Abstract
Background
Safflower (Carthamus tinctorius L.) is a difficult crop to genetically transform being susceptible to hyperhydration and poor in vitro root formation. In addition to traditional uses safflower has recently emerged as a broadacre platform for the production of transgenic products including modified oils and pharmaceutically active proteins. Despite commercial activities based on the genetic modification of safflower, there is no method available in the public domain describing the transformation of safflower that generates transformed T1 progeny.
Results
An efficient and reproducible protocol has been developed with a transformation efficiency of 4.8% and 3.1% for S-317 (high oleic acid content) and WT (high linoleic acid content) genotypes respectively. An improved safflower transformation T-DNA vector was developed, including a secreted GFP to allow non-destructive assessment of transgenic shoots. Hyperhydration and necrosis of Agrobacterium-infected cotyledons was effectively controlled by using iota-carrageenan, L-cysteine and ascorbic acid. To overcome poor in vitro root formation for the first time a grafting method was developed for safflower in which ~50% of transgenic shoots develop into mature plants bearing viable transgenic T1 seed. The integration and expression of secreted GFP and hygromycin genes were confirmed by PCR, Southern and Western blot analysis. Southern blot analysis in nine independent lines indicated that 1-7 transgenes were inserted per line and T1 progeny displayed Mendelian inheritance.
Conclusions
This protocol demonstrates significant improvements in both the efficiency and ease of use over existing safflower transformation protocols. This is the first complete method of genetic transformation of safflower that generates stably-transformed plants and progeny, allowing this crop to benefit from modern molecular applications.
Background
Safflower is a versatile crop with several desirable traits, multiple applications and is well adapted to semi-arid conditions in the tropics and subtropics. Safflower is grown for its edible oil (high oleic and high linoleic varieties), high-protein seed cake, animal meal, bird seed and for traditional medicine [1,2]. Apart from these traditional uses safflower has recently emerged as a broadacre platform for the production of transgenic products, including modified oils such as gamma-linolenic acid [3] and pharmaceutically active proteins including human insulin and apolipoprotein [4,5]. Safflower has become an industrial crop production platform based on low out-crossing and weediness habits, a different appearance from other oilseed crops such as canola and excellent agronomic traits such as taproot architecture that accesses sub-soil water reserves [6].
Despite commercial activities based on the genetic modification of safflower, there is no method available in the public domain describing the generation and analysis of transgenic T1 progeny. The lack of a reliable regeneration of transgenic T1 progeny in safflower not only limits its potential as an industrial crop production platform but also the application of modern molecular techniques to investigate and improve this economically-important plant. Undoubtedly safflower is a difficult crop to genetically engineer, and a large body of literature describes a series of limitations in tissue culture approaches [7-9]. Firstly, under in vitro conditions safflower shoots are susceptible to hyperhydration, a physiological disorder with aberrant morphology, characterized by swollen translucent leaves and brittle stems. Hyperhydration is likely caused by two important factors - the gelling agent of the media and high humidity in culture vessels. The risk of hyperhydration increases in post-transformation selection because of prolonged culture on antibiotic selection media. Therefore the use of antibiotic selection agents in safflower transformation methods presents a dilemma: antibiotic selection agents are required to reduce the number of 'false positives' yet also contribute to the occurrence of hyperhydration. One possible alternative to antibiotic selection agents in the culture media is the use of fluorescent proteins, such as GFP, as a visual marker to aid in the identification of transgenic shoots [10].
A second major limitation in safflower transformation is the poor formation of roots from transformed shoots, a vital developmental step towards the generation of mature T0 plants and seeds. In vitro induced roots are often very weak and frequently fail to survive the transfer from tissue culture media to soil [11]. Faced with the dual issues of hyperhydration and poor in vitro root formation an alternative method [12] of safflower transformation was developed, called in planta transformation. In this approach the embryo of the young seedling is pierced and co-cultivated with an Agrobacterium harboring the T-DNA vector. Over time the embryo develops into mature safflower plants containing segments of transformed sectors, so called chimeric T0 plants. Although this approach overcomes both hyperhydration and in vitro root formation there is little convincing evidence that this in planta method reliably generated transformed T0 plants or, most critically, transformed T1 progeny. Poor in vitro root formation was also reported in other dicot crops and has been addressed using grafting techniques to enhance plant recovery from somatic embryos and genetically transformed shoots [13-16] including sunflower [17,18].
Here we report the development of a protocol for the reliable production of genetically-transformed safflower. We used antioxidants (L-cysteine, ascorbic acid) during co-cultivation and selection to reduce necrosis, iota-carrageenan with modified gelling strength to control the hyperhydration and a non-cytotoxic GFP visual marker system to identify GM-shoots. GM-shoots were grafted, using a novel in vivo grafting technique, to recover and establish mature transgenic safflower plants. Various molecular techniques demonstrate that this method generated mature T0 plants and viable stably-transformed T1 seed. This complete report of a reliable method for the genetic transformation of safflower will help this important crop to benefit from advances in plant biotechnology.
Results and Discussion
1. Construction and testing of a binary vector suitable for safflower transformation
The binary vector pORE3 [19] was modified so that the kanamycin resistance gene, npt-II, was replaced with the hygromycin resistance gene, hph, from the pVEC8 vector series [20], creating pORE6. This version of hph contains a catalase-1 intron rendering the gene inactive in Agrobacterium. In our hands this intron-interrupted version of hph greatly reduced bacterial overgrowth in post-transformation selection and regeneration experiments, similar to previously reported results [20]. Preliminary experiments in safflower transformation experiments indicated that a GFP located within the cytoplasm was cytotoxic (Additional file 1), an effect that was previously overcome by targeting the GFP to the endoplasmic reticulum of the cell [10]. To overcome cytotoxicity effects a GFP located to the apoplast was designed following the protocol [21] where the secretion peptide of conglycinin was translationally-fused to the N-terminal of GFP and a four glycine amino acid stretch was fused to the C-terminal of GFP. The secreted GFP was placed under the control of the 35S promoter, generating the plant binary vector pCW265 (Figure 1). Safflower was transformed with pCW265 and the location of this secreted GFP was confirmed as apoplastic (Additional file 2). pCW265 contains a multiple cloning site region to allow convenient ligation of genes of pathways of interest for expression in safflower.
Figure 1.
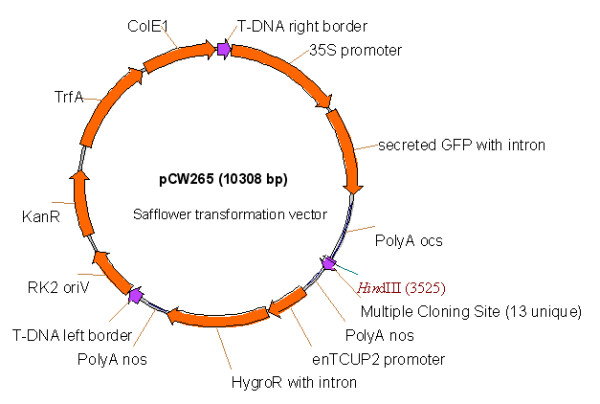
Schematic representation of the T-DNA region of the T-DNA binary vector developed for the transformation of safflower. Secretion of the GFP into the apoplast is required for healthy safflower development. Both the GFP and hygromycin CDS regions include a catalase-1 intron to abolish expression in Agrobacterium. The multiple cloning site (MCS) includes unique sequences for digestion by BstEII, SbfI, HindIII, XbaI, XmaI, SmaI, SalI, ClaI, EcoICRI, SacI, Acc65I, Spe1 and EcoRV. A complete explanation of the features of the vector backbone are found in Coutu et al., [19] and in the text.
2. Optimized Agrobacterium pre-treatments and cell density for T-DNA transfer into safflower
Preliminary transformation experiments with pCW265 determined that the optimal cell density of Agrobacterium for infection was OD600 = 0.4 for both genotypes of safflower (Figure 2A) and this density was used throughout all subsequent experiments. Acetosyringone (AS) [22] and spermidine (SPR) [23] are potent inducers of virulence (vir) genes of Agrobacterium and are used in many plant species for successful transformation. The effect of AS and SPR on T-DNA delivery into safflower cotyledons was tested (Figure 2B) by scoring the percentage of calli with a GFP fluorescence signal after co-cultivation with Agrobacterium harbouring pCW265 and two weeks culture on S2 selection media (Table 1). A combination of 100 μM of AS and 1.0 mM of SPR was found to be optimal for both genotypes, with as many as 70-90% of explants scoring positive for secreted GFP expression. A further increase in SPR concentration from 1 mM to 1.5 mM resulted in reduced T-DNA delivery in both genotypes. Our results suggest that the combination of AS and SPR as pretreatments for Agrobacterium improved the transformation efficiency in safflower.
Figure 2.
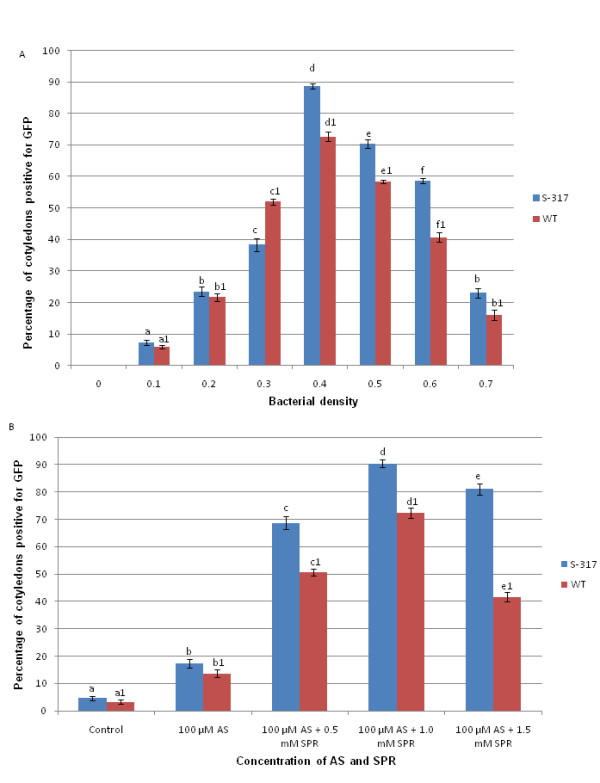
Optimization of Agrobacterium infection pre-treatments for T-DNA transfer into safflower cotyledon explants. (A) Effect of bacterial density on transient expression of GFP in cotyledons of S-317 and WT. (B) Acetosyringone and spermidine concentration in inoculation media (Winnans AB salts) and its effect on GFP expression two weeks after Agrobacterium co-cultivation (OD600 = 0.4). Each value is the mean ± SE of three independent experiments, each with 30 cotyledons. Significant differences between treatments are indicated by different letters.
Table 1.
Composition of media for transformation of safflower from cotyledon explants.
| Component | S1 Co-cultivation | S2 Callus induction | S3 Shoot initiation | S4 Shoot out-growth | S5 Shoot elongation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| S-317 | WT | S-317 | WT | S-317 | WT | S-317 | WT | S-317 | WT | |
| Media | MS[33] | MS | MS | MS | MS | MS | MS | MS | MS | MS |
| Growth regulators | ||||||||||
| TDZ | - | 1 | - | 1 | - | - | - | - | - | - |
| NAA | 0.1 | 0.1 | 0.1 | 0.1 | - | - | - | - | - | - |
| BA | 1 | - | 1 | - | 1 | - | - | - | - | - |
| KN | - | - | - | - | - | 1 | - | - | - | - |
| 2iP | - | - | - | - | - | - | - | 0.5 | - | 0.5 |
| GA | - | - | - | - | - | - | - | - | 0.1 | 0.1 |
| Antioxidants | ||||||||||
| L-cysteine | 50 | 50 | - | - | - | - | - | - | - | - |
| Ascorbic acid | 15 | 15 | - | - | - | - | - | - | - | - |
| PVP | - | - | - | - | - | - | - | - | 500 | 500 |
| Antibiotics | ||||||||||
| Hygromycin | - | - | 20 | 20 | 20 | 20 | 18 | 18 | - | - |
| Cefotaxime | - | - | 250 | 250 | 250 | 250 | 250 | 250 | 250 | 250 |
| Timentin Others | - | - | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 |
| Iota-carrageenan | - | - | 1500 | 1500 | 1500 | 1500 | 1500 | 1500 | - | - |
| AgNo3 | - | - | 3 | 3 | 3 | 3 | 1.5 | 1.5 | 1.5 | 1.5 |
| Sucrose | 30000 | 30000 | 30000 | 30000 | 30000 | 30000 | 20000 | 20000 | 20000 | 20000 |
| Agar | 9000 | 9000 | 9000 | 9000 | 9000 | 9000 | - | - | - | - |
| Phytagel | - | - | - | - | - | - | 5000 | 5000 | 5000 | 5000 |
Amounts indicate the mg l-1 of each component. S-317 and WT refer to high oleic acid content seeds and high linoleic acid content seeds, respectively.
3. Necrosis is significantly reduced by antioxidants L-cysteine and ascorbic acid
It has been demonstrated previously that co-cultivation of safflower explants with Agrobacterium decreases regeneration frequency compared with non-transformed controls and the addition of AS further reduced safflower regeneration because of the increased bacterial infectivity and resulting hypersensitive response [8]. Necrosis at the proximal end of the cotyledons following co-cultivation with Agrobacterium was observed in the present study. As poor regeneration from transformed cells ultimately limits the production of transgenic plants, we focused on improving the regeneration by reducing necrosis of the cotyledon through the addition of antioxidants L-cysteine and ascorbic acid (Figure 3). The best concentration of antioxidants was 15 mg l-1 ascorbic acid and 50 mg l-1 L-cysteine which reduced necrosis of cotyledons by half in the two genotypes tested. The antioxidants may help the post-infection survival of competent cells in and around the proximal end of the cotyledon, thereby increasing the number of transformed cells surviving to form calli and shoots. The combination of L-cysteine and ascorbic acid in co-cultivation medium may be also minimize cell death caused by the hypersensitive response of the cotyledon cells due to Agrobacterium infection.
Figure 3.
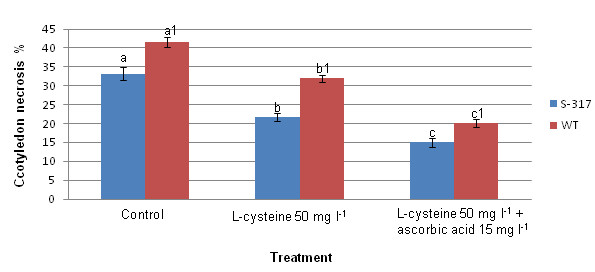
The effect of L-cysteine and ascorbic acid on cotyledon necrosis. Each value is the mean ± SE of three independent experiments, each with 30 cotyledons. Significant differences between treatments are indicated by different letters in each group of bars.
4. Plant regeneration without hyperhydration
Safflower cotyledon explants were chosen as the target tissue for genetic modification. In the WT genotype MS media with 1 mg l-1 TDZ and 0.1 mg l-1 NAA was found optimal for induction multiple shoots with little or no callus formation. In contrast, the S-317 genotype required MS media with 1 mg l-1 BA and 0.1 mg l-1 NAA for the induction of multiple shoots. With the tested combination of hormones the frequency of shoot regeneration ranged from 85-90% with as many as 8-10 shoots emerging from each cotyledon explant. Such high efficiencies of shoot regeneration with similar phytohormone regimes has been reported [24].
A major problem faced both in this and other safflower transformation studies is the hyperhydration of transgenic shoots which result in the loss of a large proportion of transgenic shoots [7,8,25]. Different concentrations of agar and iota-carrageenan were tested for their ability to reduce symptoms of hyperhydricity in two safflower genotypes that had previously been inoculated with Agrobacterium (Figure 4A). Increasing the agar concentration from 8 g l-1 to 9 g l-1 reduced the incidence of hyperhydration from 20% to 4% in S-317 and from 30% to 9% in WT. When iota-carrageenan was added at 1.5 g l-1 to MS media with 9 g l-1 agar, hyperhydration was further reduced to 0.33% and 4.0% in S-317 and WT cotyledons, respectively. Further increases in the concentration of iota-carrageenan from 1.5 g l-1 to 2.5 g l-1 abolished hyperhydration but also severely impaired the growth (data not shown). The use of 9 g l-1agar and 1.5 g l-1 iota-carrageenan was effective in controlling the hyperhydration in both genotypes of safflower without inhibiting growth and shoot proliferation, and these conditions were maintained up to the shoot out-growth stage (S4 media, Table 1). Figure 4B-D shows an example of a hyperhydrated shoot, regenerated on MS media with 8 g l-1 agar, and a healthy shoot, regenerated on modified MS media containing the optimised agar and iota-carrageenan concentrations. A similar reduction in hyperhydration was reported in Eucalyptus shoots using iota-carrageenan [26]. The mechanism by which iota-carrageenan reduces hyperhydration is unclear but may be related to polysaccharides, such as galactans, as part of their structure that may play a role in reducing osmotic stress in the cell wall.
Figure 4.
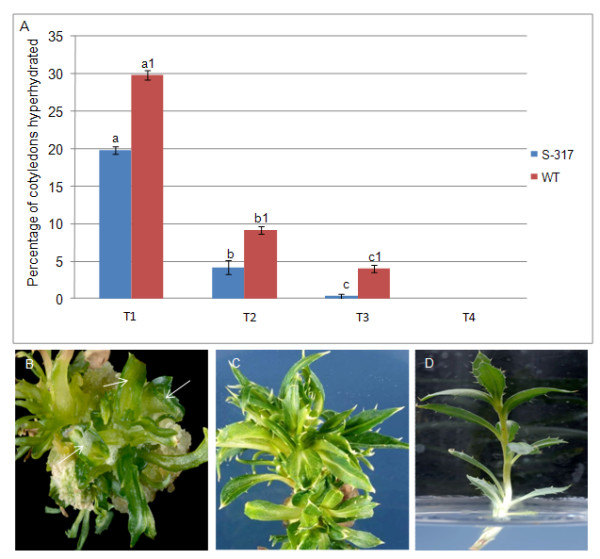
The effect of iota- carrageenan and modified agar concentration on hyperhydration of safflower shoots. (A) Media composition and percentage of cotyledons hyperhydrated after co-cultivation with Agrobacterium and 3-4 weeks on selection medium (Table 1). T1 = S2 + 8 g l-1 Agar; T2= S2+9 g l-1 Agar; T3 = S2 + 9 g l-1 Agar +1.5 g l-1 carrageenan; T4 = S2 + 9 g l-1 Agar +2.5 g l-1 carrageenan. Each value is the mean ± SE of three independent experiments, each with 30 cotyledons. Significant differences between media composition are indicated by different letters in each group of bars. (B) An example of hyperhydrated shoots grown on MS media with 8 g l-1 agar and the arrow indicates the abnormal appearance of leaf. (C) Proliferation of normal and healthy shoots on MS media with 9.0 g l-1 agar and 1.5 g l-1 iota-carrageenan; (Note the sharp margins on leaves compared to the image in panel B). (D) A single healthy shoot on elongation media, S5, and ready for grafting.
5. Visual screening of transgenic shoots using a secreted GFP
In order to reduce the prolonged culture on antibiotic selection media and to identify transformed shoots at early stages of regeneration, a secreted GFP marker was developed for use in the present study. The presence of the secreted GFP was monitored at nearly all developmental stages (Figure 5) of the regeneration and was detectable as early as 5-7 days post-infection and reached maximum expression within 2-3 weeks. Strong expression of the secreted GFP in leaf guard cells (Figure 5E) was most commonly used to detect transgenic shoots. As soon as a shoot was confirmed as a transgenic, via detection of the secreted GFP, the shoot was transferred to media lacking hygromycin to further reduce the risk of hyperhydration.
Figure 5.
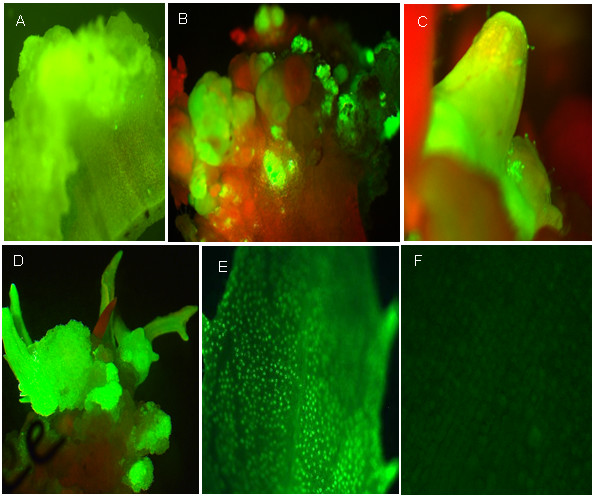
The use of a GFP visual marker to rapidly identify transgenic callus and shoots of safflower. (A) GFP expression in proximal end of the cotyledon two weeks after Agrobacterium infection. (B) Shoot bud initiation from the proximal end of cotyledon in the presence of 18 mg l-1 hygromycin after four weeks of co-cultivation (C-D) Transformed shoots regenerating on selection media in presence of 18 mg l-1 hygromycin (E) Visualization of GFP signals in guard cells of a transgenic leaf. (F) A non-transformed leaf/shoot showing no GFP signal and low background fluorescence.
6. Grafting technique to recover transgenic shoots into mature plants
Despite generating healthy elongated transgenic shoots (Figure 4B) we routinely failed to regenerate roots from transformed shoots, similar to reports in previous studies [7-9]. In most of the cases on rooting media, callus formation was observed from the base of the shoot yet no root formation was observed even after 4-6 weeks of culture with different hormone regimes. Our failures to generate rooted safflower plants forced the development of a grafting procedure to rescue transgenic shoots to allow their development into mature genetically-modified safflower plants.
Scions isolated from elongated shoots were grafted onto rootstocks to overcome the poor de novo root formation in safflower (Figure 6). When scions were 1.5-2.0 cm long and rootstocks three weeks old, our grafting protocol resulted in approximately 50% success rate as assessed by their continued growth into mature safflower plants (Figure 6). Unsuccessful grafts were obvious after 3-5 days, whilst successful grafts flourished and new leaves were formed within two weeks. The most critical factor for a successful grafting was the length of the scion and age of the rootstock. Scions of 0.5-1 cm long had a lower survival rate (15 to 20%) than larger ones with size of 1.0-2 cm (45 to 50%). Similarly, high survival rates were reported with larger scions (0.6-1.5 cm) in grafted cotton [15,27]. Small scions became dehydrated despite the presence of the silicone tube and parafilm around the graft point. With smaller scions it was also difficult to make good contact between the scion and rootstock. The effect of the rootstock genotype on the survival of grafts was also evaluated however the overall scion and rootstock varietal differences were statistically not significant (Table 2). In our glasshouse conditions grafted safflower plants matured after 3 months and produced between 2 and 5 flower heads (Figure 6 F), each head bearing between 5 and 30 T1 seeds.
Figure 6.

Grafting of transgenic shoots on to seedling rootstocks of safflower. (A) Rootstock seedling with well established root system and the transgenic scion, upper left corner. (B) V-shaped transgenic scion to be grafted onto a prepared seedling rootstock. (C) Rubber ring holding the scion and rootstock. (D) Graft union protected by parafilm. (E) Well established graft union. (F) Grafted safflower plant during seed development with multiple flower heads.
Table 2.
Effect of rootstock genotype on the survival of grafted S-317 safflower shoots
| Rootstock Genotypea | Number of shoots grafted b | Mean number of shoots survived per treatment c ± S.E | Percentage of success |
|---|---|---|---|
| WT | 16 | 6.6 ± 0.3 d | 39% |
| S-317 | 16 | 8.3 ± 0.6 d | 52% |
a 3 week old root stock;
b Scion size approximately 1.5-2.0 cm
c Graft survival measured as new leaves formed within 3 weeks of grafting
d Each treatment has 16 scions with 3 replications. Means were compared using the Multiple Pair wise Comparison Procedure (Holm-Sidak method) and found to be not significantly different.
The survival of transgenic shoots and plants throughout the entire transformation protocol is presented in Table 3, a data set generated from 20 different infections that produced 110 mature GM safflower plants. A combination of improvements were required to generate healthy and elongated GFP positive shoots, including optimised T-DNA transfer conditions and reduced hyperhydricity and necrosis. Grafting was, however, absolutely required to rescue GM-shoots to allow their development into mature plants. Grafting is a reasonably robust method to recover these transgenic shoots, with an approximate 40 to 50% success rate (Table 2 and 3). Notably, such young grafts always survived to develop into mature seed-setting plants, suggesting that our protocol has addressed many issues regarding the acclimatisation of safflower from tissue culture conditions to soil grown conditions.
Table 3.
Survival of transgenic shoots and plants through critical steps of the transformation protocol
| Genotype | Cotyledons co-cultivated with Agrobacteriuma | Shoots ready to graft obtainedb | Successful graftsc | Mature plants obtainedd | Final transformation efficiencye |
|---|---|---|---|---|---|
| S-317 | 1206 | 115 | 58 | 58 | 4.8% |
| WT | 1660 | 95 | 52 | 52 | 3.1% |
| combined | 2866 | 210 | 110 | 110 | 3.8% |
a Summary of a total of 20 independent experiments
b Shoots are positive for GFP and at least 3 cm in length
c Successful grafts as judged by new leaf growth within 2 weeks of grafting, yet still growing in controlled tissue culture conditions
d Mature plants grown in soil and in a glasshouse
e Transformation efficiency calculated as the (mature plants/infected cotyledons)x100
7. Molecular characterization of transgenic plants
We report here the generation of 110 grafted T0 plants, each scored as positive for a GFP signal, and the presence of T-DNA sequences tested by PCR amplification on genomic DNA, and a sample of these are shown in Figure 7. The analysis of PCR amplification products revealed in all cases the presence of DNA encoding both the secreted GFP and hygromycin resistance genes in all transgenic lines and its absence in the untransformed negative control, indicating the selection procedure with hygromycin and GFP were optimal. Southern blot analysis was used to determine to further confirm T-DNA integration into T0 lines and also as an estimate of the number of T-DNA insertions per line. Total genomic DNA was digested with HindIII, an enzyme that cleaves only once in the T-DNA region and hybridized with a radioactive GFP probe. Different integration patterns of T-DNA and insertion numbers were found in T0 plants indicating independent insertion events had been generated. The number of T-DNA insertions ranged from 1-7 and a sample of these patterns is shown in Figure 8. The GFP expression was further confirmed via Western blot on 11 T0 transgenic plants (Figure 9A and 9B), all showing the expected ~30 kDa protein band. In contrast, no corresponding bands were detected in the negative control plants.
Figure 7.
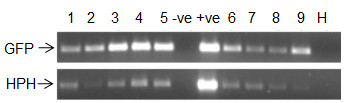
PCR amplification of transgenes from T0 safflower lines. Lanes 1-5 are PCR products generated from five genetically-modified S-317 lines, and lanes 8-11 are generated from four genetically-modified WT lines. -ve = Negative control non-transformed lines of S-317. +ve = Positive control (plasmid DNA from pCW265), H = sterile distilled water.
Figure 8.
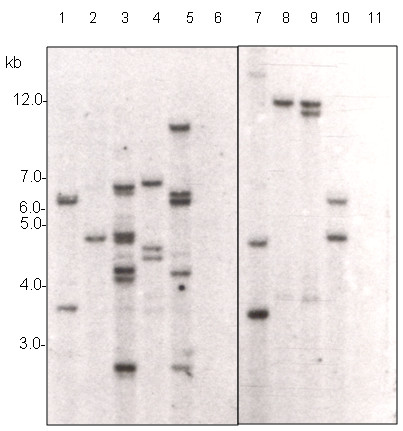
Southern-blot analysis of nine independent lines of transgenic safflower. Genomic DNA was prepared from the leaves of independent T0 lines from two varieties; S317 or WT. gDNA was digested with the restriction enzyme HindIII and probed using a labeled GFP sequence. Lane 1-5 transgenic plants from S-317 and lane 7-10 transgenic plants from WT. Lanes 6 and 11 are non-transformed S-317 and WT plants, respectively
Figure 9.
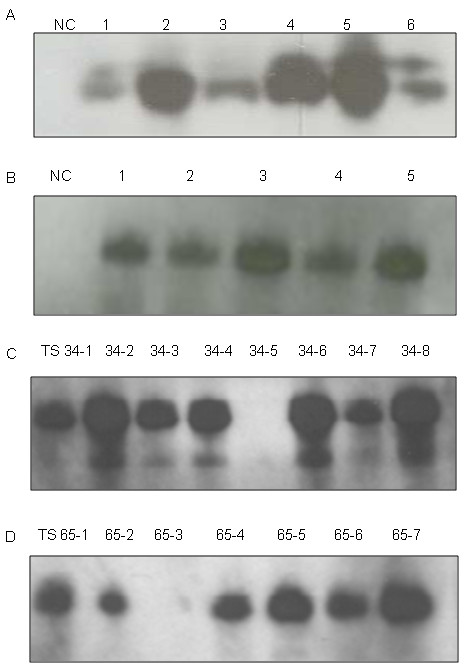
Western blot analysis of GFP expression in T0 and T1 progeny of transgenic safflower plants. (A) Lane 2-7 T0 plants from WT genotype. (B) Lane 2-6 T0 plants from S-317 genotype, Non-transgenic safflower were used as a negative control (NC). (C) Lane1-8 T1 transgenic line TS-34 and randomly selected progeny (WT genotype). (D) Lane 1-7 transgenic line TS-65 and randomly selected progeny (S-317 genotype).
A key omission in all safflower protocols published to date is the lack of evidence for the production of stably-transformed progeny with Mendelian inheritance patterns. To confirm that the transgenes were transmitted to the next generation, T0 plants were grown to maturity, self pollinated to produce T1 seeds, and progeny grown to produce T1 plants. Siblings from 6 independent T1 lines, each line containing single copy T-DNA insertion patterns as determined by Southern blot analysis, were scored for GFP expression using either Western blot analysis (Figure 9C and 9D) or epifluorescence microscopy analysis (Table 4). In our limited sampling of two lines of T1 plants (TS34 and TS65) by Western blot the expected ~30 kDa protein band of GFP was observed in most progeny and with GFP negatives presumably due to null segregating siblings. Fluorescent microscopy allowed more progeny to be screened in a further four lines (TS72, TS57, TS82 and TS88) and each line displayed the transgene inheritance ratios of 3:1 (ratio of GFP positive to negative) that may be expected for simple Mendelian inheritance patterns of a dominant marker (Table 4).
Table 4.
Inheritance patterns of GFP in T1 progeny of four transgenic safflower lines
| Transgenic line | Observed a | Expected b | Chi-square | P value | ||
|---|---|---|---|---|---|---|
| GFP+ | GFP- | GFP+ | GFP- | |||
| WT-TS-72 | 13 | 3 | 12 | 4 | 0.330 | 0.563 |
| S-317-TS-57 | 17 | 7 | 18 | 6 | 0.220 | 0.637 |
| S-317-TS-82 | 18 | 4 | 16.5 | 5.5 | 0.545 | 0.460 |
| S-317-TS-88 | 33 | 13 | 32.5 | 13.5 | 0.026 | 0.871 |
a GFP expression was scored using fluorescence microscopy on germinating seed.
b Expected ratio if a dominant trait at a single locus is inherited by progeny (3:1 segregation ratio).
These molecular analyses confirm that grafted transgenic T0 safflower plants produce transgenic T1 safflower progeny that display classic inheritance patterns. Our method has so far produced over 110 mature genetically modified safflower plants, and the progeny of each of these lines have been screened as positive for GFP fluorescence, confirming their transgenic nature. Southern blot analysis also indicated that this transformation scheme produces relatively simple T-DNA insertion events in safflower, a feature that becomes important in gene regulatory procedures. Overall these results represent the first evidence for a method to reliably generate transgenic safflower seed.
Conclusions
Here we have developed an efficient genetic transformation method for both high oleic and high linoleic safflower varieties and the overall scheme is presented in Figure 10. We have made improvements in the transformation vector, Agrobacterium pre-treatments and regeneration conditions that allow the streamlined production of transgenic shoots. Despite these improvements the development of a grafting method to overcome the poor de novo rooting was critical for the production of mature GM safflower plants that produce GM safflower progeny. This protocol can now allow safflower to expand as an industrial crop platform and also offers the chance to modernise this ancient crop to benefit from advances in plant molecular biology.
Figure 10.

Schematic representation of the complete protocol for the generation of GM safflower.
Methods
Plant material
Seeds of safflower (Carthamus tinctorius L.) cultivars S-317 (supplied by Devexco International; high oleic content seed ~70% oleic acid) and a non-commercial variety WT (high linoleic type ~ 70% linoleic acid) were used in the present study. Seeds were soaked in warm (40°C) 10% Decon90 (Decon Laboratories Ltd.) for 30 min with shaking. After discarding the detergent solution, Carbendzim 2% w/v (Sigma) was added and agitated for 30 min and subsequently rinsed with sterile distilled water. Further sterilization was carried out with 0.2% HgCl2 solution for 15 min. Traces of HgCl2 were removed by several rinses with sterile distilled water. Sterilized seeds were germinated aseptically on half-strength MS media in tissue culture pots (200 ml) and incubated at 24°C in semi-light (50-100 Lux) for 6-8 days. Excised cotyledons from 6-8 day old seedlings were used in the present study.
Bacterial strains and plasmids
The npt-II resistance gene from pORE3 [19] was removed using FseI and AscI digestion and replaced with a PCR amplified version of the intron-interrupted hygromycin resistance gene of the pVEC8 vector series [20] creating pORE6. The coding region of the secreted GFP from pCW141 [28] was PCR amplified to generate an entry clone flanked by AttL sites, pCW227, and a recombinase reaction used to introduce this into a compatible binary vector, pCW001, under the control of the CaMV35S promoter and ocs polyadenylation signal, producing pCW229. The entire 35Sp-SecretedGFP-ocs region was PCR amplified and introduced into the SfoI site of pORE6 to create pCW265. Thus-pCW265 is a T-DNA binary vector with hygromycin as a plant selectable marker, a secreted GFP to provide a non-toxic visual selectable marker and 13 unique restriction sites to allow the insertion of new sequences of interest (Figure 1). The multiple cloning site, including the HindIII site used for southern digest analysis, lies 2700 bp from the left border and 3400 bp from the right border. pCW265 was transformed into Agrobacterium tumefaciens strain AGL1 [29] via electroporation.
Transformation and plant regeneration
Preparation of Agrobacterium and explant co-cultivation
Three days prior to transformation, 25 μl of an Agrobacterium AGL1 cells harboring plasmid pCW265 from -80°C was added to 25 ml of solid LB media with kanamycin 50 mg l-1 and rifampcin 25 mg l-1 and grown for two days at 28°C. One full loop (3 mm) bacteria culture was scraped from a 2-day old plate and suspended in 10 ml liquid LB with kanamycin 50 mg l-1 and rifampcin 25 mg l-1 and grown overnight at 28°C with agitation at 150 rpm. The bacterial cells were pelleted, re-suspended in 10 ml of Winans' AB medium (pH 5.8) and re-grown with kanamycin 50 mg l-1 and rifampcin 25 mg l-1 for 16 hrs with 100 μM AS. Such cultures were adjusted to OD600 nm = 0.4 and 1 mM SPR added for the final 2 hours of growth before explant co-cultivation. Freshly isolated cotyledons (~25) were infected with 10 ml Agrobacterium culture for 10 min, including a gentle inversion every 2 minutes during the infection. Explants infected with Agrobacterium were blotted on sterile filter paper to remove excess Agrobacterium and transferred to co-cultivation media (Table 1). All the plates were sealed with parafilm and incubated in the dark at 23-24°C for 48 hrs.
Selection and plant regeneration
Two days after Agrobacterium infection explants were washed with sterile distilled water containing 500 mg l-1 cefotaxime and 50 mg l-1 timentin for 10 min and further rinsed in sterile distilled water for 10 min to remove the antibiotics and then blot dried on sterile filter paper and transferred onto petri dishes (90 mm in diameter) containing 25 ml of selection media (S2, Table 1). Un-inoculated explants were cultured in the same way as explants inoculated with Agrobacterium as a routine control experiment. After 8-10 weeks, hygromycin resistant and GFP positive shoots were excised and cultured on shoot elongation media. Regenerated shoots exhibiting green fluorescence were considered putatively transgenic; shoots showing red chlorophyll auto-inflorescence were designated as escapes and discarded.
Recovery of transgenic plants
Plants destined to become rootstocks were sterilized as described above. Healthy seedlings were selected and transferred to prepared plastic bags (10 × 6 cm) containing seedling raising mix (50% perlite/vermiculite, 30% river sand and 20% top soil) with added fertilizer (1 g l-1) Aboska brand: N (14.2%), P (6.4%), K (5%) and CaCO3 (2 g l-1) and grown at 24°C in 16/8 hr (day/night) photoperiod for another 2 weeks until ready for grafting. Such plastic bags were found to be ideal for a reduced fungal contaminant of seedlings and subsequent transfer into pots at later stages.
Suitably elongated shoots (~3 cm long and with 2-3 true leaves) were collected and used for grafting (Figure 6A). Just prior to grafting, rootstock seedlings were decapitated (Additional file 3), with roots still in soil, by a single horizontal slice through the stem approximately 6-8 cm above the cotyledonary node. A second vertical cut 3-5 mm deep was made with a sterile micro-scalpel and a silicone tube (3-4 mm internal diameter) placed over the rootstock. The base of the suitable scion was cut to prepare a matching v-shape (Figure 6B) and inserted in the prepared rootstock. The silicon ring was moved up and adjusted to hold both scion and root stock (Figure 6C) and the graft point was wrapped with parafilm (Figure 6 D). Grafted plantlets were covered with 250 ml tissue culture pots to prevent desiccation (Additional file 3), and grown at 24°C with 2000 lux 16/8 hr (day/night) photoperiod for 1-2 weeks or until 3-4 new leaves had grown from the scion. At this point the covering pots were removed from successful grafts and plants were grown in the same conditions for a further two weeks. Unsuccessful grafts were discarded. Grafted plantlets with 5-6 new leaves were transferred to bigger pots (25 cm) with seedling raising mix and grown in a glasshouse facility in Canberra, ACT, until maturity, usually requiring 3-4 months. The procedure of transferring the grafted seedling from the plastic bag to a larger pot (25 cm) is also shown in Additional file 3, highlighting the excellent growth of roots from the rootstocks and the placement of the grafted plants into the soil.
Detection of GFP via microscopy
GFP expression in putative transformed shoots grown under selective conditions was observed with a Leica dissecting microscope equipped with an epifluorescence attachment. Two different sets of filters were used: GFP2- a 510 nm long pass emission filter (transmitting red and green light) with a 480/40 nm excitation filter and GFP3- a 525/50 nm emission filter (transmitting only green light) with a 470/40 nm excitation filter.
Experimental design and data analysis
All treatments were calculated as the mean and standard error (SE) of at least three replicates with a total data points typically with n = 30 except where indicated in the text. All treatments were compared using the Holm-Sidak test for Multiple Pairwise Comparisons in Sigma Plot (Version 11) using a level of significance (α) set at 0.05. Treatments considered to be not significantly different are marked with the same letter within the appropriate subgroup of comparisons (WT or S317 genotype).
DNA extraction and PCR
Total genomic DNA was isolated from GFP-positive and hygromycin resistant young leaf tissue using the caesium chloride gradient method as described by [31]. PCR amplification was carried out to confirm genomic integration of the GFP and hph genes using either GFP-specific primer pairs GFP-F 5' ACACCCTGGTGAACCGCATCG and GFP-R 5' CGGCGGTCACGAACTCCAGC and HPH-F 5' AAAAGCCTGAACTCACCGC and HPH-R 5' TCGTCCATCACAGTTTGCC. The thermal profile of the PCR was: initial denaturation at 94°C for 15 min, 35 cycles of 92°C for 30s, 52°C for 30s, 72°C for 1 min, and finally 72°C for 10 min. Amplified products were size fractionated on 0.8% w/v agarose gel in TAE buffer. Gel electrophoresis was carried out at 80 volts for 40 min before DNA bands were visualized with a BioRad QuantiOne UV transilluminator and software.
Southern blot analysis
Total genomic DNA was isolated from transgenic young leaves using caesium chloride method following the standard procedure of [32]. 5 μg of DNA was digested with HindIII, electrophoresed through 1.0% agarose gel and DNA transferred onto Hybond N+ membranes (Amersham, UK). A purified 966 bp DNA fragment containing the GFP gene was excised from pCW229 by digestion with NotI and was 32P-labelled using the Megaprime DNA labeling system (Amersham) and used as a probe.
Westerns blot analysis
GFP was detected in extracts of safflower leaves using an earlier reported protocol [30]. Briefly, 100 mg fresh safflower leaf was crushed in 200 μL laemelli buffer (2% SDS, 100 mM Tris-base, pH 7.2, 20% glycerol, 60 mM dithiothreol; 0.02% bromophenol blue) and heated at 95°C for 5 min before centrifuging at room temperature at 13000 g for 5 min. 10 μL of supernatant was loaded onto a 12.5% polyacrylamide gel (12.5% PAGE-Sprint Buffer System; Amresco laboratory supplies) and resolved using 200 volts for 60 min. Fermentas Page Ruler Plus was used as an indicator of protein size standards. Protein gels were semi-dry electroblotted at 150 mA for 2 hours onto PVDF membranes (Sigma) and probed for GFP using an anti-GFP monoclonal primary antibody (1/5000 dilution, Clontech), a goat anti-mouse HRP-secondary antibody (1/5000 dilution, Promega) and enhanced chemical luminescence (Lightning ECL, Amersham).
Abbreviations
TDZ: Thidiazuron; BA: Benzylamineo purine; 2iP: 6-(c,c-Dimethylallylamino) purine; NAA -α: Naphthalene acetic acid; GFP: Green Fluorescent Protein; Hph: Hygromycin; SPR: Spermidine; AS: Acetosyringone; MS: Murashige and Skoog media; PVP: Polyvinylpyrrolidone; AgNO3: Silver nitrate; KN: Kinetin/6-furfurylaminopurine.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BS, SPS, AGG and CCW designed the experiments. BS and LH performed all the tissue culture and transformation experiments. LH did the grafting and Southern blot analysis. CCW constructed the vector pCW265 and performed Western blot analysis. BS and CW wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Differences in the appearance of transformed shoots with cytoplasmic GFP and a secreted GFP in safflower. (A) Cytoplasmic GFP (B) Secreted GFP. Note that although the fluorescence is much brighter in cytoplasmic versus the secreted GFP, the cytoplasmic GFP causes tissue swelling before death.
Localisation of the secreted GFP in safflower. (A) GFP expression in transgenic leaf tissue where each pavement cell is lined with a GFP signal. (B) GFP expression in individual cells of calli that are typically rounder than those in leaf cells in Panel A.
Grafting of transgenic shoots and their transfer to larger pots. (A) Decapitated seedling (roots are still in the soil). (B) An example of a grafted shoot covered with plastic container to maintain the humidity for 2 weeks. (C) Removal of grafted seedling from plastic bag without disturbing the root. (D-E) Gentle transfer of grafted seedling to the pot.
Contributor Information
Srinivas Belide, Email: Srinivas.Belide@csiro.au.
Luch Hac, Email: Luch.Hac@csiro.au.
Surinder P Singh, Email: Surinder.Singh@csiro.au.
Allan G Green, Email: Allan.Green@csiro.au.
Craig C Wood, Email: Craig.Wood@csiro.au.
Acknowledgements
We thank Nathalie Niesner, Smitha Louis, Sapna Vibhakaran Pillai and Amratha Menon for their excellent technical assistance. We also thank Grains Research and Development Corporation (GRDC) for financial assistance.
References
- Dajue L, Mundel HH. Safflower. Carthamus tinctorius (L.) Promoting the conservation and use of underutilized and neglected crops. Institute of Plant Genetics and Crop Plant Research, Gaterslaben/International Plant Genetic Resources Institute, Rome, Italy; 1996. p. 83. [Google Scholar]
- Zhang LP. In: Proceedings of the IVth International Safflower Conference. Corleto A, Mundel HH, editor. Adriatica Editrice, Bari. Italy; 1997. Safflower: a versatile plant; pp. 329–331. [Google Scholar]
- Knauf VC, Shewmaker C, Flider FJ, Emlay D, Rey E. Safflower with elevated gamma-linilenic acid. 2006. WIPO Application No. WO/2006/127789.
- Moloney MM, Boothe J, Keon R, Nykiforuk C, Rooijen V. Methods for the production of insulin in plants. 2004. WIPO Application No. WO/2004111244 A2.
- Nykiforuk CL, Shen Y, Murray E, Booth J, Busseuil D, Rhéaume E, Tardif J-C, Reid A, Moloney M. Expression and recovery of biologically active recombinant apolipoprotein AIMilano from transgenic safflower (Carthamus tinctorius) seeds. Plant Biotechnol J. 2011;9:250–263. doi: 10.1111/j.1467-7652.2010.00546.x. [DOI] [PubMed] [Google Scholar]
- Markley N, Nykiforuk C, Boothe J, Moloney M. Producing proteins using oilbody-oleosin technology. BioPharm Inter. 2006;9:6–12. [Google Scholar]
- Ying MC, Dyer WE, Bergman JW. Agrobacterium tumefaciens mediated transformation of safflower (Carthamus-tinctorius L.) cv centennial. Plant Cell Rep. 1992;11:581–585. doi: 10.1007/BF00233097. [DOI] [PubMed] [Google Scholar]
- Orlikowska TK, Cranston HJ, Dyer WE. Factors influencing Agrobacterium tumefaciens mediated transformation and regeneration of the safflower cultivar centennial. Plant Cell Tissue Organ Cult. 1995;40:85–91. doi: 10.1007/BF00041122. [DOI] [Google Scholar]
- Rao SK, Rohini VK. Gene transfer into Indian cultivars of safflower (Carthamus tinctorius L.) using Agrobacterium tumefaciens. Plant Biotechnol. 1999;16:201–206. [Google Scholar]
- Haseloff J, Kirby R, Siemering Douglas CP, Sarah H. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Pro Nat Acad Scie USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sri Shilpa K, Dinesh Kumar V, Sujatha M. Agrobacterium mediated genetic transformation of safflower (Carthamus tinctorius L.) Plant Cell Tissue Organ Cult. 2010;103:387–401. doi: 10.1007/s11240-010-9792-7. [DOI] [Google Scholar]
- Rohini VK, Rao KS. Embryo transformation, a practical approach for realizing transgenic plants of safflower (Carthamus tinctorius L.) Annals of Bot. 2000;86:1043–1049. doi: 10.1006/anbo.2000.1278. [DOI] [Google Scholar]
- De Pasquale F, Giuffrida S, Carimi F. Mini grafting of shoots, roots, inverted roots, and somatic embryos for rescue of in vitro Citrus regenerants. J the American Society for Horti Sci. 1999;124:152–157. [Google Scholar]
- Prakash O, Sood A, Sharma M, Ahuja PS. Grafting micropropagated tea Camellia sinensis (L.) O. Kuntze shoots on tea seedlings - a new approach to tea propagation. Plant Cell Rep. 1999;18:883–888. doi: 10.1007/s002990050679. [DOI] [Google Scholar]
- Jin SX, Liang SG, Zhang XL, Nie YC, Guo XP. An efficient grafting system for transgenic plant recovery in cotton (Gossypium hirsutum L.) Plant Cell Tissue Organ Cult. 2006;85:181–185. doi: 10.1007/s11240-005-9068-9. [DOI] [Google Scholar]
- Dutt M, Grosser JW. An embryogenic suspension cell culture system for Agrobacterium mediated transformation of citrus. Plant Cell Rep. 2010;29:1251–1260. doi: 10.1007/s00299-010-0910-0. [DOI] [PubMed] [Google Scholar]
- Christiane F, Paul K, Giinther H. Protoplasts from cotyledon and hypocotyl of sunflower (Helianthus annuus L.): shoot regeneration and seed production. Plant Cell Rep. 1992;11:632–636. doi: 10.1007/BF00236388. [DOI] [PubMed] [Google Scholar]
- Weber S, Friedt W, Landes V, Molinier J, Himber C, Rousselin P, Hahne G, Horn R. Improved Agrobacterium-mediated transformation of sunflower (Helianthus annuus L.): assessment of macerating enzymes and sonication. Plant Cell Rep. 2003;21:475–482. doi: 10.1007/s00299-002-0548-7. [DOI] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD. pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res. 2007;16:771–781. doi: 10.1007/s11248-007-9066-2. [DOI] [PubMed] [Google Scholar]
- Wang MB, Upadhyaya NM, Brettell R, Waterhouse PM. Intron-mediated improvement of a selectable marker gene for plant transformation using Agrobacterium tumefaciens. J Genet Breed. 1997;51:325–334. [Google Scholar]
- Nishizawa K, Maruyama N, Satoh R, Fuchikami Y, Higasa T, Utsumi S. AC-terminal sequence of soybean beta-conglycinin alpha subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 2003;34:647–659. doi: 10.1046/j.1365-313X.2003.01754.x. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Nester EW, Zambryski PC. A plant-cell factor induces Agrobacterium tumefaciens vir gene expression. Pro Nat Acad Sci USA. 1986;83:379–383. doi: 10.1073/pnas.83.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Rajam MV. Polyamines enhance Agrobacterium tumefaciens vir gene induction and T-DNA transfer. Plant Sci. 2005;168:475–480. doi: 10.1016/j.plantsci.2004.09.018. [DOI] [Google Scholar]
- Radhika K, Sujatha M, Nageshwar Rao T. Thidiazuron stimulates adventitious shoot regeneration in different safflower explants. Biologia Plant. 2006;50:174–179. doi: 10.1007/s10535-006-0003-7. [DOI] [Google Scholar]
- Sujatha M, Dinesh kumar V. In vitro bud regeneration of Carthamus tinctorius and wild Carthamus species from leaf explants and axillary buds. Biologia Plant. 2007;51:782–786. doi: 10.1007/s10535-007-0160-3. [DOI] [Google Scholar]
- Whitehouse AB, Marks TR, Edwards GA. Control of hyperhydricity in Eucalyptus axillary shoot cultures grown in liquid medium. Plant Cell Tissue Organ Cult. 2002;71:245–252. doi: 10.1023/A:1020360120020. [DOI] [Google Scholar]
- Luo J, Gould JH. In vitro shoot-tip grafting improves recovery of cotton plants from culture. Plant Cell Tiss Org Cult. 1999;57:211–213. doi: 10.1023/A:1006393009094. [DOI] [Google Scholar]
- Petrie J, Shrestha P, Liu Q, Mansour P, Wood CC, Zhou XR, Nichols P, Green A, Singh SP. Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods. 2009;6:1–9. doi: 10.1186/1746-4811-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA-transformation competent Arabidopsis genomic library in Agrobacterium. Biotech. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Winans SC, Kerstetter RA, Nester EW. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriology. 1988;170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press, Cold Spring, Harbor, NY; 1989. [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Pro Nat Acad Sci USA. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differences in the appearance of transformed shoots with cytoplasmic GFP and a secreted GFP in safflower. (A) Cytoplasmic GFP (B) Secreted GFP. Note that although the fluorescence is much brighter in cytoplasmic versus the secreted GFP, the cytoplasmic GFP causes tissue swelling before death.
Localisation of the secreted GFP in safflower. (A) GFP expression in transgenic leaf tissue where each pavement cell is lined with a GFP signal. (B) GFP expression in individual cells of calli that are typically rounder than those in leaf cells in Panel A.
Grafting of transgenic shoots and their transfer to larger pots. (A) Decapitated seedling (roots are still in the soil). (B) An example of a grafted shoot covered with plastic container to maintain the humidity for 2 weeks. (C) Removal of grafted seedling from plastic bag without disturbing the root. (D-E) Gentle transfer of grafted seedling to the pot.


