Phosphorylation of human Treslin by Cdk is essential for its association with TopBP1 and for the initiation of DNA replication in S phase.
Abstract
Treslin, a TopBP1-interacting protein, is necessary for deoxyribonucleic acid (DNA) replication in vertebrates. Association between Treslin and TopBP1 requires cyclin-dependent kinase (Cdk) activity in Xenopus laevis egg extracts. We investigated the mechanism and functional importance of Cdk for this interaction using both X. laevis egg extracts and human cells. We found that Treslin also associated with TopBP1 in a Cdk-regulated manner in human cells and that Treslin was phosphorylated within a conserved Cdk consensus target sequence (on S976 in X. laevis and S1000 in humans). Recombinant human Cdk2–cyclin E also phosphorylated this residue of Treslin in vitro very effectively. Moreover, a mutant of Treslin that cannot undergo phosphorylation on this site showed significantly diminished binding to TopBP1. Finally, human cells harboring this mutant were severely deficient in DNA replication. Collectively, these results indicate that Cdk-mediated phosphorylation of Treslin during S phase is necessary for both its effective association with TopBP1 and its ability to promote DNA replication in human cells.
Introduction
In eukaryotic cells, replication of the genome involves a highly orchestrated assembly of proteins onto the DNA (Méndez and Stillman, 2003; Sclafani and Holzen, 2007). First, the origin recognition complex binds to sequences of DNA that will function as initiation points for replication. After this binding, the origin recognition complex appears to serve as a landing pad for the replication proteins Cdc6 and Cdt1. Thereupon, these proteins mediate loading of the minichromosome maintenance complex onto the DNA, which results in formation of the prereplication complex (Diffley, 2004). The minichromosome maintenance proteins are critical elements of the helicase activity that separates the two strands of DNA for replication (Pacek et al., 2006; Ilves et al., 2010).
Full assembly of the replicative helicase and its ultimate activation require the participation of additional structural and regulatory proteins, including two conserved kinases. Studies in yeast have established the paradigm for this process (Sclafani and Holzen, 2007; Remus and Diffley, 2009; Tanaka and Araki, 2010). In particular, numerous replication proteins, including Dpb11, Sld2, Sld3, and Cdc45, also bind at or near replication origins. In the course of events leading to replication, the Dbf4-dependent kinase and Cdk carry out key phosphorylations of proteins in this network (Araki, 2010; Labib, 2010). The role of S phase Cdk (S-Cdk) activity appears to be especially pivotal in this scheme (Botchan, 2007; Tanaka et al., 2007a,b; Zegerman and Diffley, 2007). It has been demonstrated that S-Cdk phosphorylates Sld2 and Sld3. These modifications enable docking of Sld2 and Sld3 with two distinct pairs of BRCA1 C terminus (BRCT) repeats within Dpb11. These steps eventually lead to the recruitment of Cdc45 as an activator of the replicative helicase. Functional studies have established that these phosphorylations of Sld2 and Sld3 are minimally necessary for S-Cdk to promote the initiation of S phase (Tanaka et al., 2007b; Zegerman and Diffley, 2007).
A question of ongoing interest has been whether these regulatory events operate in a conserved manner in more complex organisms. In vertebrate cells, the closest relative of Dpb11 is a protein known as TopBP1 (Garcia et al., 2005). TopBP1 is a more elaborate protein that contains eight BRCT repeats, as opposed to only four in Dpb11. Nonetheless, TopBP1 also facilitates the incorporation of Cdc45 into the replicative machinery and is thus necessary for DNA replication (Van Hatten et al., 2002; Hashimoto and Takisawa, 2003). On the other hand, a conclusive assignment of vertebrate proteins as functional analogues of Sld2 and Sld3 has not been straightforward.
The vertebrate RecQ4 protein displays some limited homology with yeast Sld2 and is necessary for DNA replication, but this protein differs from Sld2 in several respects (Sangrithi et al., 2005; Matsuno et al., 2006). Recently, a protein called Treslin (also known as Ticrr) has emerged as an excellent candidate for a vertebrate counterpart of Sld3 (Kumagai et al., 2010; Sansam et al., 2010). This protein associates specifically with TopBP1 and is an essential replication protein. Two other proteins, namely GEMC1 and DUE-B, have also been proposed as candidates for vertebrate Sld3 (Balestrini et al., 2010; Chowdhury et al., 2010). However, among these proteins, only Treslin displays any, albeit limited, homology with yeast Sld3 (Fu and Walter, 2010; Sanchez-Pulido et al., 2010). In this paper, we investigate this issue further by asking whether phosphorylation of Treslin by S-Cdk is critical for its replication-initiating function. We have found that phosphorylation of a single conserved site in Treslin by S-Cdk regulates both its binding to TopBP1 and its ability to promote DNA replication. Thus, these studies establish that there is a crucial regulatory relationship between S-Cdk activity and Treslin for promoting DNA replication in vertebrate cells.
Results
Treslin associates with TopBP1 in a cell cycle–regulated manner in human cells
Previously, we demonstrated that the interaction between Treslin and TopBP1 in Xenopus laevis egg extracts depends on S-Cdk activity. The evidence was that treatment of egg extracts with the Cdk inhibitor p27 caused a substantial reduction in the binding of TopBP1 to Treslin (Kumagai et al., 2010). However, it has also been reported that treatment of asynchronous human cells with the Cdk inhibitor roscovitine does not inhibit this interaction (Sansam et al., 2010). The cell cycle in X. laevis egg extracts contains only two phases (S and M), which is characteristic of early embryonic cell cycles. On the other hand, somatic cell cycles also contain G1 and G2 phases. Therefore, we hypothesized that it may be more difficult to detect cell cycle–regulated changes in the binding of TopBP1 to Treslin in asynchronous somatic cells.
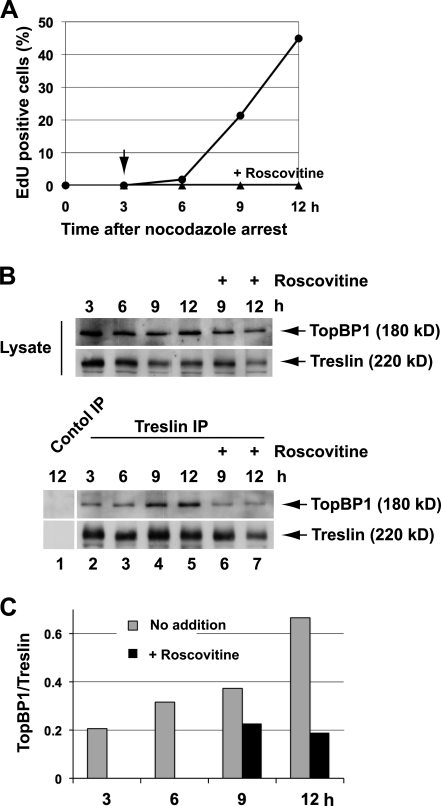
To address this possibility, we synchronized human U2OS cells by treating them with nocodazole and using the mitotic shake-off procedure (Fig. 1). We observed that the cells entered S phase at 9–12 h after release from the nocodazole-induced mitotic arrest, as indicated by the incorporation of EdU into the DNA (Fig. 1 A). As expected, roscovitine completely blocked entry into S phase. Next, we measured the binding of Treslin to TopBP1 as a function of time during the cell cycle (Fig. 1, B and C). We found that there was some binding throughout the time course. However, there was a significant increase in binding of Treslin to TopBP1 at 9–12 h after removal of nocodazole during the time of replication. Significantly, this increase was abrogated by treatment with roscovitine (Fig. 1, B and C). On the other hand, we found it difficult to observe an effect of roscovitine on the binding of Treslin to TopBP1 in asynchronous cells (unpublished data). From these studies, we conclude that there is some basal association of Treslin and TopBP1 throughout much, if not all, of the cell cycle, but this interaction increases substantially at S phase. This increase is sensitive to roscovitine, which suggests that it depends on Cdk activity. These results are generally consistent with what we observed previously in X. laevis egg extracts. Although p27 clearly inhibited the binding of TopBP1 to Treslin in egg extracts, it did not completely abolish the interaction (Kumagai et al., 2010). Overall, our results indicate that the interaction between Treslin and TopBP1 is regulated by Cdk activity in multiple systems.
Figure 1.
Treslin and TopBP1 interact in a Cdk-regulated manner in human U2OS cells. (A) U2OS cells were arrested at M phase with nocodazole, isolated from plates by shake-off, and released into fresh nocodazole-free medium. 25 µM roscovitine was added to one of two duplicate cultures at 3 h after the release (arrow). Cells were harvested every 3 h. DNA replication was monitored by incorporation of EdU in cells incubated in the absence (circles) or presence (triangles) of roscovitine. Results are from one representative experiment. A second experiment gave similar results. (B) Cell lysates were immunoprecipitated with control or anti-Treslin antibodies as indicated. Cell lysates (top) and immunoprecipitates (IP; bottom) were immunoblotted for TopBP1 and Treslin. (C) Quantitation of the results from the bottom portion of B. Results are from one representative experiment. A second experiment gave similar results.
Mapping of the regions in Treslin that bind to TopBP1
Having established that binding of Treslin to TopBP1 is under the control of Cdk activity in both X. laevis egg extracts and human cells, we sought to understand the mechanism of this regulation. Evidence has been presented that TopBP1 in egg extracts is not a direct functional target of Cdks (Hashimoto and Takisawa, 2003). Moreover, it is well established in budding yeast that Sld3, the putative functional analogue of Treslin, is directly phosphorylated in a functionally critical manner by S-Cdk (Tanaka et al., 2007b; Zegerman and Diffley, 2007). Therefore, we postulated that Treslin might also be a direct substrate of S-Cdk. To address this question effectively, we felt that it would be important to identify first what region of Treslin is responsible for interacting with TopBP1.
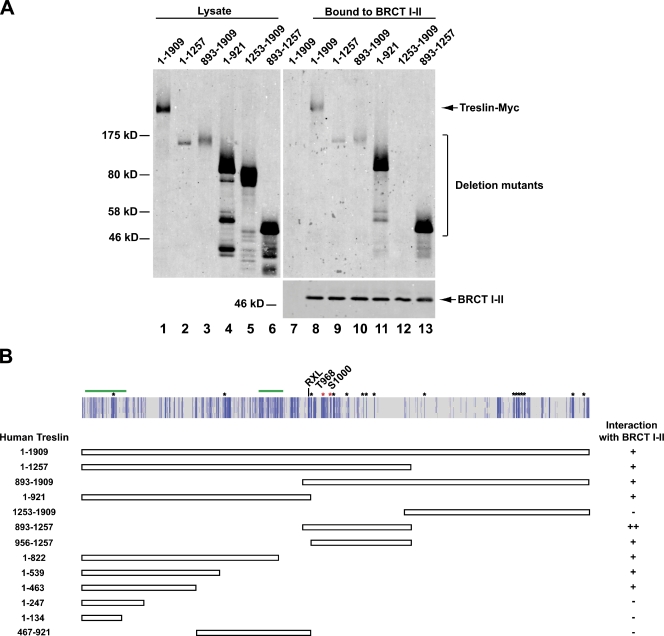
To elucidate the region of Treslin that associates with TopBP1, we developed a binding assay in human cells. For this purpose, we expressed Myc-tagged constructs of Treslin in human U2OS cells. Next, we prepared lysates from the cells and incubated the lysates with an exogenously added FLAG-tagged fragment of TopBP1 containing BRCT domains I–II. As we previously demonstrated, this region of TopBP1 is both necessary and sufficient for binding Treslin (Kumagai et al., 2010). Finally, we reisolated the TopBP1 fragment with anti-FLAG antibody beads and immunoblotted with anti-Myc antibodies. As shown in Fig. 2 A, full-length Treslin-Myc bound well to the BRCT I–II fragment. Initially, we found that a large N-terminal fragment of Treslin (residues 1–1,257) bound well to TopBP1, whereas a C-terminal fragment (residues 1,253–1,909) was defective for binding (Fig. 2, A and B). We proceeded to dissect the N-terminal fragment in further detail. We eventually found that a fragment containing residues 893–1,257 bound quite well to TopBP1. Truncation mutants with further deletions from either end of this fragment (e.g., residues 956–1,257 or 893–1,052) bound noticeably less well to TopBP1 (Fig. 2, A and B; and not depicted). Thus, most, if not all, of the 893–1,257 region is necessary for optimal binding.
Figure 2.
Deletion studies of the regions in Treslin that interact with the BRCT I–II domains of TopBP1. (A) Full-length Treslin-Myc (lanes 1, 7, and 8) and deletion mutants containing the indicated portions of Treslin (lanes 2–6 and 9–13) were transiently expressed in U2OS cells. Cell lysates (lanes 1–6) were incubated in the absence (lane 7) or presence (lanes 8–13) of a FLAG-tagged fragment of TopBP1 containing BRCT domains I–II. The incubations also contained anti-FLAG antibodies bound to protein G beads. The beads were reisolated, washed, and processed for immunoblotting with anti-Myc (top) and anti-FLAG antibodies (bottom). (B, top) Results of multiple sequence alignments of full-length Treslin using the Kalign program. Dark blue lines represent amino acids that are identical in Treslin from human, X. laevis, chicken, and zebrafish. Light blue lines represent amino acids that are identical in two or three out of these four species. Asterisks denote SP/TP motifs that have been conserved in all four species. Red asterisks indicate SP/TP motifs that are conserved in yeast Sld3 (Fig. 3 A). RXL indicates the location of a putative cyclin-interacting motif. Green lines indicate homology detected by Sanchez-Pulido et al. (2010) between Treslin and Sld3. (bottom) Summary of the abilities of various fragments of Treslin to interact with the BRCT I–II domain of TopBP1. Quantitation was performed from several experiments by measuring with the Odyssey imaging system the amounts of fragments of Treslin that bound to the BRCT I–II fragment of TopBP1. The designations on the right are as follows: −, <1% input Treslin bound to TopBP1; +, 3–6% input bound; and ++, >10% input bound.
In the course of these experiments, we also found that there is another distinct TopBP1-interacting domain in the more N-terminal region of Treslin. For example, a fragment containing residues 1–822, which does not overlap at all with the 893–1,257 fragment, also bound significantly to the TopBP1 (Fig. 2, A and B). However, the binding of this fragment was noticeably weaker than the 893–1,257 fragment. In further analyses, we observed that shorter N-terminal fragments (e.g., residues 1–463) bound as well as the 1–822 fragment. However, mutants with more extensive deletions, such as the 1–247 and 1–143 fragments, could not bind to TopBP1. Overall, we conclude that binding to TopBP1 involves at least two discontinuous regions of Treslin that contain residues 1–463 and 893–1,257. However, the latter fragment binds more strongly and thus is presumably relatively more important in the overall interaction with TopBP1.
X. laevis and human Treslin undergo phosphorylation on a conserved Cdk site
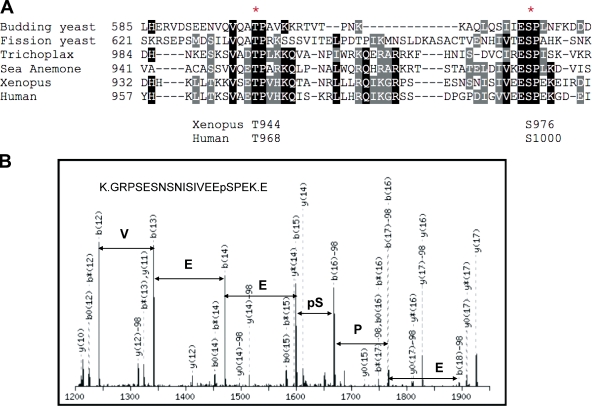
Next, we searched for Cdk phosphorylation sites in Treslin. Toward this end, we examined the sequence of Treslin for well-conserved potential Cdk phosphorylation sites (especially in the region corresponding to the 893–1,257 fragment of human Treslin). It is well known that Cdks have a marked preference to phosphorylate Ser-Pro/Thr-Pro (SP/TP) motifs. As depicted in Fig. 3 A, we noticed that Treslin contains two especially well-conserved potential Cdk phosphorylation sites (at T944 and S976 in X. laevis and at T968 and S1000 in humans). These sites are relatively close to one another. Moreover, we noted that the sequence in this area of Treslin displays some similarity with the yeast Sld3 proteins. This area of homology is distinct from those previously identified (Sanchez-Pulido et al., 2010).
Figure 3.
Analysis of the phosphorylation of Treslin. (A) Amino acids 893–1,257 of human Treslin and the corresponding portions of X. laevis Treslin and Treslin homologues from sea anemone (NCBI Protein database accession no. XP_001628252) and Trichoplax adhaerens (NCBI Protein database accession no. XP_002111061) were aligned with the C-terminal half of budding yeast and fission yeast Sld3 using the T-Coffee (Tree-based Consistency Objective Function for Alignment Evaluation) program. Only the most similar portion of the alignment is shown. Identical and similar residues are shaded in black and gray, respectively. The asterisks denote residues that are homologous with T600 and S622 of budding yeast Sld3 (corresponding to T968 and S1000 of human Treslin). (B) Part of the MS/MS spectrum corresponding to a peptide containing phosphorylated S976 of X. laevis Treslin. MS analysis was performed as described in Materials and methods.
In parallel, we also examined the phosphorylation of Treslin by mass spectrometry (MS). For this purpose, we initially analyzed X. laevis Treslin that had been isolated from S phase X. laevis egg extracts (see Materials and methods). Among several phosphorylated peptides, we detected three peptides containing phosphorylated S976 in X. laevis Treslin: K.GRPSESNSNISIVEEpSPEK.E, K.GRPSESNSNISIVEEpSPEKEIR.D, and N.ISIVEEpSPEK.E. In Fig. 3 B, we have presented a part of the MS/MS spectrum acquired from a doubly charged precursor with m/z 1069.9827. The hit had a peptide ion score of 71, and its peptide mass matched with better than 1 ppm accuracy to amino acids 961–979 from Treslin (K.GRPSESNSNISIVEEpSPEK.E). Thus, S976 of Treslin undergoes phosphorylation in egg extracts. On the other hand, we were not able to observe phosphorylation of T944, even though we could detect overlapping peptides that cover the sequence in this region (unpublished data).
S1000 of human Treslin is necessary for binding to TopBP1
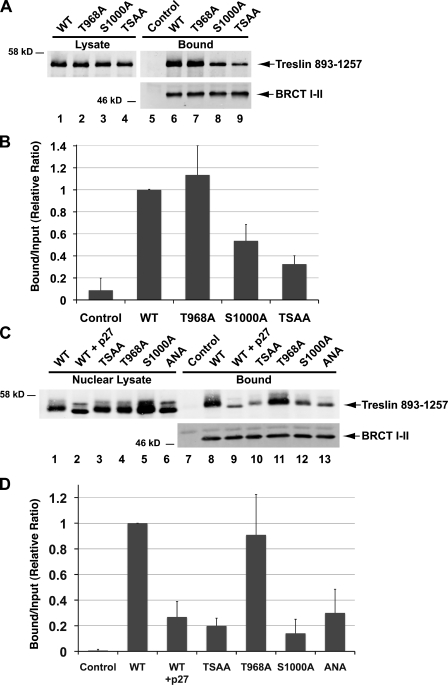
Based on this information, we decided to prepare point mutants of Treslin. Initially, we chose to mutate sites within the human protein to take advantage of the functional assays that we had developed in human cells. Toward this end, we mutated T968, S1000, or both to Ala in the context of the 893–1,257 fragment of human Treslin. We expressed these polypeptides in U2OS cells and assayed binding to the BRCT I–II fragment of TopBP1. As shown in Fig. 4 (A and B), the S1000A mutant of Treslin showed significantly reduced binding to TopBP1. In contrast, the T986A mutant bound as well as the wild-type fragment of Treslin to TopBP1. The double mutant (which we named TSAA) displayed a similar level of binding as the single S1000A mutant.
Figure 4.
The S1000A mutation affects the interaction of Treslin (893–1,257) with the BRCT I–II fragment of TopBP1. (A) Wild-type (WT) Treslin (893–1,257)-Myc (lanes 1, 5, and 6) and versions of this fragment containing the point mutations T968A (lanes 2 and 7), S1000A (lanes 3 and 8), and T968AS1000A (lanes 4 and 9) were expressed transiently in U2OS cells. Interaction with the BRCT I–II fragment of TopBP1 was assessed as described in Fig. 2 A. (B) Quantitation of the data as shown in A. Results (mean ± SD) were compiled from three independent experiments. (C) A His-tagged form of human Treslin (893–1,257)-Myc (lanes 1, 2, and 7–9) and versions of this fragment containing the point mutations T968AS1000A (lanes 3 and 10), T968A (lanes 4 and 11), S1000A (lanes 5 and 12), and R913AL915A (lanes 6 and 13) were expressed in bacteria and purified with nickel agarose beads. The fragments were incubated in X. laevis egg extracts containing sperm chromatin. In some cases, the extracts also contained p27 (lanes 2 and 9). After a 60-min incubation, nuclei were isolated from the extracts, and nuclear lysates were prepared as described in Materials and methods. Nuclear lysates (lanes 1–6) were incubated without (lane 7) or with the FLAG-tagged BRCT I–II fragment of TopBP1 (lanes 8–13) in the presence of anti-FLAG antibody beads. The beads were isolated and immunoblotted with anti-Myc (top) and anti-FLAG antibodies (bottom). (D) Quantitation of the data as shown in C. Results (mean ± SD) were compiled from three independent experiments.
To provide further validation of these results, we made use of the X. laevis egg extract system. For this purpose, we prepared wild-type and mutant forms of the human 893–1,257 fragment in bacteria and then incubated these polypeptides in S phase extracts of X. laevis eggs containing reconstituted nuclei. Subsequently, we prepared nuclear lysates from the extracts and incubated these lysates with the FLAG-tagged BRCT I–II fragment of TopBP1. Finally, we reisolated the fragment of TopBP1 with anti-FLAG antibody beads and detected binding of Treslin with anti-Myc antibodies. As shown in Fig. 4 (C and D), the wild-type and T968A versions of the fragment bound very well to TopBP1. In contrast, the S1000A mutant displayed severely reduced binding to TopBP1. For comparison, the Cdk inhibitor p27 caused a similar degree of inhibition in the binding of the wild-type 893–1,257 fragment to TopBP1.
It is known that many substrates of Cdks possess an amino acid sequence that allows docking with cyclin (Schulman et al., 1998; Loog and Morgan, 2005). This sequence is known as the RXL (Arg-X-Leu) or Cy (cyclin binding) motif. We noted that human Treslin contains an RXL motif (RNL) at positions 913–915, relatively close to S1000 (Fig. 2 B). This motif is well conserved in homologues of Treslin in other species. Accordingly, we mutated this sequence by changing both R913 and L915 to Ala (to create the ANA mutant). We found that the ANA mutant of the 893–1,257 fragment bound as poorly to TopBP1 as the S1000A mutant did (Fig. 4, C and D).
Treslin is a direct substrate of S-Cdk
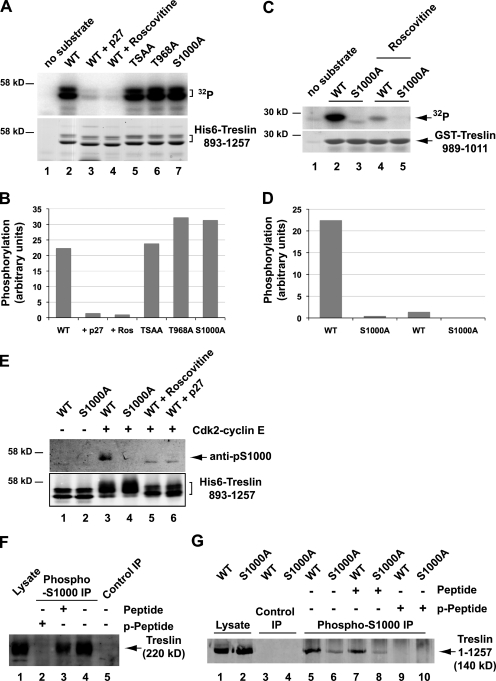
Next, we asked whether Treslin (particularly, S1000 of Treslin) could serve as a direct substrate of S-Cdk activity. For this purpose, we prepared human recombinant Cdk2–cyclin E by expression in Sf9 insect cells. Next, we incubated wild-type and mutant forms of the 893–1,257 fragment with this kinase in the presence of γ-[32P]ATP. We observed that Cdk2–cyclin E could phosphorylate the 893–1,257 fragment very well (Fig. 5, A and B). The Cdk inhibitors p27 and roscovitine completely blocked the phosphorylation. Nonetheless, the T968A, S1000A, and TSAA mutants were still good substrates of Cdk2–cyclin E.
Figure 5.
Phosphorylation of Treslin on S1000 by Cdk2–cyclin E. (A) Cdk2–cyclin E was incubated in the absence (lane 1) or presence of bacterially produced wild-type (WT) His6-Treslin (893–1,257; lanes 2–4) or the TSAA (lane 5), T968A (lane 6), or S1000A (lane 7) forms of this fragment. Incubations were performed in kinase buffer containing [32P]ATP. The incubations in lanes 3 and 4 also contained 2 µM GST-p27 and 25 µM roscovitine, respectively. Samples were subjected to SDS-PAGE and processed for phosphorimaging (top) and Coomassie blue staining (bottom). (B) Quantitation of 32P incorporation in A. Results are from one representative experiment. A second experiment gave similar results. (C) Cdk2–cyclin E was incubated with no substrate (lane 1), GST-Treslin(989–1,011) (lanes 2 and 4), or GST-Treslin (989–1,011)-S1000A (lanes 3 and 5) in a buffer containing [32P]ATP. The incubations were performed in the absence (lanes 1–3) or presence of 25 µM roscovitine (lanes 4 and 5). Samples were subjected to SDS-PAGE and processed for phosphorimaging (top) and Coomassie blue staining (bottom). (D) Quantitation of 32P incorporation in C. Results are from one representative experiment. A second experiment gave similar results. (E) Wild-type Treslin (893–1,257)-Myc (lanes 1, 3, 5, and 6) and its S1000A mutant (lanes 2 and 4) were incubated in the absence (lanes 1 and 2) or presence of Cdk2–cyclin E in a kinase buffer containing 1 mM ATP (lanes 3–6). The incubations in lanes 5 and 6 contained 25 µM roscovitine and 2 µM GST-p27, respectively. The reaction was stopped with SDS sample buffer. Samples were immunoblotted with anti–phospho-S1000 (top) and anti-Myc antibodies (bottom). (F) U2OS cell lysates (lane 1) were immunoprecipitated (IP) with either anti–phospho-S1000 antibodies (lanes 2–4) or control antibodies (lane 5). The immunoprecipitates were performed in the presence of no additional peptide (lanes 4 and 5) or 1 µg/ml of the either the immunizing phosphopeptide (lane 2) or the nonphosphorylated form of the same peptide (lane 3). Samples were immunoblotted with anti-Treslin antibodies. (G) Lysates from stable U2OS T-REx cell lines (lanes 1 and 2) expressing either wild-type Treslin (1–1,257)-Myc (lanes 1, 3, 5, 7, and 9) or the S1000A mutant of this fragment (lanes 2, 4, 6, 8, and 10) were immunoprecipitated with control (lanes 3 and 4) or anti–phospho-S1000 antibodies (lanes 5–10) in the presence of no additional peptide (lanes 3–6) or 1 µg/ml of either immunizing phosphopeptide (lanes 9 and 10) or nonphosphorylated peptide (lanes 7 and 8). Samples were processed for immunoblotting with anti-Myc antibodies.
The 893–1,257 fragment contains 23 SP/TP motifs. Therefore, we reasoned that remaining motifs might be phosphorylated during the in vitro kinase assays. To address more precisely the issue of whether S1000 could serve as a substrate, we prepared a fusion of GST with residues 989–1,011 of Treslin, which contain only this potential Cdk site. We observed that Cdk2–cyclin E could phosphorylate the wild-type version of this peptide effectively (Fig. 5, C and D). In contrast, the corresponding S1000A mutant of this polypeptide was not phosphorylated significantly by Cdk2–cyclin E. Inclusion of roscovitine in the kinase assay blocked phosphorylation of the wild-type polypeptide. From these experiments, we conclude that S1000 of Treslin is a direct target of S-Cdk activity. It appears that S-Cdk can phosphorylate additional sites in the 893–1,257 fragment of Treslin, at least in cell-free incubations. However, these phosphorylations do not appear to have a determinative role in the binding of TopBP1.
We also addressed the issue of whether S1000 of human Treslin is actually phosphorylated in human cells. Significantly, this residue has been identified as an in vivo site of phosphorylation in a survey of the phosphoproteome in a human Jurkat T cell leukemia line (Mayya et al., 2009). To complement this information, we also generated antibodies against a peptide from Treslin containing phosphorylated S1000 (anti–phospho-S1000 antibodies). These antibodies recognized the wild-type version of the 893–1,257 fragment of Treslin that had been incubated in vitro with Cdk2–cyclin E but did not react with the S1000A mutant of this peptide (Fig. 5 E). This reactivity of the antibodies with this peptide was abolished by inclusion of roscovitine or p27 during the incubation with the kinase.
We also used the anti–phospho-S1000 antibodies to analyze Treslin in human cells. These antibodies proved to be too weak for detection of Treslin by immunoblotting of cell lysates. However, we were able to immunoprecipitate the endogenous Treslin in U2OS cells effectively with the anti–phospho-S1000 antibodies (Fig. 5 F). Inclusion of the phosphopeptide used for generation of the antibodies efficiently abolished this immunoprecipitation, whereas the nonphosphorylated version of the same peptide did not block this pull-down. As described in further detail in the next section, we also created stable U2OS T-REx cells that can express wild-type and S1000A versions of the 1–1,257 fragment of human Treslin. We found that the anti–phospho-S1000 antibodies could efficiently immunoprecipitate the wild-type fragment of Treslin but not the S1000A mutant (Fig. 5 G). Collectively, our findings and a previously published study (Mayya et al., 2009) indicate that S1000 of human Treslin is phosphorylated in vivo.
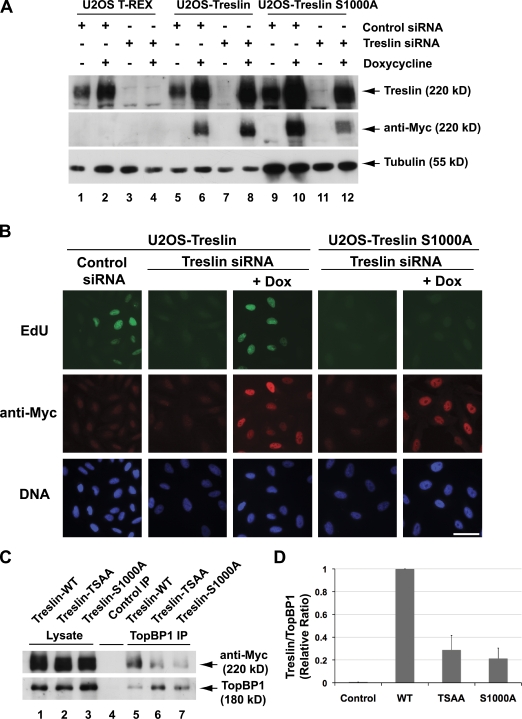
Human cells harboring S1000A mutant forms of Treslin are defective for DNA replication
We proceeded to examine whether phosphorylation of Treslin on S1000 has a functionally important role in DNA replication (Fig. 6, A and B). For these experiments, we used U2OS cell lines capable of expressing recombinant Treslin in a doxycycline-inducible manner. Previously, we used this strategy to engineer cell lines capable of expressing an siRNA-resistant form of Treslin that could rescue the siRNA-mediated ablation of endogenous Treslin. In these experiments, we used an untagged form of Treslin. To facilitate the analysis of mutants, we generated similar lines that could express a C-terminally Myc-tagged form of siRNA-resistant Treslin (Treslin-Myc). As shown in Fig. 6 (C and D), wild-type Treslin-Myc bound well to the endogenous TopBP1 in U2OS cells. In contrast, the S1000A mutant of the full-length Treslin-Myc interacted poorly with TopBP1. Likewise, the double TSAA mutant showed similarly weak binding.
Figure 6.
S1000 of Treslin is essential for DNA replication. (A) Control U2OS T-REx cells (lanes 1–4) and U2OS T-REx cells harboring wild-type (lanes 5–8) or S1000A Treslin-Myc (lanes 9–12) were incubated for 24 h in the absence (lanes 1, 3, 5, 7, 9, and 11) or presence of doxycycline (lanes 2, 4, 6, 8, 10, and 12). Cells were transfected with either control siRNA (lanes 1, 2, 5, 6, 9, and 10) or Treslin siRNA (lanes 3, 4, 7, 8, 11, and 12) and incubated for another 72 h. Cell lysates were immunoblotted with anti-Treslin (top), anti-Myc (middle), and anti-tubulin antibodies (bottom). (B) Cells were incubated with 10 µM EdU for 1 h. Samples were stained with Click-iT reagent to detect newly synthesized DNA (top), anti-Myc antibodies to detect Treslin (middle), and Hoechst 33342 dye to detect total DNA (bottom). Bar, 50 µM. (C) Expression of wild-type (WT) Treslin (lanes 1, 4, and 5), Treslin-T968AS1000A (lanes 2 and 6), and Treslin-S1000A (lanes 3 and 7) was induced with 1 µg/ml doxycycline for 24 h. Cell lysates were prepared and immunoprecipitated (IP) with either control (lane 4) or anti–human TopBP1 antibodies (lanes 5–7). Samples were immunoblotted with anti-Myc (top) or anti-TopBP1 antibodies (bottom). (D) Quantitation of the binding of various forms of Treslin to TopBP1 as shown in C. Results (mean ± SD) were compiled from three independent experiments.
As previously described (Kumagai et al., 2010), cells treated with Treslin siRNA in the absence of doxycycline showed greatly reduced DNA replication (Figs. 6 B and 7 C). Induction of wild-type Treslin-Myc with doxycycline resulted in substantial restoration of DNA replication. In parallel, we also induced expression of the S1000A or TSAA mutants of Treslin-Myc in the Treslin-ablated cells. In these cases, there was essentially no rescue of DNA replication (Figs. 6 B and 7 C; and not depicted).
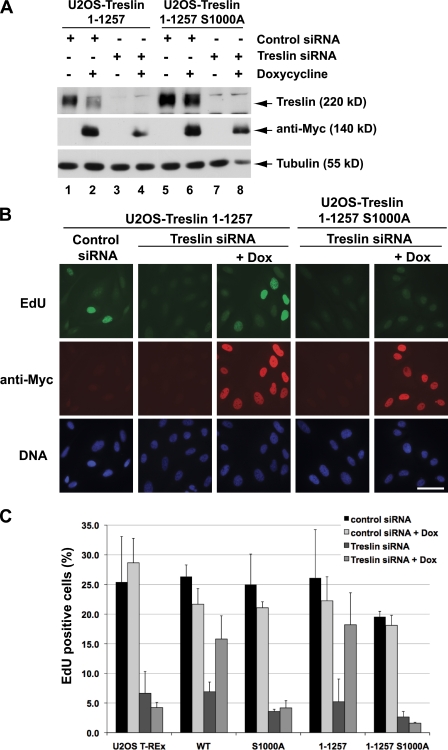
Figure 7.
A C-terminal domain of Treslin is dispensable for DNA replication. (A) U2OS cells harboring wild-type (lanes 1–4) or S1000A Treslin (1–1,257)-Myc (lanes 5–8) were incubated for 24 h in the absence (lanes 1, 3, 5, and 7) or presence of doxycycline (lanes 2, 4, 6, and 8). Cells were transfected with either control siRNA (lanes 1, 2, 5, and 6) or Treslin siRNA (lanes 3, 4, 7, and 8) and incubated for another 72 h. Cell lysates were immunoblotted with anti-Treslin (top), anti-Myc (middle), and anti-tubulin antibodies (bottom). (B) Cells treated with siRNA were incubated with 10 µM EdU for 1 h and then processed for staining with Click-iT reagent (top), anti-Myc antibodies (middle), and Hoechst dye DNA (bottom). Bar, 50 µM. (C) Quantitation of the data from Figs. 6 B and 7 B. Results (mean ± SD) were compiled from three independent experiments.
To pursue these findings further, we also engineered cells that harbor inducible forms of the 1–1,257 fragment of Treslin, which is sufficient for binding TopBP1. We observed that the wild-type version of this fragment rescued DNA replication very effectively in Treslin siRNA–treated cells (Fig. 7, A–C). Indeed, the rescue appeared to be somewhat more effective than what we observed with full-length Treslin. Nonetheless, the S1000A mutant of the 1–1,257 fragment was still unable to rescue DNA replication in the siRNA-ablated cells. Collectively, these results indicate that phosphorylation of S1000 plays an important role in DNA replication in human cells.
Treslin interacts with Cdc45 in vertebrate cells
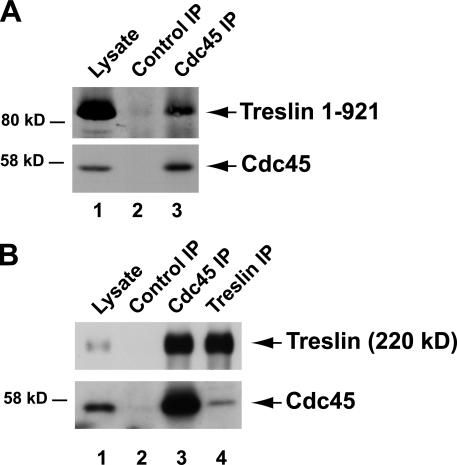
A key characteristic of budding and fission yeast Sld3 is that this protein interacts specifically with Cdc45 (Kamimura et al., 2001; Yabuuchi et al., 2006). To address whether Treslin interacts with Cdc45, we performed binding experiments in human cells and X. laevis egg extracts. Because commercially available antibodies were relatively ineffective for detecting endogenous Cdc45 in human U2OS cells, we decided to express recombinant Cdc45 transiently in these cells. We also coexpressed various fragments of Treslin in these cells. As shown in Fig. 8 A, we could immunoprecipitate recombinant Cdc45 efficiently with anti-Cdc45 antibodies. We found that the 1–921 fragment of Treslin was coimmunoprecipitated quite well with these antibodies. Significantly, this fragment largely excludes the principal TopBP1-interacting, Cdk-regulated domain of Treslin identified in this study. In control experiments, there was no Cdc45 or Treslin in immunoprecipitates with control antibodies. We could not observe good immunoprecipitation of full-length Treslin with Cdc45 (unpublished data).
Figure 8.
Treslin interacts with Cdc45 in human cells. (A) Treslin (1–921)-Myc and human Cdc45 were transiently expressed in U2OS cells. Cell lysates (lane 1) were immunoprecipitated (IP) with either control antibodies (lane 2) or anti–human Cdc45 antibodies (lane 3) and immunoblotted for the Myc tag (top) or Cdc45 (bottom). (B) Benzonase-digested nuclear lysates (lane 1) from interphase X. laevis egg extracts were immunoprecipitated with control (lane 2), anti-Cdc45 (lane 3), and anti-Treslin antibodies (lane 4). The samples were immunoblotted for Treslin (top) and Cdc45 (bottom).
Because we could not detect a complex between endogenous Cdc45 and Treslin in human cells, we decided to pursue this issue with X. laevis egg extracts. For this purpose, we prepared lysates of nuclei that had been incubated for 45 min in interphase egg extracts and then digested these lysates with the nuclease Benzonase. Thereafter, we immunoprecipitated the clarified digests with anti-Cdc45 and anti-Treslin antibodies and finally performed immunoblotting of the immunoprecipitates. As shown in Fig. 8 B, we could readily detect Treslin in anti-Cdc45 immunoprecipitates. There was also a significant, albeit lower, amount of Cdc45 in the anti-Treslin immunoprecipitates. It is possible that the anti-Treslin antibodies, which were raised against the N-terminal region of the protein, may occlude the Cdc45-interacting region of Treslin. Collectively, these experiments indicate that vertebrate Treslin, like yeast Sld3, interacts specifically with Cdc45. Moreover, Cdc45 appears to associate with a portion of these proteins that lies mostly upstream of the region that associates with Dpb11/TopBP1 in a Cdk-regulated manner (Tanaka et al., 2007b).
Discussion
It is well established that the Cdks are universal regulators of cell cycle transitions, but enumeration of the critical targets of these enzymes has been an ongoing challenge (Morgan, 1997). In the case of the G1/S transition, S-Cdk activity promotes the firing of early replication origins at the threshold of S phase (Araki, 2010). Subsequent waves of origin firing eventually result in complete replication of the genome. Recent advances in the budding yeast system have indicated that phosphorylation of Sld2 and Sld3 by S-Cdk plays a pivotal role in the triggering replication (Tanaka et al., 2007b; Zegerman and Diffley, 2007). Indeed, these modifications comprise the minimal set of phosphorylations by S-Cdk in controlling initiation of replication.
Phosphorylation of Sld2 and Sld3 promotes their association with distinct regions of Dpb11, another important replication protein. In particular, phosphorylation of Sld2 on T84 enables interaction with the C-terminal pair of BRCT repeats within Dpb11. Likewise, phosphorylation of Sld3 on T600 and S622 allows docking with the two N-terminal BRCT domains of Dpb11. An important question is whether this type of regulation exists in multicellular organisms, especially vertebrates. Assessment of this issue has been somewhat problematic because putative homologues of these proteins in higher eukaryotes appear to be quite different, at least superficially, from their yeast counterparts. The best candidate for vertebrate Sld2 is RecQ4 (Sangrithi et al., 2005; Matsuno et al., 2006). This protein shows some sequence similarity with Sld2, associates with TopBP1, and is necessary for replication. However, RecQ4, in contrast to Sld2, is dispensable for functional incorporation of Cdc45 into the replication machinery. Moreover, the binding of RecQ4 to TopBP1 is apparently not dependent on Cdk activity.
Recently, several candidates have emerged as potential vertebrate analogues of Sld3, namely Treslin, GEMC1, and DUE-B (Balestrini et al., 2010; Chowdhury et al., 2010; Kumagai et al., 2010; Sansam et al., 2010). All of these proteins associate with TopBP1 and are necessary for recruitment of Cdc45 to chromatin. GEMC1 appears to be a bona fide Cdk target, and the association of Treslin with TopBP1 depends on Cdk activity. However, among the three proteins, only Treslin appears to possess any sequence homology with Sld3, although this similarity is low and limited to a few patches within the proteins (Sanchez-Pulido et al., 2010).
In this study, we have sought to gain further insight into this issue by asking whether Treslin is a direct regulatory target of Cdk activity. Because human and X. laevis Treslin each contains dozens of potential Cdk phosphorylation sites, we attempted to narrow down potentially important regions of Treslin by mapping its TopBP1-interacting region. We found that the binding of TopBP1 to Treslin is relatively complex in that it involves two discontinuous regions of Treslin. However, one of these regions appears to play a relatively greater role in the interaction. Within this region, we focused our attention on two well-conserved and closely spaced SP/TP sites. Our results have indicated that phosphorylation of one of these sites (S1000 of human Treslin) is critical for both binding of TopBP1 and DNA replication. The sequence around this site shows some resemblance to the region in budding yeast Sld3 that contains the critical sites for S-Cdk. This region was not identified in previous sequence comparisons between Treslin and Sld3 (Sanchez-Pulido et al., 2010). Mutagenesis of the second residue in Treslin did not appear to affect binding to TopBP1 (and we have not detected phosphorylation of this site in mass spectrometric analyses). Thus, S1000 stands out as an especially important site of phosphorylation in Treslin.
Another important function of yeast Sld3 involves its interaction with Cdc45 (Kamimura et al., 2001; Tanaka et al., 2007b). In this study, we have established yet another similarity between Treslin and Sld3 by showing that Treslin interacts well with Cdc45. In yeast, Sld3 and Cdc45 can associate initially with chromatin in the absence of S-Cdk activity (Kamimura et al., 2001; Yabuuchi et al., 2006). Likewise, Treslin binds robustly to chromatin in egg extracts that have been treated with the Cdk inhibitor p27 (Kumagai et al., 2010). However, there appear to be some differences between the yeast and vertebrate systems. In particular, association of Cdc45 with chromatin in X. laevis egg extracts does depend on Cdk activity.
Overall, it appears that vertebrate Treslin and budding yeast Sld3 share many of their most important functional characteristics. Both proteins are able to associate with chromatin in the absence of S-Cdk activity. Each protein interacts specifically with Dpb11/TopBP1, and this interaction requires Cdk-dependent phosphorylation. Both polypeptides associate with Cdc45 and are necessary for incorporation of Cdc45 into the replicative helicase. Finally, both proteins are essential for DNA replication. Interestingly, Treslin and Sld3 have retained these key properties despite significant divergence at the primary sequence level. Nonetheless, we believe that it is reasonable to consider Treslin as the genuine functional counterpart of budding yeast Sld3 in vertebrate cells.
A question for the field in the future is how the Cdk-dependent association of Treslin and TopBP1 eventually leads to integration of Cdc45 into the replicative helicase. Although we have an increasingly complete accounting of the proteins that are necessary for replication in vertebrates, the actual biochemical functions of many of these proteins remain mysterious. Delineation of the regulatory relationship between Treslin and TopBP1 is a step forward toward a complete molecular understanding of replication in vertebrates.
Materials and methods
Cell culture and synchronization
U2OS cells were cultured in DME supplemented with 10% FBS, 50 U/ml penicillin, and 50 µg/ml streptomycin. For mitotic arrest, cells were treated with 0.2 µg/ml nocodazole for 18 h. Mitotically arrested cells were shaken off plates, washed twice with DME, and plated in fresh medium. For experiments involving inhibition of Cdk2, 25 µM roscovitine was added to cultures. Conditions for treatment of U2OS cells with control and Treslin siRNA (Stealth siRNA; Invitrogen) were previously described (Kumagai et al., 2010). For the experiments in this study, we used Treslin siRNA #1 (5′-GACCUGAGAGAAGAUUCAGAAGUUA-3′).
Plasmids
pcDNA5/TO-Treslin-Myc (encoding Treslin with a Myc3–nuclear localization sequence tag at the C-terminal end) was created from pcDNA5/TO-Treslin, which contains silent mutations that render Treslin resistant to the siRNA used in this study. The sequence encoding the Myc3–nuclear localization sequence tag was produced by PCR-based methods using DNA polymerase (Phusion; New England Biolabs, Inc.) and ligated to the 3′ end of the Treslin cDNA. Deletion mutants were created by PCR-based methods. Point mutations were created with the QuikChange kit using Pfu DNA polymerase (Agilent Technologies). All mutations were confirmed by DNA sequencing. A human Cdc45 cDNA was isolated by PCR from a human placental cDNA library and cloned into pcDNA5/TO.
Production of U2OS cell lines expressing Treslin and its mutants
U2OS T-REx cells (obtained from T.C. Spelsberg, Mayo Clinic, Rochester, MN) were maintained in the presence of 5 µg/ml Blasticidin S. These cells were transfected with pcDNA5/TO encoding various siRNA-resistant forms of wild-type and mutant Treslin. Cells were selected with 200 µg/ml Hygromycin B and 5 µg/ml Blasticidin S. Single colonies that expressed the desired protein upon addition of doxycycline were isolated and used for analysis.
Antibodies
Anti–human Treslin antibodies were previously described (Kumagai et al., 2010). Anti-Myc (ab9106), anti-FLAG (M2), anti–human TopBP1 (A300-111A), anti–human Cdc45 (H-300), and anti-tubulin antibodies (DM1A) were obtained from Abcam, Agilent Technologies, Bethyl Laboratories, Inc., Santa Cruz Biotechnology, Inc., and EMD, respectively. Anti–human Treslin phospho-S1000 antibodies were produced in rabbits against the phosphopeptide CGVVEE(pS)PEKGD in a commercial facility (Covance), affinity purified with the phosphopeptide, and absorbed with the nonphosphorylated version of this peptide.
Immunoprecipitation
U2OS cells were harvested and lysed in a buffer containing 20 mM Hepes-KOH, pH 7.5, 500 mM NaCl, 0.5% Triton X-100, 5 mM EDTA, 1 mM DTT, 10 mM β-glycerolphosphate, 1 mM NaF, 0.1 mM vanadate, 0.6 µM tautomycin, 10 µg/ml each of pepstatin, chymostatin, and leupeptin, and 1 mM PMSF. Lysates were cleared by centrifugation at 16,000 g for 10 min. Supernatants were diluted with 2.3 vol of 20 mM Hepes-KOH, pH 7.5, and cleared by centrifugation at 16,000 g for 10 min. Cell lysates were immunoprecipitated in the presence of 50 µg/ml ethidium bromide for 1.5 h with either anti–human Treslin or anti–human TopBP1 antibodies bound to protein A Dynabeads (Invitrogen). After the incubation, beads were washed three times with Hepes-buffered saline (HBS; 10 mM Hepes-KOH, pH 7.5, and 150 mM NaCl) containing 0.5% Triton X-100.
For immunoprecipitation of proteins from chromatin in X. laevis egg extracts, extracts were incubated with demembranated sperm nuclei (4,000 per microliter) for 45 min. Nuclei were isolated by centrifugation for 5 min at 6,100 g through a sucrose cushion containing 1 M sucrose dissolved in 20 mM Hepes-KOH, pH 7.5, 80 mM KCl, 2.5 mM potassium gluconate, and 10 mM magnesium gluconate. Pellets were resuspended in the 1-M sucrose cushion and centrifuged again. The pellets containing nuclear fractions were resuspended in 10 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 10 mM β-glycerolphosphate, 1 mM NaF, 0.1 mM sodium vanadate, 1 mM DTT, 0.6 µM tautomycin, 0.5 mM PMSF, and 10 µg/ml each of pepstatin, chymostatin, and leupeptin. 1 U/µl Benzonase (Sigma-Aldrich) was added to the nuclear fraction, and the mixture was incubated on ice for 30 min. The incubation was diluted with the same volume of 20 mM Hepes-KOH, pH 7.5, containing 100 µg/ml ethidium bromide and centrifuged at 16,000 g for 10 min. The supernatants were immunoprecipitated for 90 min with control, anti–X. laevis Cdc45, or anti–X. laevis Treslin antibodies.
Immunoblotting analysis
For quantitation of most protein interaction assays, immunoblots were subjected to dual-color analysis using the Odyssey Imaging System (LI-COR Biosciences). Myc-tagged versions of Treslin were probed with rabbit anti-Myc antibodies, and FLAG-tagged fragments of TopBP1 were detected with mouse anti-FLAG antibodies. The secondary antibodies used were Dylight 800–conjugated goat anti–rabbit antibodies (Thermo Fisher Scientific) and IRdye 680–conjugated goat anti–mouse antibodies (LI-COR Biosciences). For other immunoblotting analyses, an enhanced chemiluminescence detection kit (Thermo Fisher Scientific) was used with either HRP-conjugated secondary antibodies (Bio-Rad Laboratories) or the Clean-Blot reagent (Thermo Fisher Scientific). In these cases, quantitation was performed with the ImageJ program (National Institutes of Health).
Rescue of Treslin siRNA–treated cells
Expression of siRNA-resistant Treslin was induced in the U2OS-Treslin-Myc cell lines with 5–10 ng/ml doxycycline for 24 h before transfection with either 25 nM control siRNA or Treslin siRNA in the presence of Lipofectamine RNAiMAX (Invitrogen). Protein expression and DNA replication were analyzed 72 h after siRNA transfection. For replication assays, cells were incubated for 1 h in the presence of 10 µM EdU before detection with Click-iT reagent (Invitrogen) in conjunction with Alexa Fluor 488 azide. Expressed proteins were detected by immunofluorescence using anti-Myc and Alexa Fluor 594–conjugated anti–rabbit antibodies. Samples were mounted in Vectashield (Vector Laboratories) and viewed at RT. Microscope images were observed under an Axioplan microscope (Carl Zeiss) equipped with a Plan Neofluar (40×/0.75 NA) objective. The images were captured with an RT slider charge-coupled device camera (SPOT; Imaging Solutions) using SPOT software and imported into Photoshop (Adobe).
Interaction of Treslin and BRCT domains of TopBP1
Lysates from U2OS cells containing Myc-tagged full-length Treslin or its fragments were prepared as described in the Immunoprecipitation section for immunoprecipitation experiments. In most cases, Treslin or its fragments were expressed transiently after transfection with pcDNA5/TO vectors. In some cases, the expression of Treslin-Myc was induced with 1 µg/ml doxycycline in stable U2OS T-REx clones. Lysates from 105 cells were mixed with 0.1 µg FLAG-tagged BRCT I–II fragment from TopBP1 and subjected to immunoprecipitation for 2 h with anti-FLAG antibodies bound to protein G Dynabeads. Beads were isolated with a magnet and washed three times with HBS containing 0.5% Triton X-100. For binding experiments in X. laevis egg extracts, bacterially produced forms of Treslin were incubated for 60 min in extracts containing 4,000 demembranated sperm nuclei per microliter in the absence or presence of 2 µM GST-p27. Nuclei were isolated by centrifugation through 1 M sucrose and lysed in a buffer containing 450 mM NaCl as previously described (Kumagai et al., 2010). Lysates were diluted to reduce the concentration of NaCl, incubated in the absence or presence of the FLAG-tagged BRCT I–II fragment from TopBP1, and immunoprecipitated with anti-FLAG antibodies bound to protein G Dynabeads.
Production of recombinant proteins
Wild-type and mutant forms of human Treslin (893–1,257)-Myc were amplified by PCR and cloned into pET30 that had been digested with NcoI and NotI. The resulting plasmids were used for expression of His6-tagged versions of the fragments in Escherichia coli Rosetta (DE3)pLysS cells (EMD). E. coli expressing the fragments were resuspended in 0.2 M sodium borate, pH 8.2, containing 0.5 M NaCl, 0.5% NP-40, 5 mM EGTA, and 1 mM PMSF and then were sonicated. The lysates were clarified by centrifugation for 10 min at 22,000 g and incubated with nickel agarose beads for 2 h. The proteins were eluted with 150 mM imidazole in HBS. To produce a GST fusion protein containing amino acids 989–1,011 of human Treslin and the S1000A mutant of this peptide, the appropriate DNA was amplified by PCR and cloned into pGEX4T-3 that had been digested with BamHI and EcoRI. These GST fusion proteins were prepared in bacteria as described previously (Lee et al., 2005). GST-p27 was prepared in the same manner.
In vitro phosphorylation assays
Human Cdk2 and His6-tagged human cyclin E proteins were expressed separately in baculovirus-infected Sf9 cells. Lysates from Cdk2-expressing cells were mixed with His6–cyclin E that had been bound to nickel agarose beads. The mixtures were incubated at RT in the presence of 0.5 mM ATP and 10 mM MgCl2 for 20 min as previously described (Shou and Dunphy, 1996). The active Cdk2–cyclin E complex was isolated from the nickel beads by elution with 150 mM imidazole in HBS. For typical assays, Cdk2–cyclin E was incubated for 5 min with 1 µg substrate in a volume of 20 µl containing 10 mM Tris-HCl, pH 7.5, 1 µCi γ-[32P]ATP, 10 µM ATP, 10 mM MgCl2, and 1 mM DTT. For reactions lacking γ-[32P]ATP, the concentration of ATP was increased to 1 mM, and the incubation was performed for 30 min. Reactions were stopped by the addition of SDS gel sample buffer. Samples were subjected to SDS-PAGE and analyzed by phosphorimaging or immunoblotting, as appropriate.
MS analysis
Treslin was isolated by incubating a FLAG-tagged truncation of TopBP1 containing BRCT domains I–VI in X. laevis nuclear lysates (Kumagai et al., 2010). This fragment was bound to anti-FLAG antibodies that had been coupled to protein G Dynabeads. After washing the beads three times with 20 mM Hepes-KOH, pH 7.5, 80 mM NaCl, 2.5 mM EGTA, and 0.1% NP-40 and once with 20 mM Hepes-KOH, pH 7.5, the proteins were eluted with SDS sample buffer. After gel separation, the selected gel band was in-gel digested with trypsin as previously described (Shevchenko et al., 2006). Recovered peptides were extracted with two changes of 5% formic acid and 50% acetonitrile, and pooled extracts were dried down in a vacuum centrifuge. Dried peptide extracts were redissolved in 15 µl of 5% acetic acid, sonicated, and analyzed by liquid chromatography MS/MS.
Liquid chromatography MS/MS was performed on a nanoLC system (Ultimate3000; Dionex) interfaced online with a hybrid mass spectrometer (LTQ Orbitrap; Thermo Fisher Scientific) via the robotic nanoflow ion source TriVersa (Advion BioSciences) as previously described (Junqueira et al., 2008). To minimize column memory, several washing steps and blanks were introduced before each run. Spectra acquired during the last blank run were searched against a protein sequence database.
Acquired MS/MS spectra were first converted to the Mascot generic format and then searched against a full protein database (National Center for Biotechnology Information) and against the sequence of Treslin using MASCOT software (v.2.2.0; Matrix Science) under the following settings: mass tolerance was set as ±10 ppm for precursors and ±0.5 D for fragments; variable modification was Propionamide (C), Carbamidomethyl (C), and Oxidation (M); and no enzyme specificity (for the search against Treslin sequence only) or trypsin (for the search against the full National Center for Biotechnology Information database). All hits with peptide ion scores >25 were then manually evaluated.
Acknowledgments
We are grateful to laboratory members for comments on the manuscript. We would also like to thank Rochelle Diamond for assistance with flow cytometry, Alex Varshavsky for use of the Odyssey system, and Lydia Dennis for technical assistance.
This work was supported by National Institutes of Health grants GM043974 and GM070891 to W.G. Dunphy.
Footnotes
Abbreviations used in this paper:
- BRCT
- BRCA1 C terminus
- HBS
- Hepes-buffered saline
- MS
- mass spectrometry
- S-Cdk
- S phase Cdk
- SP/TP
- Ser-Pro/Thr-Pro
References
- Araki H. 2010. Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell Biol. 22:766–771. 10.1016/j.ceb.2010.07.015 [DOI] [PubMed] [Google Scholar]
- Balestrini A., Cosentino C., Errico A., Garner E., Costanzo V.. 2010. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat. Cell Biol. 12:484–491. 10.1038/ncb2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M. 2007. Cell biology: a switch for S phase. Nature. 445:272–274. 10.1038/445272a [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Liu G., Kemp M., Chen X., Katrangi N., Myers S., Ghosh M., Yao J., Gao Y., Bubulya P., Leffak M.. 2010. The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol. Cell. Biol. 30:1495–1507. 10.1128/MCB.00710-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. 2004. Regulation of early events in chromosome replication. Curr. Biol. 14:R778–R786. 10.1016/j.cub.2004.09.019 [DOI] [PubMed] [Google Scholar]
- Fu Y.V., Walter J.C.. 2010. DNA replication: metazoan Sld3 steps forward. Curr. Biol. 20:R515–R517. 10.1016/j.cub.2010.05.033 [DOI] [PubMed] [Google Scholar]
- Garcia V., Furuya K., Carr A.M.. 2005. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst.). 4:1227–1239. 10.1016/j.dnarep.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Takisawa H.. 2003. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 22:2526–2535. 10.1093/emboj/cdg238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves I., Petojevic T., Pesavento J.J., Botchan M.R.. 2010. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 37:247–258. 10.1016/j.molcel.2009.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira M., Spirin V., Santana Balbuena T., Waridel P., Surendranath V., Kryukov G., Adzhubei I., Thomas H., Sunyaev S., Shevchenko A.. 2008. Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 7:3382–3395. 10.1021/pr800140v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y., Tak Y.S., Sugino A., Araki H.. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097–2107. 10.1093/emboj/20.8.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G.. 2010. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 140:349–359. 10.1016/j.cell.2009.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24:1208–1219. 10.1101/gad.1933010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Gold D.A., Shevchenko A., Shevchenko A., Dunphy W.G.. 2005. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 16:5269–5282. 10.1091/mbc.E05-07-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M., Morgan D.O.. 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 434:104–108. 10.1038/nature03329 [DOI] [PubMed] [Google Scholar]
- Matsuno K., Kumano M., Kubota Y., Hashimoto Y., Takisawa H.. 2006. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 26:4843–4852. 10.1128/MCB.02267-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayya V., Lundgren D.H., Hwang S.I., Rezaul K., Wu L., Eng J.K., Rodionov V., Han D.K.. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2:ra46. 10.1126/scisignal.2000007 [DOI] [PubMed] [Google Scholar]
- Méndez J., Stillman B.. 2003. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 25:1158–1167. 10.1002/bies.10370 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261–291. 10.1146/annurev.cellbio.13.1.261 [DOI] [PubMed] [Google Scholar]
- Pacek M., Tutter A.V., Kubota Y., Takisawa H., Walter J.C.. 2006. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 21:581–587. 10.1016/j.molcel.2006.01.030 [DOI] [PubMed] [Google Scholar]
- Remus D., Diffley J.F.. 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21:771–777. 10.1016/j.ceb.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L., Diffley J.F., Ponting C.P.. 2010. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr. Biol. 20:R509–R510. 10.1016/j.cub.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Sangrithi M.N., Bernal J.A., Madine M., Philpott A., Lee J., Dunphy W.G., Venkitaraman A.R.. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 121:887–898. 10.1016/j.cell.2005.05.015 [DOI] [PubMed] [Google Scholar]
- Sansam C.L., Cruz N.M., Danielian P.S., Amsterdam A., Lau M.L., Hopkins N., Lees J.A.. 2010. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 24:183–194. 10.1101/gad.1860310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman B.A., Lindstrom D.L., Harlow E.. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA. 95:10453–10458. 10.1073/pnas.95.18.10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R.A., Holzen T.M.. 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41:237–280. 10.1146/annurev.genet.41.110306.130308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Shou W., Dunphy W.G.. 1996. Cell cycle control by Xenopus p28Kix1, a developmentally regulated inhibitor of cyclin-dependent kinases. Mol. Biol. Cell. 7:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Araki H.. 2010. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 119:565–574. 10.1007/s00412-010-0291-8 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Tak Y.S., Araki H.. 2007a. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2:16. 10.1186/1747-1028-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Umemori T., Hirai K., Muramatsu S., Kamimura Y., Araki H.. 2007b. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 445:328–332. 10.1038/nature05465 [DOI] [PubMed] [Google Scholar]
- Van Hatten R.A., Tutter A.V., Holway A.H., Khederian A.M., Walter J.C., Michael W.M.. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159:541–547. 10.1083/jcb.200207090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi H., Yamada Y., Uchida T., Sunathvanichkul T., Nakagawa T., Masukata H.. 2006. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 25:4663–4674. 10.1038/sj.emboj.7601347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P., Diffley J.F.. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 445:281–285. 10.1038/nature05432 [DOI] [PubMed] [Google Scholar]