Abstract
DNA sequence variants (DSVs) are major components of the “causal field” for virtually all-medical phenotypes, whether single-gene familial disorders or complex traits without a clear familial aggregation. The causal variants in single gene disorders are necessary and sufficient to impart large effects. In contrast, complex traits are due to a much more complicated network of contributory components that in aggregate increase the probability of disease. The conventional approach to identification of the causal variants for single gene disorders is genetic linkage. However, it does not offer sufficient resolution to map the causal genes in small size families or sporadic cases. The approach to genetic studies of complex traits entails candidate gene or Genome Wide Association Studies (GWAS). GWAS provides an unbiased survey of the effects of common genetic variants (common disease - common variant hypothesis). GWAS have led to identification of a large number of alleles for various cardiovascular diseases. However, common alleles account for a relatively small fraction of the total heritability of the traits. Accordingly, the focus has shifted toward identification of rare variants that might impart larger effect sizes (rare variant-common disease hypothesis). This shift is made feasible by recent advances in massively parallel DNA sequencing platforms, which afford the opportunity to identify virtually all common as well as rare alleles in individuals. In this review, we discuss various strategies that are used to delineate the genetic contribution to medically important cardiovascular phenotypes, emphasizing the utility of the new deep sequencing approaches.
Keywords: Genetics, Next-Generation Sequencing, Complex traits, Polymorphism
The human nuclear genome (henceforth “genome”) is a simple and yet a very complex structure. It is a large monotonous macromolecule comprised of 3.2 billion repeating nucleotides of adenine (A), cytosine (C), guanine (G) and thymine (T), which are arranged in a seemingly random order. Yet, these four nucleotides not only determine expression of various biological and pathological phenotypes but also serve as the platform for various genomic and environmental factors to exert their functional and biological effects. Consequently, elucidation of the molecular structure of the genome including its nucleotide sequence is fundamental to understanding the molecular pathogenesis of human diseases.
Sequencing of the human genome, however, has been a daunting task, at least until the very recent years. The Human Genome Project (HGP), which was launched in 1990 with the primary goal of deciphering sequence of the human genome took more than a decade to complete, even in a draft form, and cost close to $3 billion 1, 2. DNA sequencing technology, however, has undergone a colossal shift during the past six years. Various new techniques that sequence millions of DNA strands in parallel have been developed. The new technologies, which are collectively referred to as the “Next-Generation Sequencing” (NGS) platforms, as opposed to Sanger method 3, which was used in the HGP, have increased DNA sequencing output and have reduced the cost of DNA sequencing by ~ 500,000-fold. These advances in DNA sequencing technologies along with the rapidly declining cost of sequencing are changing the approach to genetic studies of not only single gene disorders but also common complex disorders.
Despite its apparent simplicity, the genome is a complex structure. The complexity is far beyond the primary base sequence of the genome. DNA is a large macromolecule that requires a complex system to orchestrate its compaction inside the nucleus in a manner that selected genes are accessible to specific DNA processing enzymes, such as polymerases in an orderly and dynamic fashion, as demanded by the cell in response to internal and external stimuli 4. Thus, understanding the functional content of the genome necessitate knowledge beyond the complete genome sequence. Based on today’s knowledge, only 1% of the human genome is transcribed into mRNA and translated into proteins. An additional 0.5% serves as a template for non-coding RNAs and the regulatory regions that control gene expression 5. The functions of the remaining 98.5% of the genome including functional conserved non-coding elements (CNEs), which comprise at least 6% of the genome 6, remain unknown. Hence, this large segment of the genome is referred to as “the dark matter of the genome” 5. The discoveries of non-coding RNAs, microRNAs, splice variants and regulatory elements in trans point to the complex mechanisms by which the genome governs various biological processes including phenotypic expression of diseases (Figure 1). To elucidate the determinants of any biological and clinical phenotype, a comprehensive approach that not only utilizes information content of the nucleotides sequence but also that of the transcripts; whether coding or non-coding, chromatin structure and function; and transcriptional machinery that orchestrates gene expression among the others would be necessary. The focus of this review is on strategic approaches to identify the DNA sequence variants (DSVs) that either strongly determine disease risk, as in single gene disorders, or influence susceptibility to a disease. Because the majority of the known disease-causing DSVs are located within exons, the current focus of human genetic studies is on whole exome sequencing 7. With advances in DNA sequencing technology and increasing knowledge of the non-protein coding regions of the genome, one expects a rapid shift from whole exome to whole genome sequencing as the desirable approach to identify disease-causing or disease-associated DSVs.
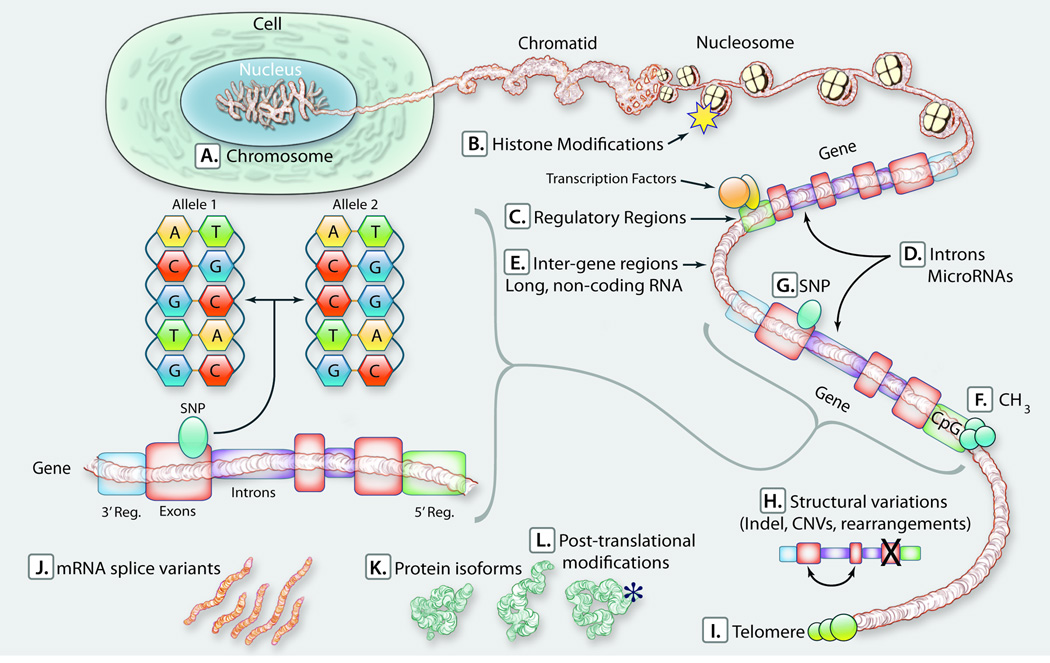
Figure 1. Genomic and genetic determinants of phenotype.
The nuclear genome is comprised of 4 nucleotides that are in tandem and randomly repeated in a complex 2-meter long polymer with a diameter of 2 nM and a volume of ~ 4 × 107 uM3,4. It is packed into the nucleus as 22 pairs of somatic and 2 sex chromosomes. Various components of the genome ranging from compactness of DNA to specific base pair changes could impart phenotypic effects. Examples are: A. Chromosomal abnormalities; B. Modifications of the octomeric histon complex, comprised of two copies H2A, H2B, H3 and H4 proteins, through methylation and acetylation; C. Changes in transcription factors; D. Expression of microRNAs from introns and inter-gene regions; E. Expression of long non-coding RNAs; F. Methylation of the CpG dinucleotides on promoters; G. SNPs; H. SVs/CNVs; I. Changes in telomere structure and function; J. Alternative mRNA splicing; K. Expression of protein isoforms; and L; Post-translation modification of proteins. It is also notable that at least 6% of the human genome is under evolutionary purifying selection, which indicates functional significance. However, the functions and biological impacts of these CNEs remain unknown 6.
To identify and characterize determinants of a phenotype, a comprehensive approach that builds all constituents of the phenotype into the modeling would be necessary. A prototypic comprehensive approach has been completed for two model organisms 100, 101. (Illustration Credit: Cosmocyte/Ben Smith).
GENETIC DIVERSITY
Catalogs of common genetic variation have been accumulated over the last 3 decades. For example the database of single nucleotide polymorphisms (dbSNP Build 132) has more than 37 million entries. The number of polymorphic variants in a single genome, however, was largely unknown until the report of J. Craig Venter’s diploid genome sequence in 2007 8. The findings were notable for presence of 4.1 million DSVs, including ~3.5 million single nucleotide polymorphisms (SNPs), which affected approximately 44% of the annotated genes. Likewise, the Venter genome contained about 10,000 non-synonymous SNPs (nsSNPs) of which ~7,000 were considered potentially harmful variants. In addition, structural variations (SV), which involved up to several million nucleotides, comprised ¾ of the variant nucleotides.
Subsequent sequencing of additional individual genomes confirmed the findings of extensive DSVs in each genome and pointed out the presence of a large number of novel variants. Today, the genome and exome sequences of a relatively large number of individuals have become available along with the HapMap and 1,000 Genomes data 9–15. The initial results of the 1,000 Genome Projects indicate that each genome has approximately 250 to 300 loss-of-function variants in the annotated genes and 50 to 100 variants that already have been implicated in inherited disorders 16. In addition, each genome has approximately 30 de novo variants, a finding that indicates a germ line mutation rate of 1×10−8 per generation 16. Furthermore, each genome has several large (>50 Kbp) and about 100 heterozygous copy number variants (CNVs) covering about 3 Mbp 17, 18. Collect3ively, the data indicate that the humans differ in about 0.12% of their genomes, or about 4 million DSVs per genome, comprised of about 3.5 million SNPs and several hundred thousand SVs including CNVs.
The functional and biological significance of the vast number of DSVs in the human genome are unknown. Nevertheless, they are expected to exert effects that follow a gradient ranging from negligible to severe 19. Among the approximately 10,000 nsSNPs in each genome, about 2/3rd are predicted by in silico analysis to be deleterious to function. Likewise, SVs that encompass several thousand to million base pairs could duplicate or delete a gene or multiple genes and hence, would be expected to hold significant clinical implications 15, 20. Nevertheless, in a given clinical phenotype, a small number of alleles are expected to exert large effects, a handful moderate effects and a very large number with modest or no effects. Presumably clinical phenotype is the consequence of the additive effects and interactions among multiple alleles with varying magnitude of effect.
GENETIC MECHANISMS OF HUMAN DISEASES
Common disease-common variant (CD-CV) hypothesis
Common cardiovascular diseases have considerable genetic components, as evidenced by familial aggregation and twin studies 21–23. The estimated heritability of common complex diseases, defined as a proportion of the phenotypic variance accounted for by genetic factors, varies from 20% to 80%, depending on the phenotype and study characteristics. In contrast to single gene disorders, wherein a single DSV imparts a large determinative effect, no single allele or locus dominates as the determinant of a complex phenotype. Accordingly, complex diseases result from the cumulative and interactive effects of a large number of loci, each imparting a modest marginal effect on expression of the phenotype (Figure 2). The CD-CV hypothesis posits that multiple common alleles, defined as alleles with a population frequency of ≥0.05, contribute to the risk of developing common diseases. The CD-CV hypothesis underpins Genome Wide Association Studies (GWAS), wherein cases and controls are genotyped for hundreds of thousands of common variants. In GWAS, linkage disequlibrium (LD) - the correlation between markers – is exploited to tag common variants that influence medically important traits. Effective tag SNPs and their underlying haplotypes in selected reference populations have been extracted from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) data set and arrayed for low cost genotyping in GWAS. SNP arrays typically offer >99% reproducibility but despite the density of SNPs on the arrays, they ‘cover’ only a fraction of the total variation in the genome. Standard SNP arrays have markers that are correlated with (and so effectively tag) 80–90% of common variation (variants with frequencies >5%), far less of the less common variation (frequencies 0.01%-5%), and are virtually useless for rare family or individual variation. One of the important limitations of GWAS is that it may not directly identify the variants that are causally linked to the phenotype. Identification of the causal variants is important to understand the molecular mechanisms involved in the pathogenesis of the phenotype, which remains one of the most important challenges for future research. Although, GWAS have been successful for identifying many loci associated with important cardiovascular diseases (http://www.genome.gov/gwastudies/) there are additional large gaps in our understanding of the genetic contribution to these conditions. Common alleles, at least from the perspective of their individual marginal effects, account for a relatively small fraction of the total heritability of those disorders. As such, SNPs indentified in GWAS of systemic hypertension, dyslipidemia, and cardiac conduction intervals account for only a small fraction of inter-individual variance 24–29. However, the attributable risk of a common allele in a population might be considerable, simply because of its high minor allele frequency (MAF).
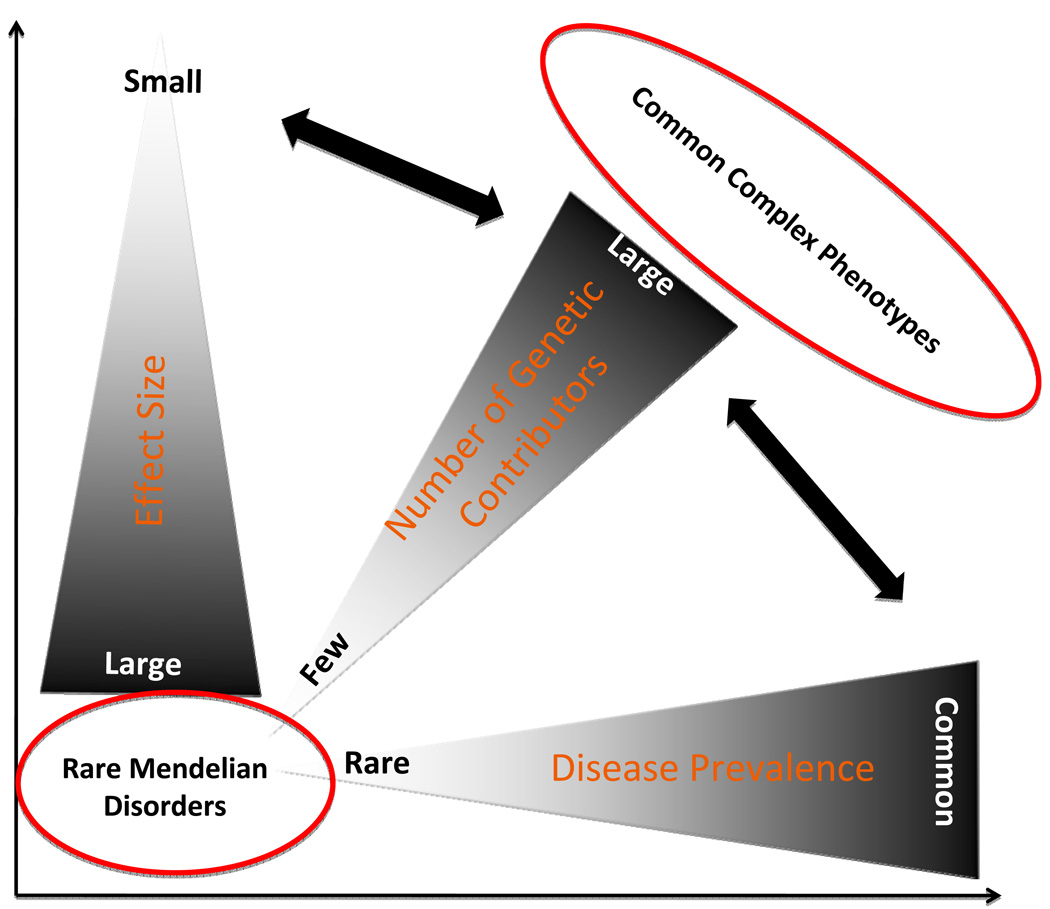
Figure 2. Gradients of disease prevalence, MAFs and effect sizes.
The prevalence of disease, number of genetic determinants and the effect sizes of the DSVs are depicted as continuums. Single gene disorders are caused by rare variants with large effect sizes. Typically, several other variants also expected to contribute to phenotypic expression of the diseases. On the opposite end of the spectrum are the common complex traits, which are caused by a very large number of genetic variants, each imparting a modest effect size.
Rare variant-common disease hypothesis (RV-CD)
The shortcomings of GWAS in explaining the heritability of common complex disease often referred to as the “missing heritability” might be in part due to the presence of rare DSVs with relatively large effects that are not tagged by the typical marker sets used in GWAS 30. Rare alleles are typically defined as those that have a MAF of ≤ 0.01 in a population. Whenever rare DSVs are unique to an individual or to a family, they are considered as “private” variants. Possible contributions of rare alleles with large effects on common diseases have led to the RV-CD hypothesis. In support of this hypothesis, uncommon and rare alleles in genes known to cause single gene disorders could contribute to susceptibility for complex phenotypes and enhance detection of otherwise clinically under-diagnosed conditions. A notable example is identification of mutations in MYH7, MYBPC3, TNNT2, TNNI3 and MYL3 genes, causal genes for autosomal dominant hypertrophic cardiomyopathy (HCM)(reviewed in 31), in a subgroup of individuals in a community-based cohort who had an increased left ventricular wall thickness 32. Similarly, multiple rare alleles in ABCA1, the responsible gene for Tangier disease 33, 34, also contribute to plasma high-density lipoprotein cholesterol (HDL-C) levels in the general population 35. Likewise, multiple rare alleles in genes not associated with single gene disorders might account for a substantial portion of heritability of complex traits. The notion that a number of rare variants might impart large effects on the phenotype is plausible. However, currently, there is insufficient data to substantiate this hypothesis and conclude that multiple rare alleles are major components of missing heritability.
Gradients of allele frequencies and effects
The CD-CV and RV-CD hypotheses represent opposite ends of the spectrum of gradients of allele frequencies and effects (Figure 2). The full allele frequency spectrum includes alleles that are exceedingly rare and even unique to an individual genome to being extremely common. Likewise, it would be expected that the effects of the alleles (also called expressivity and penetrance) vary from negligible to large and implying determinative effects on clinical traits. Alleles that have large effects are generally deleterious and cannot rise to high frequencies in the population due to negative selection. Therefore, one expects an inverse correlation between frequency and the magnitude of the effect, namely, DSVs that impart large effects tend to be rare and those that exert weak effects can be more common (Figure 2). Nevertheless, it is important to note that rare alleles can also have weak or no effect. A particular locus may contain a very large number of rare alleles, so paradoxically there may be many rare DSVs with large effect and a few common variants with weak effects. The resulting “genetic architecture” of medically important traits seems be an empiric question and one can already conclude that there is no single answer.
Cardiovascular diseases originate from the confluence of many different factors. Although a genetic variant may have only a weak effect on the process taken as a whole, it may substantially influence one of the known underlying pathways. For example, genetic effects on lipid biomarkers may often be more readily detected than their effect on myocardial infarction. In general, an important determinant of the effect of DSVs is their causal proximity to the phenotype (Figure 3). The more tightly coupled the genetic variant, in terms of biological functions, such as enzymatic activity or protein interactions, the greater the effect. Likewise, a small number of variants with relatively large effects are observed to influence mRNA expression levels of their respective genes 36. Therefore, genetic analysis of biomarkers, including transcript abundance, can be a very effective strategy for “divide and conquer” the much more complex origins of cardiovascular disease.
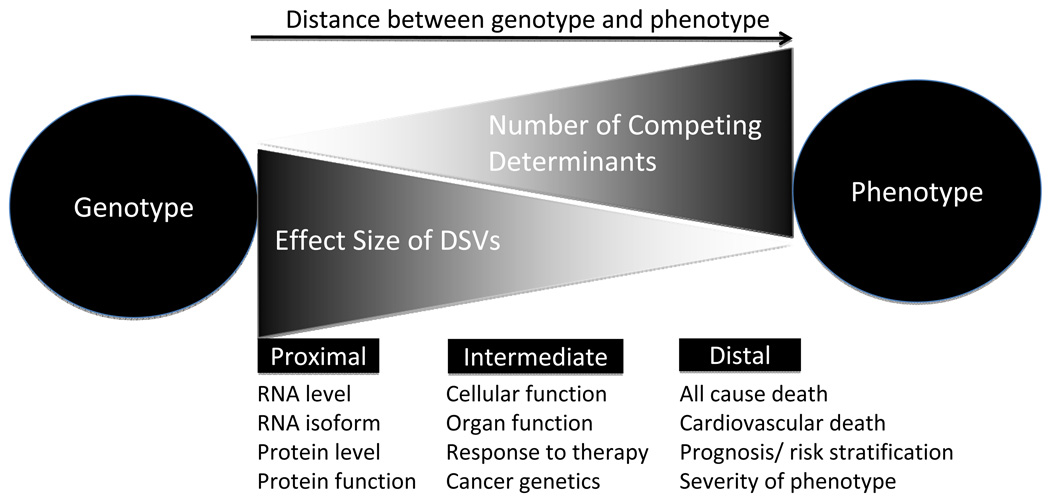
Figure 3. Relationship between effect sizes of DSVs and proximity of the phenotype.
The influence of genetic variants is expected to correlate inversely with the proximity of the phenotype to genes. The effect size is greater for the proximal phenotypes, such as mRNA levels than for distant phenotypes, such as mortality, wherein a large number of competing genetic and non-genetic determinants contribute to the phenotype.
APPROACH TO GENETIC STUDIES
Genetic factors are components of the “causal field” for virtually all medically important traits. Causal fields can be composed of necessary and sufficient factors as in the case of single gene disorders. Most often, however, there is a much more complicated network of unnecessary and insufficient components that in aggregate increase the probability of disease. These factors are contributory causes – they are non-redundant components of pathways which by themselves may be unnecessary but if altered can influence the occurrence of disease. The genotypic effect can be represented as a probability from 0–1 that it influences a particular clinically important trait. Genotypes with large effects are best represented by single gene disorders, such as familial cardiomyopathies wherein a single mutation leads inexorably to a major disease (Figure 2). Even in the single gene disorders the severity of the disease is influenced not only by the causal mutation but also by genetic modifiers and environmental factors. On the other end of spectrum of genetic effects are much weaker effects, as uncovered in GWAS of complex phenotypes such as atherosclerosis (Figure 2).
Candidate gene association studies
The approach is based on a prior knowledge of candidacy of the gene(s) of interest in the pathogenesis of the phenotype. Unless performed in a very large sample size of well-characterized populations, the approach is prone to spurious results, particularly for distant phenotypes. The approach has led to identification of a number of alleles that influence responsive to drugs, both in terms of efficacy as toxicity (pharmacogenetics). For example, DSVs in CYP2C9 and VKORC1, genes encoding cytochrome P450 isoform 2C9 and vitamin K epoxide reductase, respectively are associated with response to treatment with anti-coagulant coumadin. Similarly, DSVs in genes coding for P-450 enzymes CYP3A4, CYP3A5, and CYP2C19 are associated with responsive to treatment with anti-platelet agent clopidogrel, a pro-drug that is converted in the liver to an active metabolite 37, 38. Likewise, DSVs in α2C- and β1-adrenergic receptors are associated with the response of patients with systolic heart failure to treatment with β blockers 39, 40. Moreover, DSVs in APOE, PCSK9, and HMGCR have been implicated in response to statins 41. As regards drug toxicity, DSVs in SLCO1B1, encoding solute carrier organic anion transporter 1B1 are associated with statin-induced myopathy 42, 43. Likewise, DSVs in genes causing congenital long QT syndrome are associated with drug-induced cardiac arrhythmias 44. DSVs implicated in pharmacogenetics appear to have moderate effect sizes and hence, might have some clinical implications in guiding drug efficacy and avoiding toxicity (Figure 3).
Genome-Wide Association Studies (GWAS)
GWAS has been widely used to delineate the genetic basis of common complex disorders. GWAS are case-control studies, wherein research subjects are typed for a large number of SNPs, typically 300,000 to 1,000,000 SNPs/CNVs, and the allele or genotype frequencies are evaluated for differences between groups or for correlations with continuous traits. GWAS is primarily designed to provide an unbiased survey of the effects of common genetic variants. Markers chosen for GWAS typically have MAFs of ≥ 0.05 and selected to “tag” the most common haplotypes observed in the major continental populations. Such tagging is more complete in European and East Asian populations compared to African populations because of inherent differences in the LD patterns.
The power of the GWAS to detect the phenotype-associated alleles depends directly on the sample size of the study population, MAFs, strength of LD between the markers and the causal variants and the effect sizes of the alleles. The density of the genotyping arrays have increased significantly over time and the current versions can easily genotype as many as 2.5 million SNPs and CNVs. Collectively, these advances have enhanced the power to detect the associated alleles. During the past decade, GWAS have been completed for a very large number of cardiovascular phenotypes. The National Human Genome Research Institute (NHGRI) maintains a catalog of published GWAS that could be accessed at http://www.genome.gov/gwastudies/.
A major strength of GWAS is that it may lead to identification of novel pathways involved in the pathogenesis of the phenotype. Despite the apparent simplicity, however, the results of GWAS are subject for multiple-hypothesis testing because of typing of a very large number of SNPs, and hence, beget correction for the possibility of a random association due to multiple testing. In addition, often the same study population is analyzed for the association of the genotypes with multiple phenotypes, which also increases the likelihood of spurious associations. Therefore, statistical corrections for multiple hypotheses testing are essential. The best approach to correct for multiple hypotheses testing in GWAS remains to be established. The conventional Bonferroni method for correcting for multiple testing (p=α/n) is considered too conservative because the Bonferroni correction assumes that the tests are independent, which is not the case for GWAS markers due to residual LD in local regions of the genome. Permutation tests are probably the most robust for correcting for multiple testing but are computationally very intensive and impractical considering the very large number of genotypes (SNPs X individuals) in the GWAS. Various statistical methods have been applied to correct for multiple testing in GWAS and determine the threshold for statistical significance. Based on these calculations, a p value of < 5× 10−8 or more stringently < 1× 10−8 is considered evidence of a strong association 45.
GWAS have been extremely useful for identifying a very large number of phenotype-associated alleles, including many novel loci. The paucity of discovering functional SNPs as the associated alleles, however, is notable. Accordingly, the results of GWAS have typically not been fruitful in immediate elucidation of the responsible mechanisms behind the observed genetic association. Consequently, GWAS demand complementation with robust mechanistic studies to elucidate the biological mechanisms responsible for the genetic association.
Alleles identified in GWAS are seldom the true causative alleles but are likely in LD with the true causative alleles. Thus, extensive additional studies are typically required to complement the results of GWAS to identify the disease-causing alleles. These shortcomings render the results of GWAS in the discovery population as provisional, requiring replications in independent study populations and ultimately validation through experimentation.
The results of GWAS have minimal to modest impact, if any, on pre-clinical diagnosis, risk stratification or genetic-based prevention and treatment at an individual level. Therefore, the significance of additional mechanistic studies cannot be over-emphasized. However, the appropriate platforms to validate the results of GWAS through molecular mechanistic studies remain to be established. The challenge is best illustrated for delineation of the responsible mechanism(s) for the observed association of SNPs at 1p13 locus with plasma cholesterol levels and coronary atherosclerosis 46–48. Accordingly, the minor allele of the rs599839 SNP located in SORT1 gene at the 1p13 locus is associated with a decrease in serum low-density lipoprotein-cholesterol (LDL-C) by 0.14 mmol/L and a 9% decrease in risk of coronary atherosclerosis 47. Two recent studies attempted to elucidate the responsible mechanisms but unfortunately, reported discordant results 47, 49. Musunuru et al. fine mapped the locus and upon further analysis defined rs12740374 to be the causative SNP. The minor allele of this SNP created a C/EBP-α binding site on SORT1 promoter, which enhanced transcriptional activity. Over-expression of Sort1 in mouse live using recombinant adeno-associated viruses led to reduced plasma LDL-C levels. In contrast, siRNA-mediated suppression of expression of Sort1 in the liver had the opposite effect. Therefore, the studies identified increased expression level of SORT1 imparted by the minor allele of rs12740374 as the responsible mechanism for reduced plasma LDL-C level 49. However, studies by Kjolby et al. showed the opposite effects 50. Accordingly, over-expression of Sort1 stimulated hepatic release of lipoproteins and led to increased plasma LDL levels 50. Thus, despite the concordant and reproducible results of GWAS, studies to delineate the responsible mechanisms have not led to concordant results. The contrasting results may hint to the challenges encountered in recapitulating the results of human genetic studies of complex phenotypes in model organisms.
Identification of the responsible mechanism(s) for association of the SNPs at the 9p21 locus, which has been robustly linked to atherosclerosis 51–53, also has been challenging. The refined locus does not contain a known gene but the region contains cyclin-dependent kinase inhibitor 2A and 2B (CDKN2A, CDNK2B), methylthioadenosine phosphorylase (MTAP) and ANRIL, the later codes a long non-coding RNA. None of these genes appear to be a biologically plausible candidate gene for atherosclerosis. The cell cycle regulators CDKN2A and CDKN2B are tumor suppressor proteins and markers of cell senescence 54. Deletion of the orthologous region of 9p21 locus in the mouse genome is associated with reduced expression levels of Cdkn2a and Cdkn2b, enhanced proliferation and reduced senescence of smooth muscle cells 55. The findings suggest accelerated smooth muscle cells proliferation as the potential mechanism for the observed association of the 9p21 locus and atherosclerosis.
MTAP, which is also located at the region, codes for an enzyme that is involved in polyamine metabolism and generation of adenine and methionine. Deletion of this gene is embryonically lethal and in homozygous form is associated with reduced life span, because of severe lymphoproliferative disease resembling T-cell lymphoma 56. The third gene is ANRIL, which is expressed in cells involved in atherosclerosis, such as smooth muscle cells, endothelial cells and macrophages. It has multiple isoforms but none has an open reading frame and hence, the gene does not appear to code for a protein. Deletion of the 9p21 region involving ANRIL is implicated in melanoma and solid tumors 57.
The 9p21 locus, despite being a gene desert, is extremely rich in enhancers 58. It contains at least 33 enhancers, including one that interacts with two risk alleles for coronary artery disease (CAD) at this locus. The risk alleles disrupt the binding site for STAT1, which is the signal transducer for a variety of ligands including interferon-α, interferon-γ and cytokines. Binding of STAT1 to the wild type alleles inhibits expression of CDKN2B-AS (non-protein coding CDKN2B antisense RNA1)58. Treatment of endothelial cells carrying the risk allele with interferon-γ represses expression of CDKN2B and induces expression of CDKN2B-AS. These findings implicate augmented inflammatory response in the presence of 9p21 risk alleles to suppression of expression of CDKN2B by CDKN2B-AS RNA. Collectively, the results of the mechanistic studies link inflammation to suppression of expression of cell cycle inhibitor CDKN2B and ensuing accelerated proliferation of smooth muscle cells, as a mechanism for the observed association of the 9p21 locus with CAD in GWAS 55, 58.
GWAS, which are mainly restricted to testing the effects of the CD-CV hypothesis, have the inherent limitation of identifying alleles that typically impart minimal to modest effects (Figure 2). The common alleles, by and large, seem to explain a small fraction of heritability of the complex phenotypes 20. In a GWAS of systemic hypertension in 2,000 cases and 3,000 controls, no SNP had statistically significant association with the case-control status 59. Likewise, SNPs identified through GWAS typically exert small effect sizes on plasma LDL-C and HDL-C levels, typically ≤ 1mg/dl change in HDL-C levels 26. The proponents of GWAS have advocated the need for larger sample sizes studies. However, the sample size alone, while possibly increasing the number of associated loci, seems unlikely to explain the “missing heritability”60, 61. For example, a recent meta-analysis of 14 GWAS comprised of more than 40,000 individuals, typed for more than 500,000 SNPs, led to identification of 22 loci, as determinants of the QRS duration 28. The 22 loci collectively accounted only for a total of 5.7% (± 2.3) of the observed variance in the QRS duration. Typing of a very large number of common SNP in a very large number of individuals could increase the number of loci identified in a GWAS and hence, explain a higher fraction of heritability. In a GWAS study of 100,184 individuals of European ancestry typed or imputed for 2.6 million SNPs 22 loci were associated with plasma LDL-C, 31 with HDL-C, and 16 with triglycerides (TG) 62. These variants accounted for ~25–30% of the genetic variance for each trait. Thus, GWAS with larger sample sizes and denser SNP typing are unlikely to fully explain heritability of the complex traits.
GWAS of proximal phenotypes, such as mRNA levels might be a desirable approach because of anticipated larger effects. The approach has led to identification of several common associated variants 63, 64. Disappointedly, however, in some studies, these variants were distinct from variants that are associated with the relevant distant phenotype. Hence, there might be a discord between genetic variants that influence the proximal phenotypes, such as mRNA levels and the distal or clinical phenotypes.
The shortcoming of GWAS in elucidation the genetic determinants of the complex phenotypes may be in part due to the fact that a large number of the genetic variants in each genome are private. Consequently, the emphasis has shifted toward the RV-CD hypothesis 65. The notion is in accord with the presence of a gradient of effects that in one extreme – when the effects are the largest – results in single gene disorders with a Mendelian pattern of inheritance and on the other extreme the effects are negligible 19. Accordingly, the paradigm in the genetic studies of complex phenotype is shifting toward identification of uncommon and rare variants with large effects. The shift has been in part accelerated by the availability of the NGS platforms, which enable identification of the uncommon and rare variants through whole exomes and whole genome sequencing. Consequently and in view of precipitous drop in the cost of DNA sequencing, approaches based on whole exome and whole genome sequencing are expected to dominate genetic studies in the coming years. These studies will elucidate whether uncommon and rare variants account for a significant component of the “missing heritability” or alternative mechanisms, such as epistasis, gene-environmental interactions, and epigenetics might play larger roles.
The problem of ‘missing heritability’ has raised significant concern about the utility of GWAS in delineating the genetic basis of complex traits 66,67,68, 69. The “missing heritability” in part may reflect the definition of heritability that in narrow sense is defined as the proportion of the phenotypic variance attributable to additive genetic factors. The definition reflects a general model of gene action that has only coarse explanatory power and is subject to many difficulties of estimation 70, 71. In fact, Feldman and Lewontin anticipated most of the problems related to heritability interpretation in complex disease long before the current era of GWAS 71. Some of the possibilities that may account for the relatively modest portion of the heritability captured by GWAS are listed in Table 1.
TABLE 1.
Potential Explanations for the modest capture of Heritability by GWAS
|
In the RV-CD hypothesis population genetics theory predicts and empirical observation demonstrates that there are large numbers of rare alleles among whom some have large effects. There are many fewer common alleles with weak effects and it is these alleles that are reliably identified in GWAS. On the other hand, if one counts the absolute number of people with each kind of allele the relationship is inverted. For any particular locus there are small numbers of people with rare alleles and large numbers with common alleles. For this reason, the attributable fraction for a particular locus can be dominated by the common alleles even though the rare alleles have much larger effects 72. One way to consider the attributable fraction is the fraction of cases that would be eliminated if the allele were not present. Therefore, even weakly acting common alleles can be the most significant contributors to cardiovascular disease.
Direct DNA Sequencing
The cost of sequencing the entire human genome is expected to drop to about $1,000.00 by the end of 2011. This evolution has been made possible by switching to massively parallel sequencing platforms wherein millions of DNA strands are sequenced in parallel and simultaneously. The technologies have made it feasible to sequence two or three genomes or a dozen of exoms in a week. A major advantage of the whole genome and exome sequencing approaches is in its enabling principle that allows detection of not only the common (MAF>0.05) and uncommon (MAF<0.05 - >0.01) but also rare (MAF<0.01) and private (found only in the probands or genetically-related immediate family members) variants.
Application of the NGS extends beyond the DNA sequencing as the core genome technology and also affords the opportunity to sequence and analyze the whole transcriptome (RNA-Seq), epigenetic modifications (Methyl-Seq) and transcription factor binding sites (ChIP-Seq). The approach is quantitative and enables relatively small amount of template. In the present review the focus is on DSVs.
Next-Generation Sequencing Platforms
Sydney Brenner, Nobel Laureate in Physiology and Medicine (2002), introduced the first technique of sequencing of millions of copies of the DNA simultaneously, referred to as MPSS in 2000 73. Soon George Church and colleagues described the technique of multiplex polony sequencing 74. The first commercial NGS platform was based on pyrosequencing technique 75. However, it was soon surpassed in output by reversible dye-termination and sequencing by ligation approaches. Sequencing platforms continue to evolve at a rapid pace with enhanced capacity to generate bigger outputs and more accurate reads. Accordingly, the newer instruments can generate up to 300 Gb of throughput per sequencing run, which would be sufficient to cover 2–3 genomes and a dozen or so exomes and transcriptomes. Detailed technical review of the existing platforms is beyond the scope of the present review and can be found elsewhere 76. The two most commonly used platforms for whole exome and whole genome sequencing are the SOLiD systems (Applied Biosystems, Inc.), which is based on sequencing by ligation-based chemistry and HiSeq systems (Illumina, Inc), which utilizes reversible terminator-based sequencing by synthesis chemistry. Both platforms generate short reads that typically are 50 to 120 bases long and each can generate about 20 – 30 Gb per day. The accuracy of the sequence reads depends on various factors including depth of coverage. Overall, the systems have a high accuracy rate, typically >99.9%. However, given the vast size of the sequence output, even a very low error rate can lead to a considerable number of erroneous calls and hence, downstream work. For medical sequencing, nonetheless, it is essential to validate the variant calls either by an alternative method, such as Sanger sequencing or by repeating the deep sequencing in toto and accepting only those variants that were reproduced.
In contrast to short read NGS platforms, pyrosequencing (Roche 454 sequencing systems) can generate a read length of ~ 400 bases and more than 1 million reads per run in about 10 hours. However, the size of sequence output is much smaller and hence, the cost per base is much higher. Because of the length of the reads the system is best suited for de novo sequencing. The error rate is about 0.1%. Therefore, for medical sequencing confirmation of the variants is essential.
Newer techniques include single molecule real-time sequencing, which is also referred to Third Generation sequencing, can generate an average read length of more than 1,000 bases. However, the system at the present time has a high error rate and does not seem to be suitable for medical sequencing. Finally, NGS platforms are also available for sequencing of small genomes and targeted sequencing of relatively small regions or small number of genes.
Whole-genome Sequencing
Whole genome sequencing using NGS instruments only recently has become feasible in individual laboratories. The existing platforms afford the opportunity to sequence one to three genomes in a single run in 7–8 days. However, currently only few centers have the sequencing and bioinformatics capacity and financial means to handle large-scale whole genome sequencing projects. Technical aspects include the size of the mappable data (to the reference sequence), depth of coverage, error rate of allele calling, and the gaps in the coverage. Technical advances have made it feasible to apply whole genome sequencing to identify the genetic cause of Mendelian disorders; at least in proof-of-principle studies 77. Application of this approach to other single gene and multi-gene disorders is likely to accelerate significantly during the next few years. The advantage of whole genome sequencing is that it affords the opportunity to detect all SNPs, whether coding or non-coding, and to some extent CNVs in the genome. It also does not depend on a target capture technology, which may suffer from unequal capture of the desired genomic regions. The disadvantages are simply the limited capacity of most laboratories to handle and store terabytes of data that is generated by the sequencer and boinformatics. Various algorithms have been developed to restrict the number of candidate variants and facilitate identification of the causative variants. The key components are population MAFs (novel, known, de novo), type of the variants (deletion, frame shift, missense, splice), evolutionary conservation of the variants and expected biological effects.
Whole-exome Sequencing
The whole exome sequencing approach is designed to capture, enrich and sequence all exons in the genome. Each genome is estimated to contain approximately 300 Mbp representing ~ 180,000 exons of approximately 23,000 protein-coding genes. The focus on whole exome sequencing as opposed to whole genome sequencing stems from the existing data, which indicate that more than 2/3 of the known disease-causing genes in humans are located within exons. Steps involved in whole exome or sub-genomic sequencing include library preparation, target capture, target enrichment and sequencing. Commercially available capture technologies enable efficient capture of the exome and their sequencing. Nonetheless, the efficiency of capture could vary in different genomic regions and often 5 to 20% of the exons may not be captured and sequenced adequately to afford robust allele calling. Whole exome sequencing has been successfully applied to identify the genetic causes of rare Mendelian disorders such as Freeman-Sheldon syndrome, congenital chloride-loosing enteropathy, Kabuki syndromes and hypertension due to hyperaldosteronism 78,78–81. Figure 4 illustrates an example of heterozygous and homozygous mutation read out of a NGS platform. However, the use of whole exome sequencing to identify genetic causes of uncommon and heterogeneous Mendelian disorders could face formidable challenges, particularly in small families and sporadic cases to discern the disease causing variants from those that by chance alone are presented in the affected individuals.
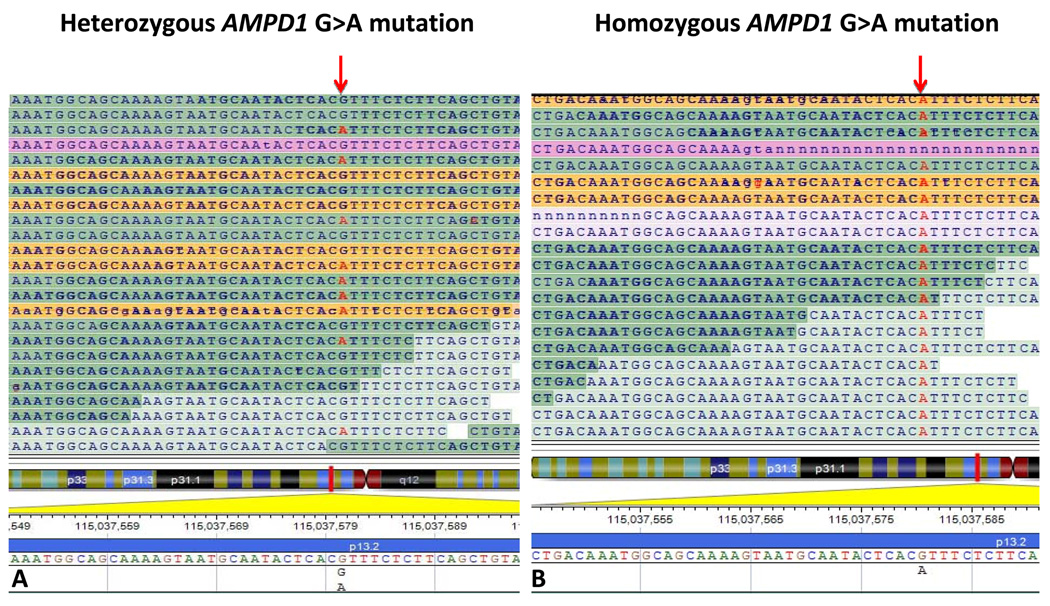
Figure 4. Detection of single nucleotide variant by whole-exome sequencing.
Panel A illustrates an example of sequence output (anti-sense strand) of a NGS machine from a family member heterozygous for G>A (c.C34T, p.Q12X) mutation in AMPD1. Panel B represents a sequence read out from another family member who is homozygous for the mutation and has skeletal myopathy. The homozygous p.Q12X mutation leads to skeletal myopathy due to AMPD deficiency, which was confirmed biochemically. An arrow indicates the mutation.
Targeted Sub-genomic Sequencing
Targeted sub-genomic sequencing is in essence similar to whole exome sequencing except that selected exons or sub-genomic regions are amplified by long-range PCR or captured using custom-made capture probes, enriched and sequenced using NGS platforms. The approach might be desirable for genetic screening through long-range PCR or capture and subsequent sequence all exons in the known genes implicated in the phenotype, such as screening of the known genes coding for sarcomeric proteins in patients with cardiomyopathies and their family members 82. This approach is not much cheaper than the whole exome sequencing approach but is clearly less demanding in terms of bioinformatics. Like all target capture and sequencing approaches, it has the problem of uneven capture and PCR amplification of the intended targets and hence, the risk of under-detection. Likewise, the approach by definition is limited to known targets and therefore, somewhat is limited in its scope. Moreover, given the feasibility and declining cost of whole exome / genome sequencing and in view of the complexity of the genetic determinants of the clinical phenotypes, sub-genomic sequencing is best suitable to specific circumstances, such as follow up studies to genetic linkage and GWAS.
Design of Genetic Studies
A phenotype is in part the consequence of effects of multiple common and rare alleles each imparting a gradient of effects. While GWAS is typically designed to identify common alleles, the NGS approach is best suited for a comprehensive detection of common as well as rare variants. The power of NGS to identify the causal variants is primarily determined by design of the study and the characteristics of the population. As in all genetic studies, robust phenotyping and family-based genetic studies are far superior to studies in isolated cases or in a cohort of sporadic cases.
Phenotyping
Robust phenotyping is an essential but often an inadequately defined component of the genetic studies. Clinical phenotyping often does not offer sufficient resolution or specificity. Even the most clinically robust phenotype, such as all cause mortality is subject to enormous etiological heterogeneity. On the other hand, discerning the etiological subtypes renders the approach to uncertainties of accurate identification of the subtypes. Likewise, phenotypic admixture is also not uncommon, as illustrated for the commonly pooled phenotypes of coronary atherosclerosis, ischemic heart disease and myocardial infarction, as a single phenotype. While these phenotypes have overlapping components, each has partially separate mechanistic basis. Likewise, clinical phenotypes are usually a continuum but often are considered categorical. For example, dichotomization of coronary atherosclerosis, a continuous phenotype, as a categorical phenotype of < or > 60% minimum lumen diameter stenosis not only is subject to the imprecision of the quantification, which could be quite large, but also inadequate represents the phenotypic burden. Phenocopy conditions also compound and confound accurate diagnosis. Collectively, the inadequacies of accurate clinical phenotyping reduce the successful elucidation of the genetic basis of various clinical phenotypes.
Phenotypic plasticity of mutations in a given gene also complicates a straightforward genotype-phenotype correlation. This is most remarkable for single gene disorders and best illustrated for LMNA, which encodes Lamin A/C, an important component of the inner nuclear lamina (reviewed in 83). A diverse array of mutations in LMNA cause at least 13 distinct phenotypes, which are collectively referred to as laminopathies (reviewed in 84). Phenotypic expression of LMNA mutations in the heart or cardiolaminopathies is notable for dilated cardiomyopathy (DCM), supraventricular bradyarrhythmias and conduction defects 85. Likewise, mutations in genes coding for sarcomeric proteins, such as MYH7 and TNNT2 can cause either DCM or HCM, which are on the opposite ends of phenotypic spectrum of cardiac responses to mutations or non-genetic factors 86–88. Similarly, mutations in SCN5A, which codes for a sodium channel are phenotypically expressed as the long QT syndrome, Brugada syndrome, AV conduction defects, atrial fibrillation and DCM 89–91. Phenotypic plasticity appears to be the expected rather than the exception for various DSVs in the same gene but not typically for the same DSV in a given gene.
Biological variability and shortcomings of the quantification methods also diminish the power to map genetic determinants of certain clinical phenotypes. The simplest example is the measurement of systolic and diastolic blood pressure values utilizing a sphygmomanometer, which is based on the detection of Korotkoff sounds. A single measurement of blood pressure is often inadequate and seldom two measurements even when measured within a short time period are identical. Likewise, biochemical phenotypes, such as plasma levels of pro-inflammatory cytokines and C-reactive protein (CRP) are quite dynamic and exhibit considerable intra-individual variability. A single measurement is usually inadequate to reflect the physiological or pathological burden of the phenotype. Physiological and technical variability are typically handled by increasing the sample size of the study population, which increases the power to detect significant effects. Nonetheless, increasing the sample size not only increases the cost but also renders the relevance of the findings to a single individual remote. Imperfectness of phenotyping is probably partially responsible for the “missing heritability” in the genetic studies of complex traits, as the identified DSVs only account for a small fraction of the heritability.
Family-based studies
Family studies provide the most robust approach for delineation of the genetic determinants of the phenotype. Diseases with the strongest familial inheritance are single gene disorders, which are uncommon and often rare. Therefore, the number of DSVs in each genome with very large effects is also expected to be low. According to 1,000 Genomes data, each genome encompasses about 250 to 300 loss-of-function variants in the annotated genes, 50 to 100 variants that already have been implicated in inherited disorders and about 30 de novo variants 16. DSVs with large effects are easier to identify and establish as determinants of a phenotype, as illustrated in familial single gene disorders. Deep sequencing approaches are expected to supplant microarray–based genotyping approaches for identification of genetic determinants of single gene disorders. Likewise, the approach might enable identification of the DSVs with moderate effects that may serve as modifier alleles in single gene disorders. The significance of the latter is noteworthy because of the influence of genetic background, namely modifier alleles, on phenotypic expression of single gene disorders 92.
Rare DSVs are also expected to contribute to phenotypic expression of common complex phenotypes, which show a familial aggregation. The stronger the evidence for a familial aggregation of a complex phenotype, the more likely is the presence of rare variants with large effects. In general, a larger number of genetically related family members provides a greater power to identify the causal and modifier variants, regardless of the approach being genotyping or deep sequencing. Likewise, family-based deep sequencing studies are most powerful when the causal variants occur de novo or are very rare. Moreover, deep sequencing is more powerful for identification of genetic causes of rare than common disorders. It is also more power for identification of genetic cases of Mendelian disorders with a recessive than those with an autosomal dominant mode of inheritance. Nevertheless, deep DNA sequencing could enable elucidation of genetic basis of single gene disorders in small families and probands, wherein the conventional genetic linkage studies are not sufficiently powerful to map the chromosomal locus. The powerful of whole genome/exome sequencing to pinpoint the causal DSVs also inversely correlates with prevalence and genetic heterogeneity of the disorder. In relatively common single gene disorders, a large number of alleles are expected to co-segregate with the phenotype and hence, discerning the true causal variants from those that segregate with the phenotype by chance alone is challenging. Moreover, unlike the candidate gene approach, which is based on a prior knowledge, whole genome/exome sequencing is free of a priori assumption. The approach has been successfully applied to identify the causal variants in rare autosomal recessive or autosomal dominant diseases, such as Kabuki, Miller syndromes and hyperaldosteronism 77, 78, 80, 93,78.
While it is desirable to sequence the genome/exome in all related family members, the approach is currently costly and the analysis is demanding. An alternative approach is to focus on family members that are more distantly related but are phenotypically affected, as such family members are expected to share a lower number of alleles. The approach by reducing the number of shared alleles is expected to enhance identification of the causal variants. Another desirable and practical approach is to sequence and contrast sequence data from family members that are on the opposite ends of phenotypic spectrum, for example, mild vs. severe phenotype. Similarly, the approach is expected to enrich the chance of identifying genetic variants that impart relatively large effect sizes. Nonetheless, the challenge of identifying the causal variants is magnified inversely with the size of the families.
Trio-based family studies
A trio in a family study refers to parents and an offspring. A deep sequencing strategy in a single trio does not offer much power except for the detection of rare and de novo variants in a biologically plausible or previously implicated gene in an affected offspring. However, sequencing of a large number of trios could afford the opportunity to apply the Transmission Disequilibrium Test (TDT) to identify the putative causal variants. TDT assess inheritance of an allele by an affected offspring from an affected parent. In a case of no association, it is random event and hence, a 50% chance. In the case of an association, the frequency of transmission deviates significantly from the chance.
Sporadic cases
Application of deep sequencing technologies in sporadic cases requires a case-control study design similar to those conducted in GWAS. Unlike GWAS, however, deep sequencing will identify rare as well as common alleles and hence, the frequencies of the alleles are compared in cases and controls in an allele or gene-centric (collapsing) approach to test for the presence of statistically significant differences. The design of a case-control study in a deep sequencing project is of utmost importance as an ill-conceived study design could lead to identification of an exceedingly large number of DSVs that differ between the cases and controls. Extensive amount of downstream analyses and experiments would then be required to discern the true associations from false. Several strategies could be utilized to strengthen the design of the case-control studies and hence, reduce the number of putative candidate variants for subsequent validation. One such approach is to focus on cases that exhibit the extreme ends of the phenotype of interest (for example, those with severe and premature disease). Likewise, it is often desirable to include a group of “super normal controls”, which have been exquisitely phenotyped to exclude potential sub-clinical phenotype and have no family history of the phenotype of interest. Moreover, inclusion of the cases that have been enriched for the genetic load, such as an inbred population, as well as prioritizing of analysis of variants that are located in the previously mapped GWAS loci for the phenotype of interest could increase the likelihood of restricting the number of putative candidates.
BIOINFORMATICS AND STATISTICAL ANALYSIS
Various filtering algorithms are applied to restrict the number of putative candidates and to identify the causal variants among the myriads of alleles identified through deep sequencing. The field of bioinformatics is rapidly evolving and considerable progress has been made in eliminating the current bottleneck in analysis of the NGS data. The approach to identify the causative alleles is logical and based on the family structure (co-segregation), zygosity, novelty, being de novo, MAFs, evolutionary conservation and known or anticipated biological effects of the variants. Table 2 lists the most likely putative disease- causing variants.
TABLE 2.
High priority variants identified after streamlining of deep sequencing output
|
The process typically involves mapping the sequence read outs to the reference sequence, which is successfully achieved for more than half of the reads. Using the mappable sequence, various bioinformatics programs are used to identify single nucleotide variants, small indels, and even CNVs and inversions, depending on the sequencing platform. The accuracy of allele calling depends in part on the coverage depth and quality of the reads. As would be expected a higher coverage depth would be required for calling heterozygous than homozygous variants. While the bioinformatics programs are evolving rapidly, various software are already available to annotate the alleles in terms of quality of the call, coverage statistics, novelty or known frequencies of the variants, type of the variants and their putative functional effects. A partial list of bioinformatics programs and their main application is shown in Table 3.
TABLE 3.
A Partial List of Bioinformatics Programs Used to Analyze Next Generation DNA Sequencing Data
| Program | Primary utility | Web address |
|---|---|---|
| SAMtools | A commonly used alignment format files 103 | http://samtools.sourceforge.net/ |
| The Genome Analysis Toolkit (GATK) | A structured programming framework designed to enable rapid development of efficient and robust analysis tools for next-generation DNA sequencers 104 | http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome Analysis_Toolkit |
| BWA | A fast light-weighted tool that aligns short sequences to a sequence database, such as the human reference genome. | http://bio-bwa.sourceforge.net/ |
| Novocraft | Commercial tools for reference alignment of paired-end and single-end for Illumina, Solid and 454 | http://www.novocraft.com/main/index.php |
| BFAST | Blat-like Fast Accurate Search Tool 105 | http://sourceforge.net/apps/mediawiki/bfast/index.php?title=Main_Page |
| Bowtie | Ultrafast, memory-efficient short read aligner 106 | http://bowtiebio.sourceforge.net/index.shtml |
| ANNOVAR | Functionally annotate genetic variants detected from diverse genomes 107 | http://www.openbioinformatics.org/annovar/ |
| SequenceVariant Analyzer (SVA) | A software to annotate, visualize, and analyze the genetic variants identified through NGS | http://www.svaproject.org/ |
| Integrative Genomics Viewer (IGV) | A high-performance visualization tool for interactive exploration of large, integrated datasets 108 | http://www.broadinstitute.org/software/igv/home |
| UCSC Genome Browser | Web-based tools to integrate, visualize and analyze genomics and clinical data 109 | http://genome.ucsc.edu/ |
Courtesy of Manuel Gonzalez-Garay, Ph.D. at Center for Cardiovascular Genetics, The University of Texas Health Science Center, Houston, TX.
Confirmation that a locus contributes to a disease must be based on statistical support and, ultimately, replication of the finding in independent cohorts of cases. There are now a handful of statistical methods that have been specifically tailored to address the comparison of rare variants between cases and controls 30, 94–97. In 2008 Leal and coworkers described the Combined Multivariate and Collapsing (CMC) method 94. CMC combines collapsing of rare variants into a single class and multiple-marker tests for common variants and has much greater power than single marker tests. In a recent innovation, Leal’s group has described the Kernel Based Adaptive Cluster (KBAC) method, which directly addresses the problem of detecting rare variant associations in the presence of functional misclassification 95. The sample risk is modeled using a mixture distribution with two components - non-causal and causal. The method uses continuous adaptive weighting in the comparison between cases and controls. As reference data sets become larger over the next few years it should be possible to apply even more sophisticated methods that model the differences in mutation rate between loci and the known functional interactions of gene products among other important parameters.
Identification of disease causing variants can be based on conservation of sequence, predicted alteration of protein structure or known functional sites, and direct experimental testing. All these methods have significant limitations in sensitivity and specificity. Therefore, multiple lines of independent verification will be required to reach conclusive evidence for a causal role of any particular rare variant.
PERSPECTIVE
The current practice of medicine and the enormous advances that have been made during the past several decades are primarily based on phenotype-based approaches. The imperfectness of clinical phenotyping begets considering a shift toward using surrogate phenotypes that are proximal to genes and hence, more likely to be subjected to larger effects (Figure 3). The significance of finding genetic determinants of the proximal phenotypes is that it not only could elucidate biological and functional significance of DSVs but also might translate and extend to clinical phenotype. For example, heterozygous loss-of-function mutations in PCSK9 lower plasma LDL-C levels and reduce the risk of coronary heart disease drastically over a 15-year period 98. However, a discord between association of a biochemical (proximal) and clinical phenotypes with DSV might be present. The discord is particularly evident when the effect size on the proximal phenotype is relatively small (for example a 1 mg/dl change in plasma HDL-C level), there is large locus heterogeneity and because of a large number of non-genetic factors contribute to the phenotype.
In view of the shortcomings of clinical phenotyping and given the technical feasibility of whole exome or whole genome sequencing, one may infer that the era of a genome/exome-based approach to identify genetic determinants of the phenotype might not be far in the future. A desirable genotype-based approach will exploit the genome/exome data from thousands of individuals to prospectively link the genotype to phenotype through a comprehensive analysis and define the genetic architecture of human diseases and traits.
Whole genome sequencing is likely to become a commodity that could be readily available at a reasonable cost and be easily accommodated into the decision making tree of health care of every individual. The challenging task will be to identify variants that are disease-causing or likely disease-causing and develop strategies to prevent and attenuate the evolving phenotype (Figure 5). Likewise, various complementary studies – genetic and biological – would be necessary to discern the associated alleles from the true disease causing variants. Moreover a better understanding of various components of the genome, such as chromatin modification, functional CNEs, transposons, large intergenic non-coding RNAs, small non-coding RNAs and primary transcripts would be essential 99. An integrated approach that utilizes genetic, genomics, transcriptomics, proteomics and metabolomics would be expected to facilitate identification and characterization of the mechanisms and involved in the pathogenesis of the phenotype.
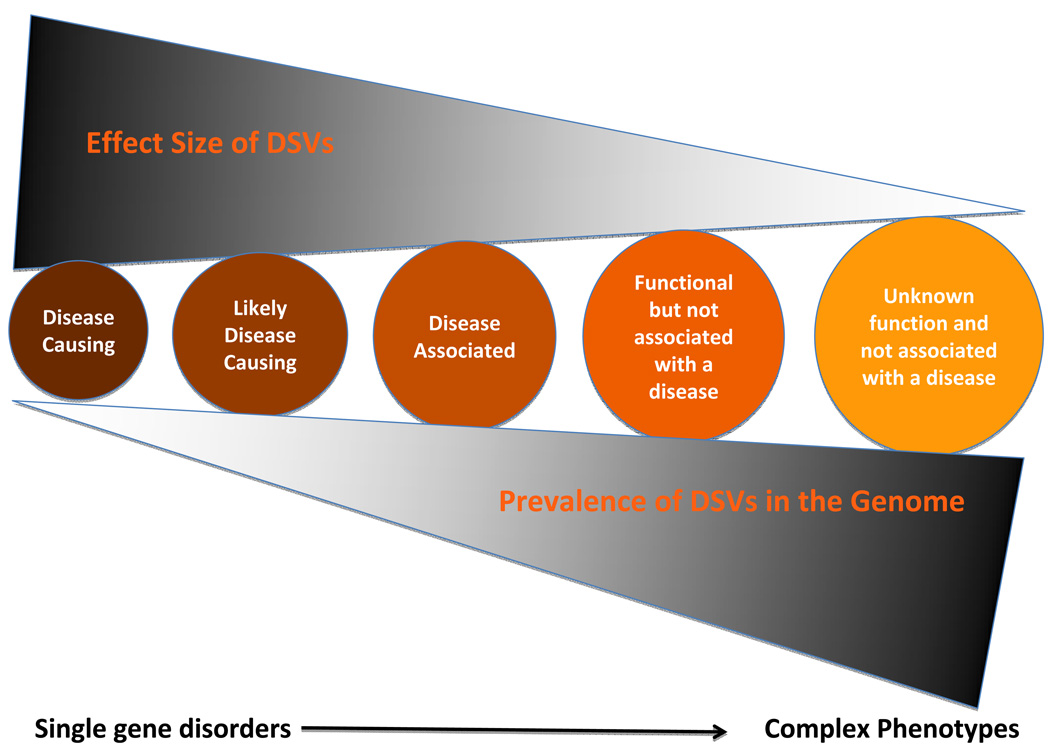
Figure 5. Clinical Implications of DSVs.
Biological and clinical significance of DSVs is expected to follow a continuum. For simplicity, we have highlighted five classes of DSVs in the continuum of their effects, in terms of their biological and clinical significance:
Disease-causing variants: Disease-causing variants when present cause a disease, albeit with variable penetrance and considerable phenotypic variability. They impart large effects and are rare in each genome. The variants co-segregate with inheritance of the phenotype in members of large families or in multiple families and are absent in the clinically unaffected family members – notwithstanding the penetrance – and in the general population. These variants are also expected to impart considerable functional and biological effects.
The disease-causing variants could provide insights into molecular pathogenesis of the phenotype and guide the development of new therapeutic and preventive targets. Likewise, they might also serve as diagnostic markers typically in familial situations and help to discern the true phenotype from phenocopy. The absence of a disease-causing variant in a family member at risk renders the likelihood of developing the disease remote. The disease-causing variants have limited utility in prognostication and risk stratification because of the complexity of determinants of the clinical phenotypes.
Likely disease-causing variants: Genetic data implies causality but the evidence is inadequate to substantiate it. Statistical evidence indicates a strong association but typically an imperfect penetrance thwarts detecting a perfect co-segregation. This is often the case for rare and private DSVs in small size families and sporadic cases. These variants are typically absent in a large number of ethnically matched independent control individuals. These variants are also expected to impart significant functional and biological effects and be more common in the genome than the disease-causing variants. Clinical implications of the likely disease-causing variants are less robust than those for the disease-causing variants.
Phenotype-associated variants: Causality is difficult to establish for this category of DSVs, particularly in sporadic cases and small families. Despite a statistical association additional functional and mechanistic studies are necessary to imply a causal role. The disease-associated variants are typically identified based on differences in MAFs frequencies in the cases and controls, such as through GWAS or candidate gene studies. These variants are often in LD with the true causal allele. The extent of LD in the genome varies but could extent to several million base pairs 102. Variants that affect structure, function and splicing of the genes carry a higher chance of being causal variants than those located in introns or inter-gene regions. Identification of these variants could provide insights into the molecular pathogenesis of the phenotype but they have no or very limited value in genetic diagnosis or risk stratification. The strength of the statistical association does not translate into clinical significance. A 5% increased in the MAF of a SNP from 0.45 to 0.50 in a large case-control study could result to exceedingly low p values and might have high attributable risk in a population but at an individual level it does not offer much clinical utility. Likewise, the clinical significance of the observed relative risks or Odds ratios should be interpreted in the context of pre-test likelihood of the clinical event. A two-fold increase in the risk of heart failure is not much clinically informative if the a priori risk of heart failure in the study population is exceedingly low.
Functional variants not associated with a clinical phenotype: The human genome contains a large number of genetic polymorphisms including insertions, deletions, non-sense variants, splice junction variants, CNVs, etc, many of which exert functional functions. Despite the evidence for biological functions, these variants are not known to influence disease-risk or be associated with any clinical phenotype. These variants have minimal clinical utility or application.
Variants with unknown significance: The vast majority of ~ 4 million DSVs in the genome probably fit into this category. Most are located in inter-gene regions and introns and are not known to convey biological functions. These variants have no known clinical utility.
Acknowledgments
RESEARCH SUPPORT
NHLBI (R01-088498); Burroughs Wellcome Award in Translational Research (#1005907), TexGen Fund from Greater Houston Community Foundation
Glossary
GLOSSARY
- DNA sequence variant (DSV)
DSV is used as a general idiom to describe all variations in the DNA sequence, whether single nucleotide polymorphisms (SNPs), copy number variants (CNVs), insertions/deletions (indels) or structural variations (SVs)
- Exome
All exons in a genome are referred to as an exome, analogous to a genome for the entire genetic material of a cell or an organism
- Single nucleotide polymorphism (SNP)
Variations in a single nucleotide sequence among individuals
- Non-synonymous single nucleotide polymorphism (nsSNP)
A change in a single nucleotide that changes the codon or amino acid sequence in the protein
- Structural variation (SV)
Typically large insertions, deletions, inversions, duplications and rearrangements in the genomes that are present in some individuals
- Copy number variant (CNV)
Each genome has two copies of DNA and hence, each gene. A structural variation that reduces or increases the two copies of a segment of DNA or a gene is referred to as a CNV
- Linkage disequilibrium (LD)
Correlation between two or more alleles that co-segregate by more than chance alone. LD typically inversely relates to the physical distance between the two alleles on a chromosome
- Haplotype
Genetic regions that are tightly correlated (because of linkage disequilibrium)
- De novo
A variant that occurs as a new genetic event in an individual
- Minor allele frequency (MAF)
The population frequency of the less common allele of an SNP
NON-STANDARD ABBREVIATIONS
- CAD
Coronary Artery Disease
- CD-CV
Common Disease-Common Variant
- CNEs
Conserved non-coding elements
- CNVs
Copy Number Variants
- dbSNP
SNP Database
- DCM
Dilated Cardiomyopathy
- DSVs
DNA Sequence Variants
- Gb
Gigabases
- GWAS
Genome Wide Association Studies
- HCM
Hypertrophic Cardiomyopathy
- HDL-C
High-Density Lipoprotein-Cholesterol
- HGP
Human Genome Project
- Indel
Insertion/deletion
- Kbp
Kilo base pair
- LD
Linkage Disequilibrium
- LDL-C
Low-Density Lipoprotein-Cholesterol
- MAFs
Minor allele frequencies
- Mbp
Million base pair
- NGS
Next Generation Sequencing
- nsSNP
Non-synonymous Single Nucleotide Polymorphisms
- RV-CD
Rare Variant-Common Disease
- SiRNA
Short-interfering RNA
- SNP
Single Nucleotide Polymorphism
- SVs
Structural variations
- TDT
Transmission Disequilibrium Test
- TG
Triglycerides
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES STATEMENT
There is no conflict of interest to declare
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la BM, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di FV, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc.Natl.Acad.Sci.U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 463:446–456. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaxter M. Revealing the dark matter of the genome. Science. 330:1758–1759. doi: 10.1126/science.1200700. [DOI] [PubMed] [Google Scholar]
- 6.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, Macdonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The Diploid Genome Sequence of an Individual Human. PLoS.Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunter C. Genomics: A picture worth 1000 Genomes. Nat Rev Genet. 2010;11:814. doi: 10.1038/nrg2906. [DOI] [PubMed] [Google Scholar]
- 10.Pennisi EGenomics. 1000 Genomes Project gives new map of genetic diversity. Science. 2010;330:574–575. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
- 11.Gamazon ER, Zhang W, Dolan ME, Cox NJ. Comprehensive survey of SNPs in the Affymetrix exon array using the 1000 Genomes dataset. PLoS ONE. 2010;5:e9366. doi: 10.1371/journal.pone.0009366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z, Zheng H, Hellmann I, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, Qin J, Ma L, Li G, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Ni P, Ruan J, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, San A, Ping C, Yang S, Chen F, Li L, Zhou K, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Yang H. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song Xz, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 14.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, Kim RW, Huntley JJ, Luo S, Schroth GP, Wu TD, Kim H, Yang KS, Park WY, Church GM, Lee C, Kingsmore SF, Seo JS. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE. Diversity of human copy number variation and multicopy genes. Science. 330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, Chinwalla A, Conrad DF, Fu Y, Grubert F, Hajirasouliha I, Hormozdiari F, Iakoucheva LM, Iqbal Z, Kang S, Kidd JM, Konkel MK, Korn J, Khurana E, Kural D, Lam HY, Leng J, Li R, Li Y, Lin CY, Luo R, Mu XJ, Nemesh J, Peckham HE, Rausch T, Scally A, Shi X, Stromberg MP, Stutz AM, Urban AE, Walker JA, Wu J, Zhang Y, Zhang ZD, Batzer MA, Ding L, Marth GT, McVean G, Sebat J, Snyder M, Wang J, Eichler EE, Gerstein MB, Hurles ME, Lee C, McCarroll SA, Korbel JO. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marian AJ. Nature's genetic gradients and the clinical phenotype. Circ Cardiovasc Genet. 2009;2:537–539. doi: 10.1161/CIRCGENETICS.109.921940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N.Engl.J.Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 22.Post WS, Larson MG, Myers RH, Galderisi M, Levy D. Heritability of left ventricular mass: the Framingham Heart Study. Hypertension. 1997;30:1025–1028. doi: 10.1161/01.hyp.30.5.1025. [DOI] [PubMed] [Google Scholar]
- 23.Adams TD, Yanowitz FG, Fisher AG, Ridges JD, Nelson AG, Hagan AD, Williams RR, Hunt SC. Heritability of cardiac size: an echocardiographic and electrocardiographic study of monozygotic and dizygotic twins. Circulation. 1985;71:39–44. doi: 10.1161/01.cir.71.1.39. [DOI] [PubMed] [Google Scholar]
- 24.Arora P, Newton-Cheh C. Blood pressure and human genetic variation in the general population. Curr Opin Cardiol. 2010;25:229–237. doi: 10.1097/HCO.0b013e3283383e2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8 Suppl 1:S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirruccello J, Kathiresan S. Genetics of lipid disorders. Curr Opin Cardiol. 2010;25:238–242. doi: 10.1097/HCO.0b013e328338574d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, van der Harst P, Muller M, Eijgelsheim M, Alonso A, Hicks AA, Padmanabhan S, Hayward C, Smith AV, Polasek O, Giovannone S, Fu J, Magnani JW, Marciante KD, Pfeufer A, Gharib SA, Teumer A, Li M, Bis JC, Rivadeneira F, Aspelund T, Kottgen A, Johnson T, Rice K, Sie MP, Wang YA, Klopp N, Fuchsberger C, Wild SH, Mateo Leach I, Estrada K, Volker U, Wright AF, Asselbergs FW, Qu J, Chakravarti A, Sinner MF, Kors JA, Petersmann A, Harris TB, Soliman EZ, Munroe PB, Psaty BM, Oostra BA, Cupples LA, Perz S, de Boer RA, Uitterlinden AG, Volzke H, Spector TD, Liu FY, Boerwinkle E, Dominiczak AF, Rotter JI, van Herpen G, Levy D, Wichmann HE, van Gilst WH, Witteman JC, Kroemer HK, Kao WH, Heckbert SR, Meitinger T, Hofman A, Campbell H, Folsom AR, van Veldhuisen DJ, Schwienbacher C, O'Donnell CJ, Volpato CB, Caulfield MJ, Connell JM, Launer L, Lu X, Franke L, Fehrmann RS, Te Meerman G, Groen HJ, Weersma RK, van den Berg LH, Wijmenga C, Ophoff RA, Navis G, Rudan I, Snieder H, Wilson JF, Pramstaller PP, Siscovick DS, Wang TJ, Gudnason V, van Duijn CM, Felix SB, Fishman GI, Jamshidi Y, Ch Stricker BH, Samani NJ, Kaab S, Arking DE. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, Ehret GB, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas RJ, Sinner MF, Gieger C, Najjar SS, Kao WH, Muhleisen TW, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel KH, Naitza S, Steinbeck G, Marroni F, Hicks AA, Lakatta E, Muller-Myhsok B, Pramstaller PP, Wichmann HE, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis GR, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010;40:360–369. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita H, Larson MG, Barr SC, Vasan RS, O'Donnell CJ, Hirschhorn JN, Levy D, Corey D, Seidman CE, Seidman JG, Benjamin EJ. Single-gene mutations and increased left ventricular wall thickness in the community: the Framingham Heart Study. Circulation. 2006;113:2697–2705. doi: 10.1161/CIRCULATIONAHA.105.593558. [DOI] [PubMed] [Google Scholar]
- 33.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat.Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 34.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, Kaminski WE, Hahmann HW, Oette K, Rothe G, Aslanidis C, Lackner KJ, Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat.Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 35.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple Rare Alleles Contribute to Low Plasma Levels of HDL Cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 36.Hao K, Chudin E, Greenawalt D, Schadt EE. Magnitude of stratification in human populations and impacts on genome wide association studies. PLoS ONE. 5:e8695. doi: 10.1371/journal.pone.0008695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, Steg PG, Ferrieres J, Danchin N, Becquemont L. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 38.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 39.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci U S A. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C–adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, Hovingh GK, Kastelein JJ. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ Cardiovasc Genet. 2009;2:173–181. doi: 10.1161/CIRCGENETICS.108.818062. [DOI] [PubMed] [Google Scholar]
- 42.Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 44.Roden DM, Viswanathan PC. Genetics of acquired long QT syndrome. J Clin Invest. 2005;115:2025–2032. doi: 10.1172/JCI25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 46.Kleber ME, Renner W, Grammer TB, Linsel-Nitschke P, Boehm BO, Winkelmann BR, Bugert P, Hoffmann MM, Marz W. Association of the single nucleotide polymorphism rs599839 in the vicinity of the sortilin 1 gene with LDL and triglyceride metabolism coronary heart disease myocardial infarction. The Ludwigshafen Risk and Cardiovascular Health Study. Atherosclerosis. 2010;209:492–497. doi: 10.1016/j.atherosclerosis.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 47.Linsel-Nitschke P, Heeren J, Aherrahrou Z, Bruse P, Gieger C, Illig T, Prokisch H, Heim K, Doering A, Peters A, Meitinger T, Wichmann HE, Hinney A, Reinehr T, Roth C, Ortlepp JR, Soufi M, Sattler AM, Schaefer J, Stark K, Hengstenberg C, Schaefer A, Schreiber S, Kronenberg F, Samani NJ, Schunkert H, Erdmann J. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 48.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Yee J, Friedlander Y, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Meigs JB, Williams G, Nathan DM, MacRae CA, Havulinna AS, Berglund G, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Daly MJ, Nemesh J, Korn JM, McCarroll SA, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Ouwehand W, Deloukas P, Scholz M, Cambien F, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Scheffold T, Berger K, Huge A, Martinelli N, Olivieri O, Corrocher R, McKeown P, Erdmann E, Konig IR, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Do R, Xie C, Siscovick D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, Jansen P, Heeren J, Willnow TE, Nykjaer A. Sort1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 12:213–223. doi: 10.1016/j.cmet.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 51.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 53.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadariya Y, Yin B, Tang B, Shinton SA, Quinlivan EP, Hua X, Klein-Szanto A, Al-Saleem TI, Bassing CH, Hardy RR, Kruger WD. Mice heterozygous for germ-line mutations in methylthioadenosine phosphorylase (MTAP) die prematurely of T-cell lymphoma. Cancer Res. 2009;69:5961–5969. doi: 10.1158/0008-5472.CAN-09-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 58.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peden JF, Hopewell JC, Saleheen D, Chambers JC, Hager J, Soranzo N, Collins R, Danesh J, Elliott P, Farrall M, Stirrups K, Zhang W, Hamsten A, Parish S, Lathrop M, Watkins HC, Clarke R, Deloukas P, Kooner JS, Goel A, Ongen H, Strawbridge RJ, Heath S, Malarstig A, Helgadottir A, Ohrvik J, Murtaza M, Potter S, Hunt SE, Delepine M, Jalilzadeh S, Axelsson T, Syvanen AC, Gwilliam R, Bumpstead S, Gray E, Edkins S, Folkersen L, Kyriakou T, Franco-Cereceda A, Gabrielsen A, Seedorf U, Eriksson P, Offer A, Bowman L, Sleight P, Armitage J, Peto R, Abecasis G, Ahmed N, Caulfield M, Donnelly P, Froguel P, Kooner AS, McCarthy MI, Samani NJ, Scott J, Sehmi J, Silveira A, Hellenius ML, van 't Hooft FM, Olsson G, Rust S, Assmann G, Barlera S, Tognoni G, Franzosi MG, Linksted P, Green FR, Rasheed A, Zaidi M, Shah N, Samuel M, Mallick NH, Azhar M, Zaman KS, Samad A, Ishaq M, Gardezi AR, Memon FU, Frossard PM, Spector T, Peltonen L, Nieminen MS, Sinisalo J, Salomaa V, Ripatti S, Bennett D, Leander K, Gigante B, de Faire U, Pietri S, Gori F, Marchioli R, Sivapalaratnam S, Kastelein JJ, Trip MD, Theodoraki EV, Dedoussis GV, Engert JC, Yusuf S, Anand SS. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 61.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schafer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, Marz W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, Hong MY, Karczewski KJ, Huber W, Weissman SM, Gerstein MB, Korbel JO, Snyder M. Variation in transcription factor binding among humans. Science. 328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McClellan J, King MC. Genomic analysis of mental illness: a changing landscape. JAMA. 2010;303:2523–2524. doi: 10.1001/jama.2010.869. [DOI] [PubMed] [Google Scholar]
- 68.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 69.Antonarakis SE, Chakravarti A, Cohen JC, Hardy J. Mendelian disorders and multifactorial traits: the big divide or one for all? Nat Rev Genet. 2010;11:380–384. doi: 10.1038/nrg2793. [DOI] [PubMed] [Google Scholar]
- 70.Layzer D. Heritability analyses of IQ scores: science or numerology? Science. 1974;183:1259–1266. doi: 10.1126/science.183.4131.1259. [DOI] [PubMed] [Google Scholar]
- 71.Feldman MW, Lewontin RC. The heritability hang-up. Science. 1975;190:1163–1168. doi: 10.1126/science.1198102. [DOI] [PubMed] [Google Scholar]
- 72.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant...or not? Human Molecular Genetics. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 73.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 74.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 75.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kircher M, Kelso J. High-throughput DNA sequencing--concepts and limitations. Bioessays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 77.Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi M, Scholl UI, Yue P, Bjorklund P, Zhao B, Nelson-Williams C, Ji W, Cho Y, Patel A, Men CJ, Lolis E, Wisgerhof MV, Geller DS, Mane S, Hellman P, Westin G, Akerstrom G, Wang W, Carling T, Lifton RP. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331:768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meder B, Haas J, Keller A, Heid C, Just S, Borries A, Boisguerin V, Scharfenberger-Schmeer M, Stahler P, Beier M, Weichenhan D, Strom TM, Pfeufer A, Korn B, Katus HA, Rottbauer W. Targeted Next-Generation Sequencing for the Molecular Genetic Diagnostics of Cardiomyopathies. Circ Cardiovasc Genet. 2011 doi: 10.1161/CIRCGENETICS.110.958322. (PMID 21252143) [DOI] [PubMed] [Google Scholar]
- 83.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes and Development. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Worman HJ, Bonne G. "Laminopathies": A wide spectrum of human diseases. Experimental Cell Research. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Curr Opin Cardiol. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watkins H, Anan R, Coviello DA, Spirito P, Seidman JG, Seidman CE. A de novo mutation in alpha-tropomyosin that causes hypertrophic cardiomyopathy. Circulation. 1995;91:2302–2305. doi: 10.1161/01.cir.91.9.2302. [DOI] [PubMed] [Google Scholar]
- 87.Seidman CE, Seidman JG. Mutations in cardiac myosin heavy chain genes cause familial hypertrophic cardiomyopathy. Mol.Biol.Med. 1991;8:159–166. [PubMed] [Google Scholar]
- 88.Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, McDonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N.Engl.J Med. 2000;343:1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- 89.Tan HL, Bink-Boelkens MT, Bezzina CR, Viswanathan PC, Beaufort-Krol GC, van Tintelen PJ, van den Berg MP, Wilde AA, Balser JR. A sodium-channel mutation causes isolated cardiac conduction disease. Nature. 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 90.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 91.Bezzina C, Veldkamp MW, van Den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, Der Hout AH, Mannens MM, Wilde AA. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ.Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 92.Daw EW, Chen SN, Czernuszewicz G, Lombardi R, Lu Y, Ma J, Roberts R, Shete S, Marian AJ. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Hum Mol Genet. 2007;16:2463–2471. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ionita-Laza I, Buxbaum JD, Laird NM, Lange C. A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet. 2011;7:e1001289. doi: 10.1371/journal.pgen.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence Variations in PCSK9, Low LDL, and Protection against Coronary Heart Disease. The New England Journal Of Medicine. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 99.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 100.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, Alves P, Chateigner A, Perry M, Morris M, Auerbach RK, Feng X, Leng J, Vielle A, Niu W, Rhrissorrakrai K, Agarwal A, Alexander RP, Barber G, Brdlik CM, Brennan J, Brouillet JJ, Carr A, Cheung MS, Clawson H, Contrino S, Dannenberg LO, Dernburg AF, Desai A, Dick L, Dose AC, Du J, Egelhofer T, Ercan S, Euskirchen G, Ewing B, Feingold EA, Gassmann R, Good PJ, Green P, Gullier F, Gutwein M, Guyer MS, Habegger L, Han T, Henikoff JG, Henz SR, Hinrichs A, Holster H, Hyman T, Iniguez AL, Janette J, Jensen M, Kato M, Kent WJ, Kephart E, Khivansara V, Khurana E, Kim JK, Kolasinska-Zwierz P, Lai EC, Latorre I, Leahey A, Lewis S, Lloyd P, Lochovsky L, Lowdon RF, Lubling Y, Lyne R, Maccoss M, Mackowiak SD, Mangone M, McKay S, Mecenas D, Merrihew G, Miller DM, 3rd, Muroyama A, Murray JI, Ooi SL, Pham H, Phippen T, Preston EA, Rajewsky N, Ratsch G, Rosenbaum H, Rozowsky J, Rutherford K, Ruzanov P, Sarov M, Sasidharan R, Sboner A, Scheid P, Segal E, Shin H, Shou C, Slack FJ, Slightam C, Smith R, Spencer WC, Stinson EO, Taing S, Takasaki T, Vafeados D, Voronina K, Wang G, Washington NL, Whittle CM, Wu B, Yan KK, Zeller G, Zha Z, Zhong M, Zhou X, Ahringer J, Strome S, Gunsalus KC, Micklem G, Liu XS, Reinke V, Kim SK, Hillier LW, Henikoff S, Piano F, Snyder M, Stein L, Lieb JD, Waterston RH. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science. 330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R, Carlson JW, Carr A, Jungreis I, Marbach D, Sealfon R, Tolstorukov MY, Will S, Alekseyenko AA, Artieri C, Booth BW, Brooks AN, Dai Q, Davis CA, Duff MO, Feng X, Gorchakov AA, Gu T, Henikoff JG, Kapranov P, Li R, Macalpine HK, Malone J, Minoda A, Nordman J, Okamura K, Perry M, Powell SK, Riddle NC, Sakai A, Samsonova A, Sandler JE, Schwartz YB, Sher N, Spokony R, Sturgill D, van Baren M, Wan KH, Yang L, Yu C, Feingold E, Good P, Guyer M, Lowdon R, Ahmad K, Andrews J, Berger B, Brenner SE, Brent MR, Cherbas L, Elgin SC, Gingeras TR, Grossman R, Hoskins RA, Kaufman TC, Kent W, Kuroda MI, Orr-Weaver T, Perrimon N, Pirrotta V, Posakony JW, Ren B, Russell S, Cherbas P, Graveley BR, Lewis S, Micklem G, Oliver B, Park PJ, Celniker SE, Henikoff S, Karpen GH, Lai EC, Macalpine DM, Stein LD, White KP, Kellis M. Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science. 330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, Carter D, Papaspyridonos M, Livingstone S, Ganske R, Lohmussaar E, Zernant J, Tonisson N, Remm M, Magi R, Puurand T, Vilo J, Kurg A, Rice K, Deloukas P, Mott R, Metspalu A, Bentley DR, Cardon LR, Dunham I. A first-generation linkage disequilibrium map of human chromosome 22. Nature. 2002;418:544–548. doi: 10.1038/nature00864. [DOI] [PubMed] [Google Scholar]
- 103.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS ONE. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2009;38:D613–D619. doi: 10.1093/nar/gkp939. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]