fluorophores, like those suspended in the cytosol. The fluorescence of randomly oriented fluorophores is unpolarized, leading to the expectation that the fluorescence intensity is the same for every polarizer setting. If this expectation is not borne out by the actual observation, the unequal intensities between settings 0°, 45°, 90°, and 135° must be caused by instrument bias, for example by varying intensity of the excitation beam as a function of beam polarization. Therefore, one can derive a correction factor based on the a priori knowledge of isotropic fluorescence from a region with random fluorophores, such as the cytosol. Using the intensity recorded at 0° polarization as a reference, we derived correction factors for instrument bias using bleach corrected intensity values:
where pointed brackets indicate intensities averaged over many pixels that comprise fluorescence from randomly oriented fluorophores. Pixel intensities in regions with fluorescence anisotropy are then multiplied by the correction factors to minimize instrument bias:
The intensity values corrected for bleaching and instrument bias were entered into the above expressions for fluorescence anisotropy.
Determining and averaging orientation angles and calculating variance
The calculation methods used to determine the net orientation angle of GFP dipoles are described in the first section of the results, summarized in Fig. S1, and described in detail (see Calculation of fluorescence anisotropy). From these calculations, a net orientation angle was determined for every camera pixel. For further analysis, pixel-based orientation values were averaged over a region that corresponded to the complete septin ring, using the atan2 function, to calculate an orientation value for the entire septin structure. Additional averaging over a population of septin rings for a given strain, again making use of the atan2 function, lead to the “offset angle” that is reported in Table I. For the ring averages and the population averages it is important to consider how much variability is present in the measurements. Therefore, we devised methods both to average the pixel values and then additionally assess how variable our calculations were both between pixels within a given ring and between rings of the same strain.
For averaging orientation values that vary between 0 and 180°, special mathematical operations need to be used. This is because values near 0 and 180° represent nearly equivalent orientations, but their standard arithmetic average is near 90°, which is perpendicular to the original orientations and therefore wrong. To correctly average a set of measured angles, each angle in the set is multiplied by 2. Next, the cosine and sine of each of the doubled angles are calculated, added, and divided by the number of angles to find the mean cosine and sine values for a given septin structure. The mean values are then used in the atan2 function, which uses xy coordinate values as input (instead of their ratio) and returns the angle between the x axis counter-clockwise to the line connecting the origin and the specified coordinate. This angle, divided by 2, is the mean dipole orientation of the initial set of angles.
Variance in the measurements across a septin ring or between rings in a population can be expressed by examining the mean cosine and sine values calculated in the previous step. Although the original cosine and sine values are xy coordinates located on the unit circle, the averaged values are located inside the circle. The length from the origin to the averaged coordinate point is a measure of the spread among the angles (Fisher, 1993). If all dipole orientations are identical and there is no angular spread, the xy coordinates calculated as the mean orientation will be on the unit circle and have the length 1. The more spread there is among the individual angles, the shorter the line to the averaged coordinates. The length of the line is calculated by the square root of the squared mean cosine plus the squared mean sine value. The so-called circular variance is 1 minus the length (Fisher, 1993). This number is subtracted from 1 to give the variance of angles about the mean, with 0 denoting a structure with no variance (perfectly ordered) and 1 denoting complete variance or randomness (absolutely no order).
This variance calculation has been performed at two different levels. First, we describe the uniformity of angles which make up a single septin structure (which we call “ring var”, Table I). In ring var, we use weighted averages of angles which are calculated using their associated polarization ratio and background corrected mean fluorescence intensity (Fig. S2). Second, we describe the uniformity of offset angles for a population of septin structures by averaging offset angles for a population of different rings (which we call “ pop var,” Table I). In summary, “ring var” captures variability in measurements within a ring and “pop var” captures variability between rings. In either case, a var value that is close to 0 arises from measurements with little variability.
Calibration of LC polarizer
The polarization of the excitation light was conditioned by a LC universal compensator (Cambridge Research and Instrumentation, Inc.), which was operated as a variable polarizer. The LC compensator/polarizer is built from a fixed linear polarizer and two variable retarder plates, made from LC devices A and B (Oldenbourg and Mei, 1995). By varying the voltages applied to the LC devices, their retardance can be changed, allowing the LC polarizer to transmit any desired polarization (Shribak and Oldenbourg, 2003). The LC polarizer was sequentially set to transmit four linear polarization states, each characterized by its angle with respect to the horizontal axis in the specimen image (0°, 45°, 90°, and 135°). The polarization states were calibrated against a linear polarizing sheet with known transmission axis that was sandwiched between a slide and cover glass and placed as a specimen in the microscope. The transmission axis of the polarizing sheet was rotated to the four desired orientations. For each orientation, the retardance of the LC devices A and B was systematically varied until a minimum amount of light passed the polarizing sheet (image on camera darkest; during calibration the emission filter was removed from the microscope). Using high numerical aperture optics (Plan-Apochromat 60× objective lens and an apochromat condenser, both oil immersion and fully illuminated 1.4 numerical aperture) we typically achieved an extinction ratio (I‖/I’) of 60. Because the intensity was at a minimum, the actual polarization state transmitted through the LC analyzer was orthogonal to the orientation of the transmission axis of the linear polarizing sheet. The sheet was rotated by 45° steps and each time the calibration procedure was repeated, resulting in four sets of retardance values for LC-A and LC-B. Both LC devices and camera were under computer control, assisting in the calibration and enabling fast retrieval of stored retardance values to switch quickly between the four linear polarization states.
Statistical error analysis
There are several sources of statistical errors in the measurements reported here. These are caused by both detection limitations brought on by hardware and limitations in intensity measurements of fluorescence signals in live cells. The ability of our analysis software to accurately determine the dipole orientation at a given camera pixel relies on achieving a sufficiently high dynamic range in the intensities recorded at different polarizer angles. Systematic variations in intensity caused by changes in polarizer angle must be larger than random variations because of shot noise caused by photon statistics. To reduce shot noise to an acceptable level, we used exposure times of around 0.3–1 s, long enough to achieve noise levels of a few percent points of the signal. The dynamic range in systematic variations depends on several factors, including the numerical aperture of the excitation and imaging optics, the amount of unpolarized background light caused by instrumental factors and disordered GFP populations, the constraint imposed by the linker sequence between the septin molecule and GFP, and finally the actual distribution of dipole orientations and its projection in the plane perpendicular to the illumination and imaging axis. For example, supplementary angles in the image plane (as discussed in this study) near 45° and 135° would appear isotropic, even if the constructs were perfectly ordered.
In addition to affecting the ability to detect preferential alignments, the dynamic range is directly proportional to the polarization ratio that is measured. Hence, like the dynamic range, the ratio is affected by instrumental factors and by the particular order taken on by the GFP molecules. Instrumental factors include the numerical aperture of the excitation and imaging optics. As described previously (Axelrod, 1979, 1989; Scalettar et al., 1988), the main effect of high NA optics on measurements of polarized fluorescence is a reduction of the measured polarization ratio. With increasing numerical aperture and beam divergence, the range of angles subtended by the excitation and imaging beam increases and is superimposed in a single measurement. In other words, the higher the NA the more “isotropic” the measurement scheme becomes and the lower the ratio that is measured for a given fluorophore distribution. Because the reduction applies nearly equally to all distributions, regardless of their fluorophore arrangements and orientations, it does not appreciably affect the patterns of anisotropy considered here. Furthermore, the azimuth measured for a given distribution is generally not affected by the numerical aperture but chiefly depends on the angle between the central rays of the illumination and imaging beams and the axes of the fluorophore distribution.
Transmission electron microscopy
Cells grown overnight at 30°C in AFM were treated with 125 µM FCF, taken from a stock solution of 250 mM FCF in 99.5% EtOH for 3 h, and processed similarly to Byers and Goetsch (1976), with modifications, to evaluate the ultrastructure of septins in A. gossypii. Cells were allowed to settle by gravity before fixation in 10–15× volume of 3% glutaraldehyde (GTA)/1% PFA/0.1% tannic acid in 0.1 M sodium cacodylate buffer, pH 7.2–7.4 (NaCac buffer) for 2 h at RT with swirling of sample every 15 min. Fix was replaced, then cells were incubated one more hour at RT and then incubated overnight in fix on a rotator at 4°C. The next day, cells were washed in PBS and resuspended in 2 ml of solution A (100 mM KPO4, pH 7.5, and 1.2 M sorbitol) with 10 µg/ml zymolyase to remove cell wall, then incubated with gentle rotation at 37°C until 75% of the hyphae are phase dark (∼15–30 min). To halt digestion, cells were spun down at 1,000 rpm for 2 min and were washed twice in 2.5% GTA/1% PFA in 40 mM KPO4, pH 6.5–6.7. Digested cells were then refixed in 3% GTA/1% PFA/0.1% tannic acid in NaCac buffer for 2 h at RT, fix was replaced, and cells put on rotator overnight 4°C. Cells were spun at 1,800–3,000 rpm for 2 min to get the sample to the bottom and washed several times in NaCac buffer over 2 h. Cells were then resuspended in 2% OsO4 in NaCac buffer for 2 h at RT. Cells were rinsed twice in dH2O (to remove all trace of NaCac buffer) and enbloc stained with 2% aqueous uranyl acetate for 1–2 h at RT (1% UA for immunolabeling) in the dark. Cells were dehydrated through a graded series of ethanols for 30 min each on rotator, and then further dehydrated in 100% ethanol on rotator, using six rinses over 6 h. For TEM, cells were then left in 100% ethanol 12–36 h, without rotation, at 4–6°C to ensure complete dehydration.
After two 30 min washes in propylene oxide (PO) at room temperature, samples were embedded in epon (LX112 kit; Ladd, Inc.). Samples were immersed in 1:1 LX112/PO for 1 h on rotator, then 1.5:1 LX112/PO with 4–5 changes over 8 h on a rotator. Samples were placed in a vacuum desiccator overnight, then transferred to BEEM capsules, filled with fresh LX112, and returned to the vacuum desiccator overnight. Polymerization was performed at 45°C for 8 h and then increased to 60°C for a further 24 h. Semithin sections (0.5 µm) were mounted on glass slides and stained with Toluidine blue. Thin sections were mounted on 400HH Cu Grids (T400H-Cu; Electron Microscopy Services), then stained with 2% methanolic uranyl acetate for 15 min and Reynold’s lead citrate for 3 min. All TEM images were taken at 100 kV on a JEOL TEM 1010 equipped with a digital camera (XR-41B; Advanced Microscopy Techniques).
For immunolabeling of septins, fixations and digestion were the same except for the following changes: Initial fixation of 3% PFA/1% GTA was used instead of 3% GTA/1% PFA. Osmium postfixation was omitted, and instead, the samples were incubated in 0.05 M glycine in NaCac buffer for 30 min after final fixation to remove excess aldehydes.
Additionally, 1% p-phenylenediamine (PPD) was included in each EtOH solution. Four additional 85% EtOH with 1% PPD rinses over 1 h were performed rather than going into 100% EtOH, and then pellets were resuspended in a 2:1 mixture of 85% EtOH with 1% PPD:LR white medium resin. This suspension was placed on a rotator for 2 h at room temperature and then resuspended in 1:1 85% EtOH with 1% PPD:LR white medium resin, rotated for 1 h, and then kept at 4°C overnight. The next day, samples were resuspended in 1:2 85% EtOH with 1% PPD:LR white medium resin rotated for 1 h at room temperature and then resuspended in 100% LR white medium resin. Solution was changed until clear, and then three more changes over 6 h were performed while rotating at room temperature before placing at 4°C overnight. Samples were warmed to room temperature and four changes of 100% LR White medium resin were made with 1 h each change. Samples were transferred to gelatin capsules and left for 4 h at room temperature, then polymerized at 50°C for 24 h. Blocks were removed from heat, capsules were opened and gelatin removed, and sections were placed onto nickel-coated grids. Grids were floated in a moist chamber with solutions spotted on clean parafilm. Grids with sections were washed in PBS for 5 min, put in block solution of PBS + 5% BSA (Aurion; Electron Microscopy Sciences), 0.1% gelatin, 10% normal goat serum, and 15 mM NaN3 for 15 min, washed twice for 5 min, incubated for 1 h in 1/25 mouse anti-HA (16B12; Covance), washed six times for 5 min, incubated for 1 h in a 1/25th dilution of goat anti–mouse 10 nm gold-conjugated secondary antibody (25169; Electron Microscopy Sciences; a gift of R. Sloboda, Department of Biological Sciences, Dartmouth College, Hanover, NH), then washed six times for 5 min with two additional washes in PBS without BSA and two final washes in distilled water. Washes and antibody dilutions were in PBS + 0.1% BSA (Aurion) at RT. Grids were stored at room temperature and stained with 2% aqueous uranyl acetate for 15 min and Reynold’s lead citrate for 15 min. All TEM images were taken at 100 kV on a TEM (TEM 1010; JEOL) equipped with a digital camera (XR-41B) and capture engine software (AMTV540; Advanced Microscopy Techniques).
Immunofluorescence
MDCK cells were fixed with 3% PFA in warm PHEM buffer (60 mM Pipes-KOH, pH 6.9, 25 mM Hepes, 10 mM EDTA, and 2 mM MgCl2) containing 0.1% Triton X-100. These conditions preserve the SEPT2-GFP signal. Primary antibodies (rabbit) to SEPT6 (5 µg/ml, a gift of M. Kinoshita, Graduate School of Science, Nagoya University, Nagoya, Japan), SEPT7 (0.5 µg/ml IBL; Code No. 18991), and secondary donkey DyLight 594 (2.5 µg/ml; Jackson ImmunoResearch Laboratories, Inc.) were diluted in PBS with 2% BSA. Coverslips were mounted with Vectashield (Vector Laboratories) and imaged with a microscope (IX-81; Olympus) equipped with a ProScanIITM motorized stage (Prior), an Orca-R2 camera (Hamamatsu Photonics), a Plan-Apochromat 60×/1.40 NA objective lens, and the SlideBookTM 5 software.
Online supplemental material
Figs. S1 and S2 provide a step-by-step overview of the methods for analysis of polarized fluorescence data and visualization of orientational and polarization ratio data. Figs. S3 and 4 show representative cells from every strain analyzed in this study, including whole and split septin rings and septin fibers. Fig. S5 includes further examples of septin fibers from MDCK cells with low expression of Sept2 and additional electron micrographs with immunolabeled septin filaments and apparent bundles of septin filaments at the cell cortex. Table S1 contains information on the dimensions of septin filaments reported in the literature and within this work. Tables S2–S4 include the cell strains, oligonucleotides, and plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012143/DC1.
Acknowledgments
We thank Nicolas Borghi for MDCK culture and transfection, Christine Field and Tim Mitchison for invaluable discussions and support, Alexander Barnett and Paige Rinker for mathematical consultations, Grant B. Harris for technical support and software development, and Sharon Bickel for critically reading the manuscript. We thank Roger Sloboda for sharing 10 nm gold conjugated anti–mouse antibodies and Makoto Kinoshita for SEPT6 antibody.
This work was supported by the National Science Foundation under grant No. MCB-0719126 to A.S. Gladfelter, the National Institute of Biomedical Imaging and Bioengineering under grant No. EB002583 to R. Oldenbourg, a Drexel CURE grant from the State of Pennsylvania Tobacco Settlement Fund, and National Institute of Neurological Disorders and Stroke grant NS48090-06A to E.T. Spiliotis.
Footnotes
Abbreviations used in this paper:
- AFM
- Ashbya full media
- FCF
- forchlorfenuron
- GTA
- glutaraldehyde
- LC
- liquid crystal
- TEM
- transmission EM
References
- Axelrod D. 1979. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 26:557–573 10.1016/S0006-3495(79)85271-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D. 1989. Fluorescence polarization microscopy. Methods Cell Biol. 30:333–352 10.1016/S0091-679X(08)60985-1 [DOI] [PubMed] [Google Scholar]
- Ayad-Durieux Y., Knechtle P., Goff S., Dietrich F., Philippsen P. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563–4575 [DOI] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M.S., Snyder M. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 5:841–851 10.1016/S1097-2765(00)80324-X [DOI] [PubMed] [Google Scholar]
- Bertin A., McMurray M.A., Grob P., Park S.S., Garcia G., III, Patanwala I., Ng H.L., Alber T., Thorner J., Nogales E. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 105:8274–8279 10.1073/pnas.0803330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A., McMurray M.A., Thai L., Garcia G., III, Votin V., Grob P., Allyn T., Thorner J., Nogales E. 2010. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J. Mol. Biol. 404:711–731 10.1016/j.jmb.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmer C., Ripp C., Bölker M. 2009. The germinal centre kinase Don3 triggers the dynamic rearrangement of higher-order septin structures during cytokinesis in Ustilago maydis. Mol. Microbiol. 74:1484–1496 10.1111/j.1365-2958.2009.06948.x [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. 1976. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69:717–721 10.1083/jcb.69.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Longtine M., Pringle J.R., Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066 10.1091/mbc.E03-04-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay B.S., Meseroll R.A., Occhipinti P., Gladfelter A.S. 2009. Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol. Biol. Cell. 20:2311–2326 10.1091/mbc.E08-12-1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMay B.S., Meseroll R.A., Occhipinti P., Gladfelter A.S. 2010. Cellular requirements for the small molecule forchlorfenuron to stabilize the septin cytoskeleton. Cytoskeleton (Hoboken). 67:383–399 10.1002/cm.20452 [DOI] [PubMed] [Google Scholar]
- Dietrich F.S., Voegeli S., Brachat S., Lerch A., Gates K., Steiner S., Mohr C., Pöhlmann R., Luedi P., Choi S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 304:304–307 10.1126/science.1095781 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 305:393–396 10.1126/science.1099892 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Gentry M.S., Hallberg R.L., Barral Y. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 4:345–357 10.1016/S1534-5807(03)00061-3 [DOI] [PubMed] [Google Scholar]
- Fisher N.I. 1993. Statistical Analysis of Circular Data. Cambridge University Press, Cambridge, England: 295 pp [Google Scholar]
- Frazier J.A., Wong M.L., Longtine M.S., Pringle J.R., Mann M., Mitchison T.J., Field C. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143:737–749 10.1083/jcb.143.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A.S., Pringle J.R., Lew D.J. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681–689 10.1016/S1369-5274(01)00269-7 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276 10.1016/0014-4827(71)90223-0 [DOI] [PubMed] [Google Scholar]
- Hu Q., Nelson W.J., Spiliotis E.T. 2008. Forchlorfenuron alters mammalian septin assembly, organization, and dynamics. J. Biol. Chem. 283:29563–29571 10.1074/jbc.M804962200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Milenkovic L., Jin H., Scott M.P., Nachury M.V., Spiliotis E.T., Nelson W.J. 2010. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 329:436–439 10.1126/science.1191054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoué S., Shimomura O., Goda M., Shribak M., Tran P.T. 2002. Fluorescence polarization of green fluorescence protein. Proc. Natl. Acad. Sci. USA. 99:4272–4277 10.1073/pnas.062065199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Shindo A., Park T.J., Oh E.C., Ghosh S., Gray R.S., Lewis R.A., Johnson C.A., Attie-Bittach T., Katsanis N., Wallingford J.B. 2010. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 329:1337–1340 10.1126/science.1191184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., Theesfeld C.L., McMillan J.N., Weaver E., Pringle J.R., Lew D.J. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049–4061 10.1128/MCB.20.11.4049-4061.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M.A., Thorner J. 2009. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 4:18 10.1186/1747-1028-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenbourg R., Mei G. 1995. New polarized light microscope with precision universal compensator. J. Microsc. 180:140–147 [DOI] [PubMed] [Google Scholar]
- Pan F., Malmberg R.L., Momany M. 2007. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103 10.1186/1471-2148-7-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E.A., Petty E.M. 2010. Conquering the complex world of human septins: implications for health and disease. Clin. Genet. 77:511–524 10.1111/j.1399-0004.2010.01392.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A.A., Kozubowski L., Goode B.L., Drubin D.G., Hartwig J.H. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell. 16:372–384 10.1091/mbc.E04-08-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J.D.R. 2001. Molecular Cloning: A Laboratory Manual. Vol 2, Chapter 8. Cold Spring Harbor Laboratory Press, Cold Spring Harbor: 1–41 [Google Scholar]
- Scalettar B.A., Selvin P.R., Axelrod D., Hearst J.E., Klein M.P. 1988. A fluorescence photobleaching study of the microsecond reorientational motions of DNA. Biophys. J. 53:215–226 10.1016/S0006-3495(88)83083-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shribak M., Oldenbourg R. 2003. Techniques for fast and sensitive measurements of two-dimensional birefringence distributions. Appl. Opt. 42:3009–3017 10.1364/AO.42.003009 [DOI] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I.G., Weyand M., Stark H., Wittinghofer A. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Spiliotis E.T., Nelson W.J. 2006. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 119:4–10 10.1242/jcs.02746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E.T., Hunt S.J., Hu Q., Kinoshita M., Nelson W.J. 2008. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J. Cell Biol. 180:295–303 10.1083/jcb.200710039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344 10.1126/science.290.5490.341 [DOI] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y., Kinoshita M., Takiguchi K. 2009. Septin-mediated uniform bracing of phospholipid membranes. Curr. Biol. 19:140–145 10.1016/j.cub.2008.12.030 [DOI] [PubMed] [Google Scholar]
- Tooley A.J., Gilden J., Jacobelli J., Beemiller P., Trimble W.S., Kinoshita M., Krummel M.F. 2009. Amoeboid T lymphocytes require the septin cytoskeleton for cortical integrity and persistent motility. Nat. Cell Biol. 11:17–26 10.1038/ncb1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabioiu A.M., Mitchison T.J. 2006. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 443:466–469 10.1038/nature05109 [DOI] [PubMed] [Google Scholar]
- Vrabioiu A.M., Mitchison T.J. 2007. Symmetry of septin hourglass and ring structures. J. Mol. Biol. 372:37–49 10.1016/j.jmb.2007.05.100 [DOI] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., Barral Y. 2008. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9:478–489 10.1038/nrm2407 [DOI] [PubMed] [Google Scholar]
- Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 242:381–391 10.1016/S0378-1119(99)00509-0 [DOI] [PubMed] [Google Scholar]